Abstract
Objective
Combined hyperlipidemia is a common disorder, characterized by a highly atherogenic lipoprotein profile and a substantially increased risk of coronary heart disease. The purpose of this study was to establish whether variations of apolipoprotein A5 (APOA5), a newly discovered gene of lipid metabolism located 30 kbp downstream of the APOA1/C3/A4 gene cluster, contributes to the transmission of familial combined hyperlipidemia (FCHL).
Methods and Results
We performed linkage and association tests on 128 families. Two independent alleles, APOA5c.56G and APOC3c.386G, of the APOA1/C3/A4/A5 gene cluster were overtransmitted in FCHL (P=0.004 and 0.007, respectively). This was paired with reduced transmission of the common APOA1/C3/A4/A5 haplotype (frequency 0.4461) to affected subjects (P=0.012). The APOA5c.56G genotype accounted for 7.3% to 13.8% of the variance in plasma triglyceride levels in probands (P<0.004). The APOC3c.386G genotypes accounted for 4.4% to 5.1% of the variance in triglyceride levels in FCHL spouses (P<0.007), suggesting that this allele marks a FCHL quantitative trait as well as representing a susceptibility locus for the condition.
Conclusions
A combined linkage and association analysis establishes that variation at the APOA1/C3/A4/A5 gene cluster contributes to FCHL transmission in a substantial proportion of northern European families.
Keywords: apolipoproteins, genes, risk factors, genetics, hyperlipoproteinemia
Combined hyperlipidemia (ie, raised cholesterol and triglyceride levels) affects 1% to 2% of individuals in Western society and is present in up to 20% of patients with premature coronary heart disease (CHD).1–3 The term familial combined hyperlipidemia (FCHL) was coined by Goldstein et al1 to describe a pattern of lipid abnormalities in 47 Seattle pedigrees, which was simultaneously observed in 2 other data sets.2,4 FCHL was originally described as a dominant disorder with incomplete penetrance until the third decade and to primarily affect blood triglyceride levels and secondarily cholesterol levels.1 This was proposed because plasma triglyceride levels were bimodally distributed in the first-degree relatives of affected probands older than age 20 years and fewer than one half of the offspring of affected family members were hyperlipidemic.1 However, subsequent segregation analyses5,6 and genome-wide linkage studies7,8 suggested a more complex inheritance pattern.
The lipid profile in FCHL is characterized by increased plasma triglyceride or cholesterol levels, decreased HDL cholesterol levels, the presence of small, dense LDL particles, and elevated apolipoprotein (apo) B levels.5,9 These lipid abnormalities may also be present in persons with the metabolic syndrome, which is a major cause of CHD morbidity and mortality worldwide.10
APOA5 is a newly identified gene on chromosome 11q23 involved in lipid metabolism11 and represents a candidate for conferring susceptibility to FCHL. APOA5 was discovered by a comparative sequence analysis of human and mouse genomic DNA sequences spanning the APOA1/C3/A4 gene cluster and subjected to functional analyses.11 In mice, apoA5 deficiency results in a 4-fold increase in plasma triglyceride levels. Conversely, transgenic mice that overexpress the human APOA5 gene have markedly lower plasma triglyceride levels. In humans, single nucleotide polymorphisms (SNPs) across the APOA5 locus were associated with increased plasma triglyceride levels in 2 independent data sets.11
APOA5 resides ≈30 kbp downstream of the APOA1/C3/A4 cluster. APOA1 and C3 have been extensively studied in mice12,13 and humans.14–16 In humans, minor alleles at the APOA1−3,031C>T and APOC3c.386C>G loci have been associated with combined hyperlipidemia or hypertriglyceridemia in a range of studies.17–21 However, several investigators have failed to replicate these results,22,23 which may reflect population-specific differences in the extent of linkage disequilibrium (LD) between markers and causal variants, combined with important differences in study design. In this study, we have used a combined linkage and association study design to establish that 2 distinct sequence alleles within the APOA1/C3/A4/A5 genomic interval contribute to the transmission of FCHL in a substantial proportion of northern European families.
