Abstract
The p53 target gene Wig-1 encodes a double-stranded-RNA-binding zinc finger protein. We show here that Wig-1 binds to p53 mRNA and stabilizes it through an AU-rich element (ARE) in the 3′ UTR of the p53 mRNA. This effect is mirrored by enhanced p53 protein levels in both unstressed cells and cells exposed to p53-activating stress agents. Thus, the p53 target Wig-1 is a previously undescribed ARE-regulating protein that acts as a positive feedback regulator of p53, with implications both for the steady-state levels of p53 and for the p53 stress response. Our data reveal a previously undescribed link between the tumor suppressor p53 and posttranscriptional gene regulation via AREs in mRNA.
The p53 tumor suppressor is a transcription factor that activates transcription of specific target genes. p53 triggers various cellular responses, most prominently cell cycle arrest, senescence, and apoptosis (1). Dysfunctional or inactive p53 leads to tumor development, which is underscored by the high frequency of p53 mutations in human tumors (www-p53.iarc.fr; p53.free.fr) (2), and by the tumor-prone phenotype of p53-null mice (3). Too much or uncontrolled p53 causes cell death, as demonstrated by the fact that mice null for the important negative p53 regulator mouse double minute 2 (Mdm2) die early during embryogenesis because of excessive p53-dependent apoptosis (4). There is also evidence from in vivo models that overactive p53 can cause premature aging (5). Because too much p53 is incompatible with life or normal life span and too little p53 promotes tumor formation, it is clear that p53 levels need to be strictly controlled. Regulation of p53 at the protein level has been studied extensively. The p53 mRNA levels affect the abundance and function of the p53 protein, because cells haploinsufficient for p53 suffer a 4-fold decrease in both mRNA and protein levels (6). However, relatively little is known about the regulation of p53 mRNA levels, although studies have demonstrated involvement of its 5′ UTR in regulating stability and translation (7–9).
Wig-1 (for WT p53 induced gene 1; also known as PAG608 and ZMAT3) is a p53-regulated gene (10–12) whose function has remained unclear. It encodes a Cys2His2-type zinc finger protein that localizes mainly to the nucleus (11, 13). The Wig-1 zinc fingers are almost perfectly conserved from fish to humans (14), and they have an unusual spacing within and between the zinc fingers. The latter feature is shared with a small group of dsRNA-binding proteins that lack consensus dsRNA-binding motifs (dsRBM). The most well-studied member of this group is JAZ, which binds to the dsRNA nuclear export receptor Exportin-5 (15) and can positively regulate p53 transcriptional activity by binding to the p53 protein (16). We previously demonstrated that Wig-1 binds dsRNA with high affinity, and that both the first and second zinc finger are necessary for binding to dsRNA in living cells (13, 17).
AU-rich elements (AREs) are regulatory elements present in the 3′ UTR of certain mRNAs, and they have been implicated in posttranscriptional gene regulation either by mediating rapid degradation of the mRNA by affecting deadenylation and/or by decreasing translation efficiency. AREs are generally found in mRNA of genes that need very precise control of expression, such as genes encoding proteins that regulate cell growth (e.g., c-myc, c-fos, c-jun, p53, and p21). The AREs may consist of the element AUUUA in various constellations or regions of Us only (18, 19). The 3′ UTR of p53 harbors one U-rich region (18 continuous Us) and one additional ARE containing the AUUUA motif (20, 21). There are several known proteins that interact with and regulate mRNA containing AREs. Most of these proteins affect the target mRNA negatively, but there are also positive regulators such as human antigen R (HuR) (18). HuR, which has many target mRNAs, binds to the p53 AREs and increases p53 mRNA stability (22) and translation (20). The importance of ARE-mediated regulation is emphasized by the dysregulation of ARE-containing mRNA in pathological conditions such as cancer, chronic inflammation, and autoimmune diseases (19, 23). ARE-interacting proteins are important nodes of regulation, because they can potentially control the levels of many different mRNAs in the cell, and they have been shown to have important roles in cancer as well as in embryonic development (19, 23).
Here, we report that the p53 target protein Wig-1 binds to p53 mRNA and stabilizes it by protecting it from deadenylation, and that this effect is mediated by the U-rich region in the 3′ UTR of p53 mRNA. Thus, Wig-1 is a previously unrecognized ARE-regulating protein that enhances p53 mRNA levels in a positive feedback loop.
Results
Wig-1 Enhances p53 Protein Levels and Augments the p53 Stress Response.
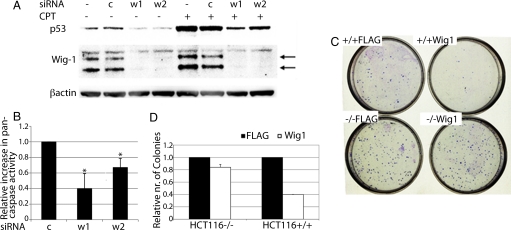
To investigate the effect of Wig-1 on p53, we assessed p53 protein levels by Western blotting after knocking down Wig-1 in U2OS cells by using two different siRNA. This knockdown resulted in a significant decrease in p53 protein levels, both in unstressed cells (by 58% and 63%, respectively, as compared with control siRNA by using quantification by densitometry) and after p53 activation with the DNA-damaging drug camptothecin (by 79% and 73%) (Fig. 1A). The p53 levels were affected in a similar way in human primary lung fibroblasts (IMR-90; Fig. S1a) and immortalized mouse fibroblasts (NIH 3T3; Fig. S1b), demonstrating conservation between cell types and species. To confirm that Wig-1 positively regulates p53 levels, we transiently expressed Wig-1 in U2OS cells (Fig. S1c respectively). Exogenous Wig-1 increased p53 protein levels by at least 80% as shown by Western blotting. Also, decreased p53 protein levels after Wig-1 knockdown in U2OS cells resulted in decreased levels of its target p21, both in unstressed cells and after treatment with camptothecin (Fig. S1d). To confirm that this effect is due to the presence or absence of the Wig-1 protein, we knocked down Wig-1 in U2OS cells by using one siRNA targeting the ORF and one targeting the 3′ UTR. Subsequently, the cells were transfected with the Wig-1 ORF or empty vector (Fig. S1e). Both Wig-1 siRNA reduced p53 protein levels to a similar degree in the presence of empty vector (by 69% and 76%, respectively), but when the Wig-1 ORF was present, only the siRNA targeting the Wig-1 ORF produced substantial down-regulation of p53 (by 65%, as compared with 27% with the siRNA targeting the 3′ UTR). Thus, the decrease in p53 levels caused by siRNA targeting endogenous Wig-1 can be reversed by overexpression of Wig-1 that is not affected by the siRNA, demonstrating that the observed effect is indeed due to reduced Wig-1 expression.
Fig. 1.
Wig-1 affects p53 protein levels and the p53 stress response. (A) Down-regulation of Wig-1 by siRNA treatment leads to decreased p53 protein levels in unstressed U2OS cells and after treatment with the p53-activating DNA-damaging drug camptothecin (CPT), as shown by Western blotting. c, control siRNA; w1, Wig-1 siRNA 1; w2, Wig-1 siRNA 2. Arrows indicate the two major Wig-1 protein species. (B) Relative increase in pan-caspase activity after mitomycin C treatment in MCF7 p53 high cells compared with p53 low cells (stably transfected with control shRNA or shRNA targeting p53, respectively). The figure shows the average of three independent experiments. Bars are shown with SEM, and asterisks indicate significant differences (P = 0.036 for Wig-1 siRNA 1; P = 0.049 for Wig-1 siRNA 2). (C and D) Colony Formation Assay in HCT116 p53 +/+ or −/− cells transfected with empty vector (“Flag”) or with Wig-1 expression plasmid (“Wig-1”) followed by CPT treatment. The figure shows one of three independent experiments, each performed in triplicate. Bars are shown with SEM.
Next, we addressed whether Wig-1-mediated modulation of p53 levels would affect the p53 response to stress. We first performed a short term apoptosis assay, in which we assessed the effect of Wig-1 knockdown on pan-caspase activation. We knocked down Wig-1 in MCF7 cells stably transfected with a shRNA targeting p53 (p53 low) or control shRNA (p53 high). The cells were subsequently treated with mitomycin C or left untreated for 24 h, and then analyzed for pan-caspase activity by staining for active caspases followed by FACS detection. Importantly, the increase in apoptosis after mitomycin C treatment was significantly lower after Wig-1 knockdown in the p53-high cells than in the p53-low cells (a reduction of 60% and 33% for the two siRNA tested, respectively, compared with control siRNA) (Fig. 1B). This experiment demonstrates that loss of Wig-1 attenuates the stress response in a p53-dependent manner.
We also tested the effect of ectopically expressed Wig-1 in a colony formation assay. We used HCT116 p53 +/+ and −/− isogenic cells to compare the effect of Wig-1 in the presence or absence of p53. We transfected the cells with Wig-1 or empty vector, treated with camptothecin to activate p53, and grew cells for 9 days before staining and counting colonies (Fig. 1 C and D). As expected, we observed much fewer colonies in camptothecin-treated HCT116 p53 +/+ cells as compared with camptothecin-treated HCT116 p53 −/− cells, consistent with a p53 response. We evaluated the effect of Wig-1 by comparing number of colonies between the Wig-1 transfected cells and the corresponding vector transfected cells. Ectopically expressed Wig-1 had a relatively minor effect in camptothecin treated HCT116 p53 −/− cells (a reduction by 16%). In contrast, in the camptothecin-treated HCT116 p53 +/+ cells, we observed a substantial inhibition of colony formation by ectopically expressed Wig-1 (a reduction by 72%). Thus, we conclude that Wig-1 potentiates the p53 response to camptothecin and mitomycin C.
Wig-1 Increases the Stability of p53 mRNA by Protecting It from Deadenylation.