Methods
Families
Family data and lipid percentile values are available at http://www.csc.mrc.ac.uk. White British probands were recruited through 5 London-based tertiary referral lipid clinics. Based on previous FCHL studies1,6 and PROCAM,24 probands were identified as potential FCHL patients on the basis of both cholesterol and triglyceride levels, which had to be higher than age- and sex-specific 95th and 90th percentile values, respectively. The diagnosis was confirmed through the presence of cholesterol or triglyceride levels higher than age- and sex-specific 90th percentile values in a blood relative.1,6 In the absence of British values, Lipid Research Clinics (LRC) percentile points were used. In LRC, male 90th and 95th percentile cholesterol values (40 to 65 years of age) range from 247.4 to 259.1 mg/dL and 265.2 to 274.1 mg/dL, respectively. Triglyceride values were 170.8 to 180.5 mg/dL (75th) and 248.7 to 254.0 mg/dL (90th). Exclusion criteria included secondary hyperlipidemia (eg, body mass index [BMI] >30 kg/m2, diabetes mellitus, untreated hypothyroidism, liver and kidney disease, drugs known to interfere with lipid metabolism), age younger than 16 years, and other forms of genetic hyperlipidemia (eg, type I and III hyperlipidemia and familial hypercholesterolemia) based on molecular diagnosis, standard clinical signs, or diagnostic criteria. The research ethics committees of participating centers approved the study design. Participants gave written informed consent. Fasting lipid and apoB levels were determined by automated methods (Beckman Instruments, Inc) using commercial kits and interassay controls.
DNA Analyses
Primer sequences for genotyping markers D11S1998 and D11SAPOC3 and sequencing of APOA1, C5, A4, and A5 exons (including 5′ and 3′ splice-site junctions and 43 to 355 bp/intron) are presented in Table I (available at http://atvb.ahajournals.org). Reactions were analyzed on an Applied Biosystems 3700 DNA sequencer. SNP genotyping was performed with a polymerase chain reaction invader assay (Third Wave Technologies) as described.11,25 Allele 1 and 2 refer to common and minor alleles, respectively. Alternative nomenclature for alleles include SNP311 (APOA5−1,131C), SNP411 (APOA5−1,2238C); APOC3S2,26 APOC33238C>G27 (APOC3c.386G), and APOA1X217 (APOA1−3,031T).
TABLE 1.
Summary of Familial Combined Hyperlipidemia Data Sets
| Breakdown of Sample | Linkage | PDT | Case-Control* |
|---|---|---|---|
| Total no. of probands | 93 | 115 | 181 |
| No. of spouses | 101 | NA | 267 |
| Total no. of subjects | 926 | 888 | 448 |
| Affected sibling/relative pairs | |||
| TG | 91/184 | NA | NA |
| CHL | 140/281 | NA | NA |
| Trios: TG/CHL/APOB | NA | 89/66/89 | NA |
| Discordant sibling pairs: TG/CHL/APOB | NA | 307/271/260 | NA |
PDT indicates pedigree disequilibrium test; TG, serum triglyceride >age-sex-specific 90th percentile values, present in 2.99±1.75 individuals/pedigree; CHL, serum cholesterol and triglyceride>age-sex-specific 90th percentile values, present in 2.38±1.39 individuals/pedigree; APOB trait, serum apoB>age-BMI-sex-specific 90th percentile values. Present in 2.78±1.75 individuals/pedigree; NA, not applicable.
The data set contained 181 probands with a definitive diagnosis of FCHL and 267 FCHL spouses from the pedigrees. No genotyping was performed on the families of 53 FCHL probands, as insufficient DNA was available from key family members.
Statistical Analyses
Nonparametric linkage analysis28 was performed on families containing an affected sibling or relative pair (excluding parent and child combinations, which provide no information in this analysis) for the following 2 correlated, standard diagnostic criteria: combined hyperlipidemia phenotype (ie, cholesterol and triglyceride levels >90th age- and sex-specific values) and triglyceride trait (ie, triglyceride >90th percentile age- and sex-specific values). Estimates of allele sharing were based on marker allele frequencies of family spouses and equal weights assigned to each family. P values were derived from the Gaussian distribution approximating the NPL distribution. Combined linkage and association analysis was performed with the pedigree disequilibrium test (PDT), a transmission disequilibrium test (TDT) for general pedigrees.29 ApoB affection status was available for 725 family members. Affected status required an age and BMI adjusted value >90th percentile values. Cutoff values were computed from a random white British control sample drawn from a west London general practice. Mean and 90th percentile values were 100.4±27.1 mg/dL and 135 mg/dL (men) and 87.6±24.5 mg/dL and 127.7 mg/dL (women), respectively. P values are based on the TDT (ie, 2 genotyped parents and affected offspring) and the discordant sibling pair components of the test. The sum PDT statistic applies more weight to larger families. The average PDT applies equal weight to families.30 The gamete-competition test,31 a generalized TDT, which accommodates pedigrees of arbitrary size and complexity, was performed on all 128 families. Haplotypes were reconstructed with Merlin (http://www.sph.umich.edu/csg/abecasis) using the most likely pattern of gene flow.32 The PDT excluded families (n=4) with >1 probable haplotype solution and parents with incomplete haplotype data. Haplotype frequencies were determined with Fugue33 and used to determine normalized disequilibrium statistic (D′) values between diallelic loci.34D′ is used as a measure of LD because it is relatively insensitive to allele frequencies. ANOVA considered probands and family spouses separately. Triglyceride and cholesterol values were log transformed and standardized for the covariates BMI, age, and sex. Interactions between covariates and genotypes were evaluated. Percentage variances were estimated from mean square errors (ie, R×100). Analyses were performed in the S-PLUS package (MathSoft Inc, 2000).