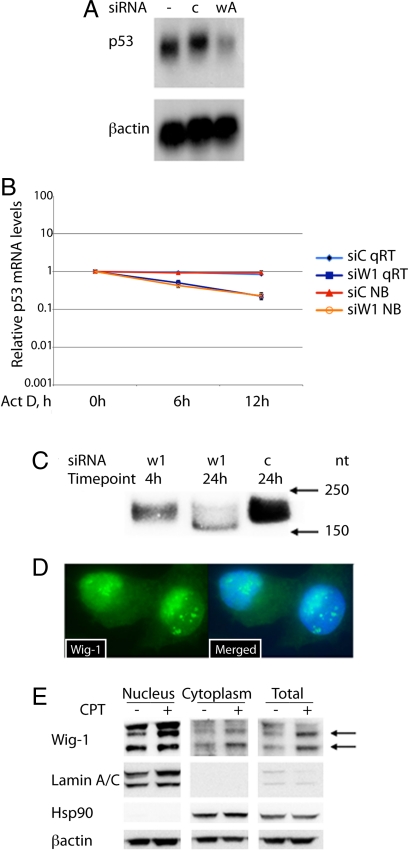
Because Wig-1 is an RNA-binding protein (13, 17), we investigated whether Wig-1 knockdown or overexpression affects p53 mRNA levels. We transfected U2OS and MCF7 cells with siRNA against Wig-1 and U2OS cells with Wig-1 expression plasmid and assessed p53 mRNA levels by Northern blotting or real-time PCR. As shown in Fig. 2A and Fig. S2 a and b, Wig-1 knockdown resulted in reduced p53 mRNA levels, whereas Wig-1 overexpression had the opposite effect.
Fig. 2.
Wig-1 knockdown decreases p53 mRNA stability through increased deadenylation. (A) Northern blot analysis shows decreased p53 mRNA levels after treatment with Wig-1 siRNA in U2OS cells. c and wA (Wig-1 siRNA), both from Ambion. (B) MCF7 cells were treated with siRNA (c and w1) and subsequently with the transcriptional inhibitor actinomycin (Act) D and harvested at 6 or 12 h. Total RNA was analyzed by Northern blotting followed by quantification using densitometry (NB) or analyzed by real-time PCR (qRT). The p53 levels were normalized to GAPDH RNA. The figure shows the mean of three independent experiments. Bars are shown with SEM (P = 0.0005 and P = 0.011 at 6 h, and P = 0.0001 and P = 0.0002 at 12 h for the Northern blotting and the real-time data, respectively). (C) Analysis of the length of the p53 poly(A) tail on Wig-1 knockdown. We prepared total RNA from siRNA-treated cells, added an adapter to the 3′ end of the mRNA, and performed cDNA synthesis followed by PCR of the end of the p53 3′ UTR (see Materials and Methods; Fig. S2f). PCR products were separated by gel electrophoresis, blotted to a membrane, and detected by using radiolabeled probes. Wig-1 knockdown resulted in decreased size of the amplified product, showing that absence of Wig-1 leads to enhanced deadenylation of the p53 mRNA. (D) Immunofluorescence staining for Wig-1 in U2OS cells demonstrates both nuclear and cytoplasmic localization of Wig-1. (E) HCT116 p53+/+ cells untreated or treated with camptothecin were fractionated into nucleus and cytoplasm and 30 μg of protein from each fraction (corresponding to ≈1% of the cytoplasmic fractions and 20% of the nuclear fractions) was analyzed by Western blotting alongside total lysate. The figure shows an increase in Wig-1 levels in both compartments after camptothecin treatment. Arrows indicate the main Wig-1 species. Lamin A/C is a nuclear marker and Hsp90 a cytoplasmic marker.
Next, we asked whether p53 mRNA stability was reduced in the absence of Wig-1. MCF7 cells transfected with siRNA targeting Wig-1 or control siRNA were treated with actinomycin D, which inhibits transcription. The p53 mRNA was analyzed by Northern blotting and real-time PCR. In cells treated with siRNA targeting Wig-1, we observed a 77–78% reduction in mRNA levels after 12 h of actinomycin D treatment, as compared with a 6–17% reduction in control siRNA treated cells (Fig. 2B; Fig. S2c). This result shows that Wig-1 increases the stability of p53 mRNA. To investigate whether Wig-1 has any effect on the transcription of p53, we examined the activity of the p53 promoter linked to a luciferase reporter gene on treatment with Wig-1 siRNA (Fig. S2d). No effect was observed. To investigate the mechanism behind the effect on mRNA stability, we looked at the length of the poly(A) tail of p53 mRNA after Wig-1 knockdown. Shortening of the poly(A) tail is one of the first and rate-limiting steps of mRNA degradation (18). Fig. 2C shows a decrease in size of the cDNA product of the very end of p53 mRNA after Wig-1 knockdown, indicating that loss of Wig-1 leads to deadenylation of the p53 mRNA. Thus, we conclude that Wig-1 stabilizes p53 mRNA by inhibiting its deadenylation.
Wig-1 Localizes to Both Nucleus and Cytoplasm and Is Increased in Both Compartments After Stress.
Because the major site for mRNA degradation is in the cytoplasm (24) and ectopically expressed Wig-1 previously has been reported to localize to the nucleus (11, 13), we examined the localization of endogenous Wig-1 by immunofluorescence staining with an antibody generated against recombinant full-length Wig-1 protein. We detected Wig-1 both in the nucleus and in the cytoplasm of U2OS cells (Fig. 2D). Also, we fractionated cells into cytoplasmic and nuclear compartments, and investigated how Wig-1 levels were affected by stress induced by camptothecin. As a p53 target gene, Wig-1 is induced in a p53-dependent manner after DNA damage (11). In accordance with this p53-dependent induction we found increased Wig-1 levels in total lysates as well as in both nucleus and cytoplasm after camptothecin treatment (Fig. 2E). Also, heterokaryon assays revealed that Wig-1 can shuttle between the nucleus and the cytoplasm (Fig. S2e)
Effect of Wig-1 on p53 Is Mediated Through the U-Rich Region in the p53 3′ UTR.
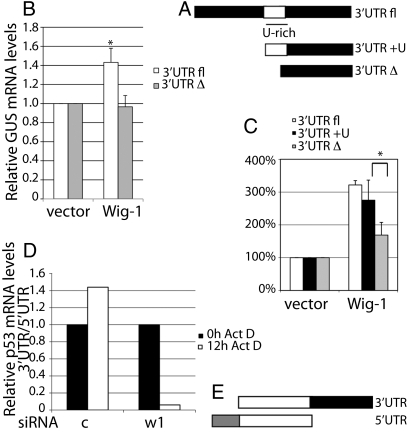
We next investigated which part of the p53 mRNA that mediates the effect of Wig-1 knockdown. The p53 constructs shown in Fig. S3a were transfected into Saos-2 (p53 null) cells that had previously been transfected with siRNA against Wig-1 or control siRNA. The expression of constructs containing the 3′ UTR of p53 was reduced after Wig-1 knockdown, whereas the constructs lacking the 3′ UTR were unaffected, as shown by Western blotting (Fig. S3b). To confirm that the 3′ UTR of p53 is required for the effect exerted by Wig-1, we used HaCaT cells stably transfected with reporter constructs containing either full-length p53 3′ UTR or the 3′ part of p53 3′ UTR (Fig. 3A) downstream of the β-glucuronidase (GUS) reporter gene. These cells were subsequently transfected with empty vector or Wig-1 expression vector (Fig. 3B). This experiment showed that ectopically expressed Wig-1 enhanced levels of the GUS reporter linked to the full-length 3′ UTR, but not the 3′ part of p53 3′ UTR. Because the U-rich region in the p53 3′ UTR, which is known to regulate mRNA stability (22), was absent in the construct with only the 3′ part of the 3′ UTR, we hypothesized that Wig-1 regulation was mediated by this region. We generated a luciferase construct that only differed by 56 nt, including the U-rich region, from the construct containing the 3′ part of the p53 3′ UTR used previously (Fig. 3A). Similarly, we made luciferase constructs containing the full-length p53 3′ UTR or the 3′ part of the 3′ UTR. HCT116 p53 −/− cells were transiently cotransfected with these reporter constructs and empty vector or the Wig-1 expression vector, and luciferase activity was assessed (Fig. 3C). Coexpression of Wig-1 increased the luciferase activity of the full-length p53 3′ UTR construct and the construct including the U-rich region, but had only a minor effect on the construct lacking the U-rich region, showing that the U-rich region is required for the effect of Wig-1 on p53 mRNA. The more potent effect in the luciferase assay is likely due to the different detection methods and the somewhat varying plasmid transfection efficiency. It is also possible that Wig-1 may have some impact on translation in addition to mRNA stability, as noted for other ARE-regulating proteins (23).
Fig. 3.
The p53 mRNA stabilization by Wig-1 requires the U-rich region of the 3′ UTR. (A) HaCaT cells stably transduced with the HRSp-GUS construct p53 3′ UTR fl or p53 3′ UTR Δ were transfected with empty vector or Wig-1 expression vector. (B) Levels of GUS and puro mRNA were determined by real-time PCR, and GUS levels were normalized to puro levels. The graph shows the average of three samples. Bars are shown with SEM, and asterisk indicates a significant difference (P = 0.044). (C) HCT116 p53 −/− cells were transiently cotransfected with luciferase reporter constructs containing the 3′ UTR regions shown in A linked to luciferase together with empty vector or Wig-1 expression vector. Three samples were measured in triplicates and bars are shown with SEM. Asterisk indicates the statistically significant difference (P = 0.016) between the effect of Wig-1 on the constructs differing in the 56 nt containing the U-rich region. (D) HCT116 p53 −/− cells were transfected with Wig-1 or control siRNA, and subsequently with the p53 constructs (E). Cells were treated with actinomycin D 24 h posttransfection, harvested at 0 and 12 h, and purified RNA was analyzed by real-time PCR. The p53 mRNA levels were normalized to GAPDH levels. Ratios between p53 mRNA levels from cells transfected with the 3′ UTR and 5′ UTR-containing constructs are shown.