Results
Increased Transmission of Minor Alleles at the APOA1/C3/A4/A5 Genomic Interval in FCHL
To evaluate the contribution of sequence variation at the APOA1/C3/A4/A5 gene locus in the transmission of FCHL, we performed nonparametric linkage analyses and a combined linkage and association test (ie, PDT) on a cohort of North European families (Table 1). In the linkage test, families were analyzed with 2 markers, D11SAPOC3, which resides within intron 3 of APOC3, and D11S1998, located ≈1.7 Mbp downstream of APOA5 (Figure, panel A). Ninety-three of the 128 families had a proband with a sibling or an informative relative pair (eg, uncle-nephew, affected cousins) for the triglyceride or the combined hyperlipidemia trait. The triglyceride trait produced NPL+ values of 1.85 (ie, LOD value, 0.74; P=0.032) and 2.0 (ie, LOD value, 0.87; P=0.023) at the D11SAPOC3 and D11S1998 loci, respectively. These values were attributable to positive linkage scores in 39 families. Corresponding NPL values for the combined hyperlipidemia trait were 0.39 and 0.47 (LOD <0.1).
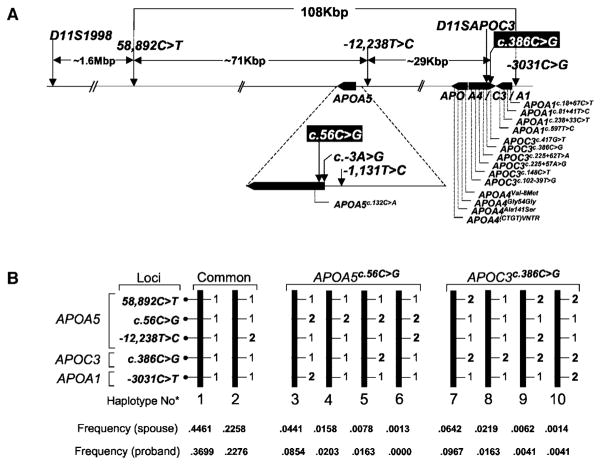
Figure 1.
Location of genetic markers and SNPs in a 108 kbp interval containing the APOA1/C3/A4/A5 gene cluster. A, Organization of the cluster (not drawn to scale) was determined from genomic (AC007707) and cDNA sequences.11 Genes are denoted by black rectangles, with attached triangles at their 3′ ends. SNPs shown in white on a black background alter the 19th codon (Ser to Trp) of APOA5 (APOA5c.56C>G) and the 40th nucleotide of the 3′ noncoding region of APOC3 (APOC3c.386C>G). The SNP at the APOA5c.−3A>G locus resides within the putative Kozak35 sequence of APOA5. Alternative nomenclature for the APOC3c.386G and APOA1−3,031T alleles include the S2 allele of APOC326 and the X2 allele of APOA1.17 B, Common, APOA5c.56G and APOC3c.386G haplotypes. 1 and 2 represents the major and minor alleles at each locus, respectively.
The PDT was performed with 7 SNPs spanning an interval of 108 kbp (Figure, panel A) on 115 nuclear families, which included 80 of the families that had been analyzed in the nonparametric linkage analyses (Table 2 and online Table II). In the PDT, genetic information is derived from an affected offspring with 2 genotyped parents, herein referred to as a trio, and/or a discordant sibling. Thirteen of the 128 FCHL families contained neither a trio nor a discordant sibling pair and were therefore excluded from the analysis. Thus, the PDT uses genetic information that is excluded in a nonparametric linkage analysis (ie, parent-child combinations and the unaffected individuals in a discordant sibling pair), whereas the PDT is unable to use certain data that are informative in a nonparametric linkage analysis. These includes data from affected sibling pairs who have an absent parent and certain affected relative pairs, such as affected cousins and uncle-nephew combinations.