To confirm that Wig-1 regulates p53 mRNA stability through the 3′ UTR, we transfected HCT116 p53 −/− cells with Wig-1 or control siRNA, and subsequently with p53 expression constructs containing the ORF and the 5′ UTR or the ORF and the 3′ UTR. Cells were treated with actinomycin D 24 h posttransfection and harvested at 0 and 12 h, followed by RNA purification and analysis by real-time PCR. In cells transfected with Wig-1 siRNA, but not with control siRNA, we observed a marked reduction in p53 mRNA levels (normalized to GAPDH) after 12 h of actinomycin D treatment (Fig. 3 D and E).
Wig-1 Binds to the 3′ UTR of p53 mRNA.
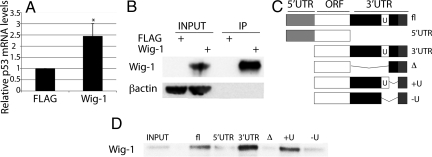
Because Wig-1 binds to dsRNA (13, 17), we investigated whether it binds directly to p53 mRNA by performing an RNA immunoprecipitation (IP) experiment. HCT116 p53 +/+ cells were transfected with FLAG-Wig-1 or empty vector, and Wig-1 was immunoprecipitated by using anti-FLAG beads. RNA was then purified and subjected to analysis by real-time PCR. The p53 mRNA was enriched >2-fold in the Wig-1-transfected lysate compared with the vector transfected lysate (Fig. 4A and B). We conclude that Wig-1 binds to p53 mRNA. Next, we performed a biotin pull-down assay to identify the region of the p53 mRNA that is required for Wig-1 binding. We labeled full-length p53 mRNA or p53 mRNA lacking various regions (Fig. 4C) with biotin, and used these labeled RNA probes to pull down FLAG-Wig-1 from U2OS cell lysates. Importantly, FLAG-Wig-1 was pulled down with the full-length p53 RNA probe and the other probes that contain the U-rich region of the 3′ UTR, but not with the probes lacking the U-rich region, demonstrating that Wig-1 binds to the U-rich region in the 3′ UTR of p53 mRNA (Fig. 4D). Next, we asked whether Wig-1 RNA binding is required for the positive regulation of p53 levels. To this end, we ectopically expressed WT Wig-1 and a zinc finger 1 point mutant (mut)Wig-1 that does not bind to RNA (17) in the HaCaT cells stably transduced with GUS reporter constructs containing p53 3′ UTR, and measured GUS mRNA levels by real-time PCR (Fig. S4a). Whereas expression of WT Wig-1 increased GUS mRNA levels, the mutWig-1 had no effect. We confirmed this result by expressing the same mutant in U2OS cells and analyzed p53 protein levels by Western blotting (Fig. S4b). Quantification using densitometry of the data in Fig. S4b showed a 123% increase in p53 levels on expression of WT Wig-1 and a 31% increase on expression of mutWig-1. This finding shows that Wig-1 RNA binding is necessary for its effect on p53.
Fig. 4.
Wig-1 binds to p53 mRNA in a U-rich region-dependent manner. (A) To establish whether Wig-1 binds directly to p53 mRNA, we performed a RNA IP experiment on lysates from cells transfected with FLAG-Wig-1 (Wig-1) or empty vector (FLAG). Wig-1 was precipitated with anti-FLAG beads, and bound RNA was purified and analyzed by real-time PCR. The graph shows the average of four independent experiments, error bars are shown with SEM, and asterisk indicates a statistically significant difference (P = 0.044). (B) Wig-1 protein levels were analyzed by Western blotting to ensure efficient pull-down. (C) To determine what region was required for Wig-1 binding to the p53 mRNA, we used the biotinylated p53 RNA probes to pull down Wig-1 from lysates of U2OS cells overexpressing FLAG-Wig-1. The constructs +U and −U differs in 70 nt containing the U-rich region. The probes were captured by streptavidin beads, and bound protein was analyzed by Western blotting (D).
Discussion
Our present results demonstrate that the p53 target Wig-1 regulates p53 at the mRNA level via an ARE in the p53 3′ UTR; thus, forming a positive feedback loop that sustains high levels of p53 expression. This regulation represents a previously undescribed mechanism for p53 regulation in several aspects. It operates through blocking deadenylation of p53 mRNA, resulting in p53 mRNA stabilization and, thus, enhanced p53 protein expression. Our data indicate that the ability of Wig-1 to maintain high p53 mRNA levels contributes significantly to the basal levels of p53, and that this regulation can augment the p53 response to cellular stress. Consistent with this idea, the DNA-damaging agent camptothecin induced Wig-1 levels in the cytoplasm, which is the primary site for mRNA degradation (24). The Wig-1-mediated augmentation of p53 mRNA levels may serve to prolong the p53 response by providing an adequate mRNA supply for de novo protein synthesis. In agreement with this notion, Wig-1 knockdown in rat neurons attenuates methamphetamine-induced p53-dependent cell death (25).
Our findings also suggest that p53-independent regulation of Wig-1 could potentiate or attenuate the p53 biological response by affecting the levels of p53 mRNA. The HuR protein has been shown to bind to the 3′ UTR of p53 mRNA and enhance p53 translation, as well as increase mRNA stability (22). Thus, both HuR and Wig-1 can bind p53 mRNA and enhance p53 protein levels. However, unlike Wig-1, HuR is not a direct transcriptional target of p53, but has been reported to translocate to the cytoplasm on UV-stress, were it stabilizes p53 mRNA (20). Interestingly, in our hands, cytoplasmic HuR levels are not increased after camptothecin treatment (Fig. S5a), in contrast to cytoplasmic Wig-1 levels that were significantly induced. This finding indicates that different stress agents may induce p53 mRNA levels through different mediators. It is also conceivable that certain types of stress can induce both HuR and Wig-1 and, thereby, trigger an even more robust increase in p53 expression.
Our observation that Wig-1 regulates p53 mRNA through an ARE suggests that Wig-1 may also target other ARE-containing mRNA. This notion is in agreement with our previous observation that Wig-1 forms RNA-containing complexes with hnRNP A2/B1, a known ARE-binding protein (26, 27). Also, we have identified several previously undescribed putative Wig-1 targets (Fig. S5b) that control cell growth, a common feature of ARE-regulated genes. These results add to the complexity of Wig-1 function, which is likely to be dependent on the cellular context. A dual role of Wig-1 in both promoting and inhibiting cell growth is supported by our previous finding that both overexpression and knockdown of Wig-1 can reduce cell growth (26).
Our findings establish a link between the p53 tumor suppressor pathway and mRNA regulation, a process increasingly acknowledged for its importance in both development and tumorigenesis, and suggest that p53 can regulate expression of specific genes via Wig-1-mediated control of mRNA stability. The identification of such p53/Wig-1 target mRNA is clearly an important task for future studies.
Materials and Methods
Cell Culture, Drug Treatment, and Colony Formation Assay.
Cells were cultured in Iscove's modified Dulbecco's medium or DMEM 1885+ (Sigma–Aldrich) supplemented with 1% l-glutamine (Sigma–Aldrich), 10% FBS (Sigma–Aldrich), and 2.5 μg/mL Plasmocin (InvivoGen) or 1% penicillin and streptomycin, or without antibiotics for transfection experiments. Stably transduced HaCaT cell lines were cultured in the presence of puromycin (Sigma–Aldrich) at a final concentration of 2.5 μg/mL for selection. For drug treatments see Table S1. For the CFA, cells were transfected in 6-well plates 24 h before drug treatment, treated, then plated on 10-cm plates in triplicate. Cells were grown in the presence of 1.5 mg/mL G418 (GIBCO) for 9 days, fixed, and stained with Giemsa stain (Sigma–Aldrich) according to manufacturer's protocol.
Transfections.
Primer and oligonucleotide sequences are shown in Table S2. Wig-1 siRNA and control siRNA from Ambion were synthesized by using the Silencer siRNA Construction kit (Ambion). Oligofectamine (Invitrogen) was used as a transfection reagent for siRNA from Ambion, whereas HiPerFect (Qiagen) was used for Qiagen siRNA according to the manufacturer's protocols. For plasmid transfections, plasmids were generated as indicated in SI Materials and Methods. For Wig-1 or p53 protein, 0.5–2 μg of DNA per well in a 6-well plate or 10 μg in a 10-cm plate was transfected by using PEI according to a standard protocol. For transfection with reporter constructs, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer's protocol.
Cellular Fractionation and Western blotting.
For buffers, see Table S3. For fractionation, cells grown on 10-cm plates were scraped in PBS, resuspended in 500 μL of ice-cold hypotonic buffer (Active Motif), incubated on ice for 15 min, lysed by addition of 25 μL of 10% Nonidet P-40, vortexed for 10 s, and centrifuged at 14,000 × g for 30 s. Supernatants were collected as cytoplasmic fraction. The pellet (nuclear fraction) was washed once in hypotonic buffer and lysed in 50 μL of WBLB. Cells for Western blotting were lysed in WBLB. Protein concentration was measured by using the Bradford reagent (Bio-Rad), and equal amounts of protein were loaded on SDS/10% polyacrylamide gels (Invitrogen) in loading buffer and reducing agent (Invitrogen) and run in Mops buffer (Invitrogen). Proteins were blotted to Hybond-ECL membranes (Amersham Biosciences) and probed with antibodies against p53 (DO1, 1:1,000, PharMingen; or FL-393, 1:500, Santa Cruz Biotechnology), Wig-1 (1:1,000) (17), β-actin (1:5,000; Sigma–Aldrich), p21 (1:1,000; Cell signaling), Hsp90 (1:15,000; SantaCruz), or Lamin A/C (1:2,000; SantaCruz). Primary antibodies were detected by using HRP-conjugated secondary antibodies (GE Healthcare) and Super Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific) with a CCD camera (Fujifilm).