TABLE 2.
PDT Results for the APOA5c0.56C>G and APOC3c0.386C>G Loci in Familial Combined Hyperlipidaemia
| Trios |
Discordant Siblings |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transmitted |
Nontransmitted |
Affected |
Unaffected* |
|||||||
| Allele | Trait | %† | n | % | n | %‡ | n | % | n | PDT P Values, Sum/Average |
| APOA5c0.56G | TG | 10.2 | 18 | 8.5 | 15 | 10.6 | 41 | 5.7 | 22 | 0.004/0.010 |
| CHL | 9.9 | 13 | 8.3 | 11 | 11.1 | 37 | 5.1 | 19 | 0.005/0.022 | |
| APOB | 8.3 | 15 | 6.7 | 12 | 13.4 | 45 | 8.0 | 27 | 0.023/0.050 | |
| APOC3c0.386G | TG | 12.6 | 22 | 8.1 | 14 | 14.6 | 56 | 10.1 | 39 | 0.019/0.011 |
| CHL | 13.9 | 18 | 6.9 | 9 | 15.7 | 52 | 10.5 | 39 | 0.010/0.007 | |
| APOB | 11.2 | 20 | 9.6 | 17 | 13.8 | 47 | 9.7 | 33 | 0.088/0.124 | |
n=number of alleles.
Levels <age-sex-specific 90th percentile values.
Allele percentage transmitted to affected offspring calculated from total number of alleles (ie, major plus minor) transmitted to offspring. Conversely, percentage not transmitted to offspring calculated from the total of parental alleles not transmitted to offspring.
Percentage of minor alleles transmitted to affected and unaffected siblings calculated from total number of alleles (ie, major plus minor) in the discordant sibling pair data set.
TG indicates serum triglyceride>age-sex-specific 90th percentile values; CHL, serum cholesterol and triglyceride>age-sex-specific 90th percentile values; APOB, apoB>age-sex-specific 90th percentile values.
We observed increased transmission of the APOA5c.56G (P=0.022 to 0.004) and APOC3c.386G (P=0.019 to 0.007) alleles to affected offspring in the trios and to affected siblings within discordant sibling ships (Table 2 and online Table II). In the complementary gamete competition test, a P value of 0.003 was obtained for the APOA5c.56G allele (combined hyperlipidemia and triglyceride traits), compared with P values of 0.014 (combined hyperlipidemia) and 0.049 (triglyceride) for the APOC3c.386G allele.
The PDT produced some support for linkage and association of the APOA5c.56G (P=0.023 to 0.050) and APOC3c.386G alleles (P=0.088 to 0.124) with elevated levels of apoB (ie, >90th age-, BMI-, and sex-specific percentile values) (Table 2 and online Table II). This was also obtained when we used slightly lower than 90th percentile cutoff values (>120 mg/dL [women] and >130 mg/dL [men]) to define apoB-affected status, but the P values were poorer (P=0.0606 to 0.1370 for the APOA5c.56G allele and 0.116 to 0.150 for the APOC3c.386G allele).
The minor alleles at the APOA5c.56C>G (Table 3), APOC3c.386C>G (Table 3), APOA5−1,131T>C (data not shown), and APOA5c.−3A>G (data not shown) loci were more prevalent in FCHL probands versus the spouses within the families. The APOA5c.56G allele was present in 22% of probands compared with 13% of normolipidemic spouses. The values for the APOC3c.386G allele were 28% and 18%, respectively. Importantly, the case-control study (ie, probands versus family spouses) complemented the PDT data. The frequencies of the minor alleles at the APOA5c.56C>G, APOA5c.−3A>G, APOA5−1,131T>C, and APOC3c.386C>G loci in FCHL probands (Table 3 and data not shown) and affected siblings (Table 2 and online Table II) were similar (ie, 0.1215, 0.1144, 0.1111, and 0.1436, respectively, versus 0.1114, 0.1156, 0.1296, and 0.1566). Likewise, the frequencies of the APOA5c.56G and APOC3c.386G alleles in spouses (0.0712 and 0.0993, Table 3) and unaffected siblings (0.0511 and 0.1048, Table 2) were comparable.
TABLE 3.