RNA Preparations, Northern Blotting, and Quantitative Real-Time PCR.
Primer sequences are shown in Table S2. Total RNA was extracted by using TRIzol (Invitrogen) or the RNeasy kit (Qiagen). Real-time PCR performed on RNA from HaCaT cells was run on an MJ Research DNA Engine Opticon machine. The QuantiTect SYBR Green RT-PCR Kit from Qiagen was used to assemble 25-μL real-time PCRs in triplicates. Real-time PCR on RNA from the HCT116 cells was carried out in an Applied Biosystems 7500 real-time PCR machine using transcript-specific TaqMan Gene Expression Assays. For Northern blotting, 12 μg of total RNA was separated on a formaldehyde-Mops 1% agarose gel and blotted onto a Z-probe nylon membrane (Bio-Rad). Total RNA was hybridized with PCR-derived probes for p53, β-actin, or GAPDH at 42 °C using Ultrahyb solution (Ambion). Labeling and purification of probes was performed using Ready-to-go beads and ProbeQuant G-50 Micro Columns (Amersham Bioscience).
Deadenylation Assay.
Primer and probe sequences are shown in Table S2. The length of the poly(A) tail of p53 mRNA was monitored by using a PCR-based technique based on the property of yeast poly(A) polymerase to add 14 guanosine nucleotides to the 3′ end of RNA transcripts (28), thus providing a fixed-length target for reverse transcription. Total RNA (400 ng) was poly(G)-tailed with 0.5 nM GTP at 37 °C for 60 min using 300 units of yeast poly(A) polymerase (USB). Reverse transcription using the tailed primer PGRT was followed by PCR using the universal primer T7L and the gene-specific primer P53FOR1 located 90 bp upstream of the polyadenylation site in p53 mRNA. The resulting PCR product, encompassing the entire p53 mRNA polyadenylation sequence, was separated on a 15% polyacrylamide gel, and hybridized overnight with the end-labeled probe TP53P1, located 30 bp upstream of the poly(A) tract. This approach allowed visualization of p53 PCR products differing in size because of variation in the length of the polyadenylation sequence.
Luciferase Assay, Immunofluorescence Staining, and Pan-Caspase Activity Assay.
For luciferase assays, cells were harvested in lysis buffer, and luciferase activity was assessed by using the Dual Luciferase Assay Kit (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. Each transfected well was assayed in triplicates or duplicates.
For immunofluorescence staining, U2OS cells were plated on coverslips, grown for 24 h, washed in PBS, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and probed with Wig-1 rabbit polyclonal antibody (1:200) (17) in blocking buffer (Table S3) at 4 °C overnight. Next day, the coverslips were washed in PBS, incubated with secondary FITC-conjugated anti-rabbit antibody (1:200; DakoCytomation) in blocking buffer at room temperature for 60 min. Coverslips were mounted by using Vectashield H-1000 containing DAPI (Vector Laboratories) and examined under a Zeiss Axioplan 2 fluorescence microscope with an AxioCam HRm Camera (Carl Zeiss) using Axio Vision 4.2. For analysis of pan-caspase activity, cells were labeled with FAM-VAD-FLICA, which detects active caspases, according to the manufacturer's recommendations (Ichemicon). Caspase-positive cells were assessed by FACS using a FACSCalibur flow cytometer (Becton Dickinson) using the Cell Quest software.
RNA IP and Biotin Pulldown Assay.
For buffers, see Table S3. For the RNA, IP transfected cells were lysed in RIP buffer B, cleared by centrifugation at 20,000 × g 30 min at 4 °C, and filtered through a 45-μm filter. Lysates containing 30 mg of protein were subjected to IP for 2 h rotating at 4 °C on 600 μL of anti-FLAG M2 beads (Sigma–Aldrich), preequilibrated three times with RIP buffer A and once with RIP buffer B rotating at 4 °C for 10 min. Input was removed, and beads were washed twice with buffer B rotating at 4 °C for 10 min, collected between washes by centrifugation at 50 × g for 2 min at 4 °C. RNA was extracted by two rounds of phenol/chloroform/isoamyl alcohol (Ambion) and one round of chloroform (Merck) extraction, each round consisting of three times 30-s vortexing and 30-s incubation at 60 °C, followed by centrifugation at 20,000 × g for 5 min. RNA was precipitated by using 1 vol of isopropyl alcohol (Invitrogen), 0.1 vol of sodium acetate, and 1 μL of glycoblue (Ambion). Biotin probes were generated by in vitro transcription by Sp6 RNA polymerase (Roche) in the presence of biotin labeling mix (Roche) according to the manufacturer's protocol and using the indicated constructs as templates. U2OS cells transiently transfected with pCMVtag2b-hwig-1 were lysed in 100 μL of PLB, left on ice for 5 min, then frozen at −80 °C. Lysates were thawed on ice and cleared by centrifugation at 15,000 × g for 15 min at 4 °C. One hundred micrograms of protein was mixed with 5 μg of biotinylated probe in CBB. Proteins were allowed to bind probes for 30 min while rotating at room temperature and then pulled down with 10 μL of streptavidin Dynabeads (New England Biolabs) while rotating for 10 min at 4 °C. Beads were collected by using magnets, washed twice with 1 mL of CBB, resuspended in Western blot loading buffer (see above), and analyzed by Western blotting.