Frequencies of the APOA5c0.56C>G and APOC3c0.386C>G Genotypes in Familial Combined Hyperlipidaemia Probands and Spouses and Serum Triglyceride Levels
| Proband (N = 181) |
All Spouses (N = 267)* |
Normolipidemic† Spouses (N3192) | ||||
|---|---|---|---|---|---|---|
| Locus | Genotype | Frequencies (n) | TG, mg/dL‡ | Frequencies (n) | TG, mg/dL‡ | Frequencies (n) |
| APOA5c0.56C≥G | 1,1 | 0.785 (142) | 3823149§ | 0.8653 (231) | 129359 | 0.870 (167) |
| 1,2 | 0.188 (34) | 4973335 | 0.127 (34) | 128365 | 0.125 (24) | |
| 2,2 | 0.028 (5) | 9283572 | 0.007 (2) | 122 | 0.005 (1) | |
| APOC3c0.386C≥G | 1,1 | 0.724 (131) | 4263266 | 0.8163 (218) | 122354¶ | 0.859 (165) |
| 1,2 | 0.265 (48) | 3983122 | 0.169 (45) | 156375 | 0.135 (26) | |
| 2,2 | 0.011 (2) | 3413134 | 0.015 (4) | 182339 | 0.005 (1) | |
Includes all individuals, no exclusion criteria applied.
Cholesterol and triglyceride levels<age-sex-specific 75th percentile values.
Mean triglyceride levels±SD.
P<0.004 (ANOVA) for the effect of the APO5c0.56C>G genotype on log triglyceride levels.
P<0.01 for comparison between allele frequencies in probands versus all family spouses.
P<0.007 (ANOVA) for the effect of the APOC3c0.386C>G genotype on log triglyceride levels.
Minor Alleles at the APOA1/C3/A4/A5 Complex Influence Plasma Triglyceride Levels
In probands, the APOA5c.56C>G genotype and BMI accounted for 13.8% and 2.4% (P<10−5 and 0.016) of the total variance in log triglyceride levels, respectively (Tables 3 and III). We detected no significant interaction between the APOA5c.56C>G genotype and BMI, and this genotype made no contribution to the overall variance in apoB or total, LDL, or HDL cholesterol levels in FCHL probands or spouses. We also performed the ANOVA after the exclusion of 3 probands with serum triglyceride levels 3 SDs above the proband mean. The total variance in log triglyceride levels attributed to the APOA5c.56C>G genotype and BMI was 7.3% and 3.3% (P=0.004 and 0.007).
In spouses, the APOC3c.386C>G genotype and the covariates, age, and BMI accounted for 5.1% (P=0.003), 2.7% (P=0.006), and 7.9% (P=4×10−6) of the total variance in serum log triglyceride levels, respectively (Table 3). Values for the APOA5c.56C>G genotype and sex were 0.59 and 0.17, respectively. Two spouses had serum triglyceride levels 3 SDs above the spouse mean, but these did not have a disproportionate influence on the fraction of variance explained by this SNP (4.4%, P=0.007), age (2.7%, P=0.006), or BMI (8.4%, P=2.2×10−6). The APOC3c.386C>G genotype did not contribute to the variance in serum triglyceride levels in FCHL probands, which may suggest that the impact of an APOC3c.386G allele on serum triglyceride levels in certain susceptible individuals (ie, individuals with raised serum cholesterol levels and a blood relative with hyperlipidemia) is sufficiently large to raise triglyceride levels above the threshold value required for a diagnosis of FCHL. The APOC3c.386C>G genotype did not contribute to the variance in apoB or total, LDL, and HDL cholesterol levels in either FCHL probands or family spouses (online Table III).
In a complementary analysis, small decreases in the frequencies of the APOC3c.386G, APOA5−1,131C, and APOA5c.−3G alleles were observed in FCHL spouses with serum cholesterol and triglyceride levels <75th percentile age- and sex-specific values relative to all FCHL spouses (Table 3 and data not shown). This trend was not observed for the APOA5c.56G allele, indicating that this allele resides on a different haplotype than the APOC3c.386G, APOA5−1,131C, and APOA5c.−3G alleles.
Transmission of Haplotypes at the APOA1/C3/A4/A5 Genomic Interval in FCHL
The extent of allelic association in the APOA5 and APOA1/C3/A4 genomic interval was determined from FCHL spouse genotypes. The APOA5c.56G allele was in LD with the minor allele at the APOAI−3031C>T locus (D′=0.6, P<10−6) (Table IV) and was rarely observed on a haplotype containing the APOC3c.386G (Figure, panel B), APOA5−1,131C, and APOA5c.−3G (data not shown) alleles. The APOC3c.386G allele was in LD with the minor alleles at the APOA5−1,131T>C (D′=0.86, P<10−6) and APOA5c.−3A>G (D′=0.83, P<10−6) loci (online Table IV). Thus, it is clear that the APOA5c.56G and APOC3c.386G alleles define different haplotypes (Figure, panel B).
TABLE 4.
Transmission of APOA1/C3/A4/A5 Haplotypes in Families with Familial Combined Hyperlipidemia
| Trios |
Discordant Siblings |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transmitted† |
Nontransmitted |
Affected‡ |
Unaffected |
||||||||
| Haplotype Architecture* | Haplotype No. | Frequencies, Probands/Spouses | % | n | % | n | % | n | % | n | P Values, Sum/Average |
| 11111 | 1 | 0.3699/0.4461 | 34.0 | 111 | 43.9 | 143 | 39.4 | 145 | 45.9 | 168 | 0.011/0.012 |
| 11121 | 8 | 0.0163/0.0219 | 3.1 | 10 | 2.1 | 7 | 3.3 | 12 | 1.6 | 6 | 0.518/0.462 |
| 11211 | 2 | 0.2276/0.2258 | 22.7 | 74 | 21.5 | 70 | 21.7 | 80 | 23.0 | 84 | 0.965/0.793 |
| 12111 | 4 | 0.0203/0.0158 | 5.2 | 17 | 2.8 | 9 | 5.2 | 19 | 2.5 | 9 | 0.023/0.009 |
| 12112 | 3 | 0.0854/0.0441 | 6.7 | 22 | 6.13 | 20 | 4.3 | 16 | 3.0 | 11 | 0.275/0.448 |
| 12121 | 5 | 0.0163/0.0078 | 1.5 | 5 | 0.0 | 0 | 1.4 | 5 | 0.3 | 1 | 0.056/0.047 |
| 12211 | 6 | 0.0000/0.0013 | 0.3 | 1 | 0.0 | 0 | 0.3 | 1 | 0.3 | 1 | 0.371/0.371 |
| 21121 | 7 | 0.0976/0.0642 | 11.7 | 38 | 8.9 | 29 | 8.7 | 32 | 6.8 | 25 | 0.109/0.034 |
| 21221 | 9 | 0.0041/0.0062 | 0.6 | 2 | 0.0 | 0 | 0.5 | 2 | 0.5 | 2 | 0.157/0.206 |
| 21122 | 10 | 0.0041/0.0014 | 0.0 | 0 | 0.3 | 1 | 0.3 | 1 | 0.5 | 2 | 0.317/0.317 |
Haplotypes defined by five SNPs at the APOA1/C3/A4/A5 genomic interval. Allele 1 refers to the common allele in the sequence APOA558,892C>T, APOA5c0.56C>G, APOA5−12,238T>C, APOC3c0.386C>G, and APOA1−3,031C>T, and 2 corresponds to the rare allele. Haplotypes shown in bold are discussed in the text.
Allele percentage transmitted to affected offspring calculated from total number of alleles (ie, major plus minor) transmitted to offspring. Conversely, percentage not transmitted to offspring calculated from the number of parental alleles not transmitted to offspring.
Allele percentage transmitted to affected and unaffected siblings calculated from total number of alleles (ie, major plus minor) in the discordant sibling pair data set.
We performed the PDT with 5- and 7-locus (data available on request) haplotypes and obtained comparable results. The 5-locus haplotypes excluded APOA5−1,131T>C and APOA5c.−3A>G genotype data attributable to the strong LD between the minor alleles at these loci and the APOC3c.386G allele (online Table IV). In our data set, 17 5-locus haplotypes were identified (Table 4 and online Table V). Four contained the APOA5c.56G allele (haplotypes 3 through 6), whereas 5 contained the APOC3c.386G allele (haplotypes 5 and 7 through 10). The derivation of haplotypes for FCHL family members produced genotype data for several deceased family members for the first time and reduced the potential for a type 2 error in the TDT component (ie, trios) of the PDT, attributable to alleles associated with increased mortality.
The transmission of the 17 5-locus haplotypes to affected offspring in FCHL trios and to the unaffected and affected siblings in the discordant sibling pair data set was distorted at a global level (P=0.012). This was largely attributable to reduced transmission of the common APOA1/C3/A4/A5*1 (11111) haplotype to affected offspring in FCHL trios and to the affected siblings in the discordant siblings, paired with an increased transmission of the APOA1/C3/A4/A5*4 (12111) haplotype to these individuals (Table 4, Figure, panel B). The most common haplotype carrying the APOC3c.386G allele (APOA1/C3/A4/A5*7 [21121]) was also transmitted more frequently to the affected offspring in FCHL trios and to the affected siblings in the discordant sibling ships with a modest P value of 0.034 (Table 4).