Statistical Methods.
Two-tailed t tests were used to determine statistical significance.
Supplementary Material
Acknowledgments.
We thank Michael Kolbj/orn Jensen and Louise Bro for excellent technical assistance, Dr. Reuven Agami (Netherlands Cancer Institute, Amsterdam) for providing the MCF-7 p53wt/p53kd cells, and Dr. Bert Vogelstein (Johns Hopkins Oncology Center, Baltimore) for the HCT116 cells. This work was supported by grants from the Swedish Cancer Society, Swedish Medical Research Council, the Gustaf V Jubilee Fund and Karolinska Institutet (K.G.W.), the Danish Research Council for Nature and Universe, the Carlsberg Foundation, the Åse and Ejnar Danielsens Foundation, the Becket Foundation, the Einar Willumsens Foundation, and the Karen Elise Jensens Foundation (B.N.).
Footnotes
Conflict of interest statement: K.G.W. is cofounder, shareholder, and board member of Aprea AB, a company that develops p53-based cancer therapy.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900862106/DCSupplemental.
References
- 1.Vousden KH, Lu X. Live or let die: The cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: The p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 4.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 5.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 6.Lynch CJ, Milner J. Loss of one p53 allele results in four-fold reduction of p53 mRNA and protein: A basis for p53 haplo-insufficiency. Oncogene. 2006;25:3463–3470. doi: 10.1038/sj.onc.1209387. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, You S, Foster LK, Farris J, Foster DN. The rapid destabilization of p53 mRNA in immortal chicken embryo fibroblast cells. Oncogene. 2001;20:5118–5123. doi: 10.1038/sj.onc.1204664. [DOI] [PubMed] [Google Scholar]
- 8.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 9.Mahmoudi S, et al. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 10.Varmeh-Ziaie S, et al. Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene. 1997;15:2699–2704. doi: 10.1038/sj.onc.1201454. [DOI] [PubMed] [Google Scholar]
- 11.Hellborg F, et al. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene. 2001;20:5466–5474. doi: 10.1038/sj.onc.1204722. [DOI] [PubMed] [Google Scholar]
- 12.Israeli D, et al. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendez-Vidal C, Wilhelm MT, Hellborg F, Qian W, Wiman KG. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res. 2002;30:1991–1996. doi: 10.1093/nar/30.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellborg F, Wiman KG. The p53-induced Wig-1 zinc finger protein is highly conserved from fish to man. Int J Oncol. 2004;24:1559–1564. [PubMed] [Google Scholar]
- 15.Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol Cell Biol. 2004;24:6608–6619. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Y Yang M, Wu S, Su X, May WS. JAZ mediates G1 cell-cycle arrest and apoptosis by positively regulating p53 transcriptional activity. Blood. 2006;108:4136–4145. doi: 10.1182/blood-2006-06-029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendez Vidal C, Prahl M, Wiman KG. The p53-induced Wig-1 protein binds double-stranded RNAs with structural characteristics of siRNAs and miRNAs. FEBS Lett. 2006;580:4401–4408. doi: 10.1016/j.febslet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Mazan-Mamczarz K, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galban S, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23:7083–7095. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou T, et al. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 23.Steinman RA. mRNA stability control: A clandestine force in normal and malignant hematopoiesis. Leukemia. 2007;21:1158–1171. doi: 10.1038/sj.leu.2404656. [DOI] [PubMed] [Google Scholar]
- 24.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: Is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asanuma M, et al. Suppression of p53-activated gene, PAG608, attenuates methamphetamine-induced neurotoxicity. Neurosci Lett. 2007;414:263–267. doi: 10.1016/j.neulet.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Prahl M, et al. The p53 target protein Wig-1 binds hnRNP A2/B1 and RNA Helicase A via RNA. FEBS Lett. 2008;582:2173–2177. doi: 10.1016/j.febslet.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 27.Fahling M, et al. Heterogeneous nuclear ribonucleoprotein-A2/B1 modulate collagen prolyl 4-hydroxylase, alpha (I) mRNA stability. J Biol Chem. 2006;281:9279–9286. doi: 10.1074/jbc.M510925200. [DOI] [PubMed] [Google Scholar]
- 28.Fahling M, Martin G, Keller W. Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.