Haplotype APOA1/C3/A4/A5*4 (12111) contains the APOA5c.56G allele and was transmitted twice as frequently to affected siblings than nonaffected siblings, and there was a 2-fold increased transmission of this haplotype to affected offspring in the trio data set (Table 4). Two other APOA5c.56G haplotypes were transmitted more frequently to affected subjects (Table 4), but these increases corresponded to poor P values (P=0.448 and 0.056).
Characterization of the APOA5c.56G Allele
We sequenced 18 APOA5c.56G alleles to establish whether an amino acid substitution in or a change to the Kozak sequence35 of APOA1, C3, A4, or A5 might account for the increased transmission of this allele to affected subjects in FCHL. The analysis included DNAs from 5 probands and 4 unrelated first-degree relatives with combined hyperlipidemia. Fourteen additional SNPs were identified (1 in APOA5, 4 in APOA4, 5 in APOC3, and 4 in APOA1 [Figure, panel A, and online Table VI]), including 2 nonsynonymous substitutions (APOA4Ala141Ser, APOA4Val-8Met). The minor allele at the APOA4Ala141Ser locus was identified on only 1 of 18 sequenced APOA5c.56G alleles and was therefore not studied additionally. LD was evident between the minor allele at the APOA4Val-8Met locus and the APOA5c.56G allele (data not shown), but this could not be computed accurately because of the low frequency of this allele in both spouses (1.5%) and probands (2.2%). These data indicate that an unidentified amino acid substitution within APOA1, C3, A4, and A5 or a sequence variation in the Kozak sequence of these genes cannot account for the linkage and association of the APOA5c.56G allele with FCHL.
Discussion
In the present study, we provide evidence for increased transmission of 2 distinct alleles at the APOA1/C3/A4/A5 genomic interval in FCHL. First, a linkage analysis based on excess allele sharing in affected sibling and relative pairs produced a P value of 0.023 for linkage of this genomic interval to the triglyceride trait of FCHL despite limited power in our data set for identifying a disease locus inherited through 2 or more biallelic loci.36 Next, a combined test of linkage and association demonstrated preferential transmission of the APOA5c.56G (P=0.004) and APOC3c.386G (P=0.007) alleles to affected individuals in FCHL families. This was supported by haplotype data and a gamete competition test performed on all 128 families.
We performed a large number of statistical tests, raising the issue of whether to adjust for multiple testing. For 50 independent tests, the Bonferroni correction requires a nominal P value of 0.001 for a result to be significant at a global level of 5%. However, this seems inappropriate for the present study for several reasons. We studied 3 highly correlated phenotypes and used marker alleles and data sets that were interdependent. Instead, we note that the PDT is robust to population stratification, and therefore spurious associations that may arise from this source are minimized. In addition, for each test performed, it would have been desirable to calculate the posterior probability of obtaining a nominal P value given the a priori power of the study. However, this could not be meaningfully calculated because the mode of transmission of FCHL is unknown. Moreover, as spelled out by Rothman,37 type 1 errors are random and therefore patterns in results should be given more weight than isolated results with a single low P value. In the present study, the pattern of P values for the APOA5c.56G and APOC3c.386G alleles are consistent with an increased transmission in FCHL.
In FCHL spouses, the APOA5c.56G allele was underrepresented and made no apparent contribution to serum triglyceride or cholesterol levels in the heterozygote state. These data complement the findings of Talmud et al,27 who studied 2808 healthy middle-aged men (50 to 61 years) drawn from UK general practices. In this sample, heterozygote individuals with the APOA5c.56G genotype (10.5% compared with 12.7% in FCHL spouses) had only marginally higher (ie, ≈8%) serum triglyceride levels than their homozygote peers without this allele. In comparison, the 11 individuals homozygous with an APOA5c.56G genotype had average serum triglyceride levels (241.6±107.1 mg/dL) that overlapped the 90th percentile values (236.30 to 251.34 mg/dL for the same age range) used in the present study to define affected status (triglyceride trait) in relatives of a proband with combined hyperlipidemia. Similarly, in 2600 randomly selected participants of the Dallas Heart Disease Prevention Project,25 the APOA5c.56G homozygote genotype (0.71% of participants) was associated with a 3-fold increased risk of elevated triglyceride levels (>90th percentile) compared with a APOA5c.56G heterozygote genotype. Thus, the defining difference between this present study and those of Talmud et al27 and Pennacchio et al25 is that we have used a combined linkage and association analysis to show that the APOA5c.56G allele contributes to the transmission of FCHL. In addition, we show that the homozygous APOA5c.56G genotype is present in 0.7% of white British FCHL spouses compared with 2.8% of FCHL probands and that FCHL probands with the homozygous APOA5c.56G genotype had some of the highest serum triglyceride levels in our data set.
We observed strong LD between the APOA5c.56G allele and the APOA1−3,031T allele, which has been previously implicated in FCHL.18,19,21,38 In one study, the APOA1−3,031T allele segregated with FCHL in a small series of families ascertained through a proband with this same allele.19 In a second study involving patients with peripheral vascular disease, all 5 patients homozygous for the APOA1−3,031T allele had combined hyperlipemia.18 In a Dutch cohort of FCHL families, the frequency of this genotype was higher in FCHL relatives compared with FCHL spouses.21,38 Moreover, the compound APOA1−3,031T and APOC3c.386G heterozygote genotype was associated with a 12-fold increased risk of FCHL. In the present study, the frequency of the compound APOA5c.56G and APOC3c.386G heterozygote genotype is remarkably similar to that observed for the combined APOA1−3,031C and APOC3c.386G genotype in affected Dutch FCHL relatives (6%), the frequencies being 5.5% for FCHL probands and 6.2% for FCHL relatives affected with the triglyceride trait of this condition. This was paired with lower frequencies of the compound APOA5c.56G and APOC3c.386G heterozygote genotype status in both unaffected FCHL relatives (2.6%) and FCHL spouses (2.2%), consistent with the lower frequency of the APOA1−3,031T and APOC3c.386G genotype combination observed in the unaffected FCHL relatives (3%) in the Dutch study. Thus, these data support the results of the Dutch study, which indicated that both paternal and maternal chromosomes might contribute to the transmission of FCHL. However, in marked contrast to the Dutch sample, we did not detect a dramatic difference in serum cholesterol and triglyceride levels in FCHL probands (online Table VII) or affected relatives with the compound heterozygote genotype status compared with probands or affected FCHL relatives without this combination. Moreover, only 1 of the 6 spouses with this double genotype had combined hyperlipidemia, the cholesterol and triglyceride levels being 301 and 213 mg/dL, respectively. In the Dutch families, the only spouse with a compound APOA1−3,031T and APOC3c.386G heterozygote genotype had a serum triglyceride level in excess of 900 mg/dL and a moderately raised cholesterol level (277 mg/dL).
Our data suggest that the APOC3c.386G allele (or an allele in LD) at the APOA1/C3/A4/A5 genomic interval may primarily affect the transmission of FCHL as a triglyceride quantitative trait. This proposition is consistent with previous APOC3 studies14,26,38–40 and results from 4 APOA5 data sets.27,41–43 In the UK data set of Talmud et al,27 for example, the haplotype equivalent to our APOA1/C3/A4/A5*7 (21121) haplotype was associated with 1.2-fold higher mean serum triglyceride levels, whereas in the study of Ribalta et al43 involving 16 FCHL families, an APOA5 allele (ie, APOA5−1,131C), which is in strong LD with the APOC3c.386G allele, was present at a higher frequency in the affected subjects (n=42) compared with the nonaffected family members (n=61). The APOA5−1,131C allele in the Ribalta study was also associated with elevated VLDL-apoB levels. We observed a 1.3-fold increased transmission of the APOC3c.386G allele to individuals with high levels of serum apoB compared with a 1.4-fold increase for the APOA5c.56G allele. However, we found that the APOC3c.386C>G genotype, which was highly predictive of the APOA5−1,131T>C genotype, had no detectable impact on total serum apoB levels in either FCHL probands or spouses, consistent with the data of Ribalta et al43 and Talmud et al.27 We also observed that the APOC3c.386G allele is in strong LD with the minor allele at the APOA5c.−3C>G locus, which substitutes a G for A at position −3 in a nucleotide of the Kozak consensus sequence of APOA5.35 Thus, in future studies, it will be important to establish whether this variant of the Kozak sequence is fully functional and, if so, whether it acts in concert with the APOC3c.386G or the APOA5−1,131C allele to perturb serum triglyceride levels.
The APOA5c.56G allele alters codon 19 of the predicted amino-terminal signal sequence of apoAV, which substitutes a serine residue with tryptophan. The von Heijne formula44 places Ser19 of apoAV at the −5 position of the preapolipoprotein (counting from the predicted cleavage site between positions −1 and −1 of mature apoAV). According to the −3, −1 rule, this region of a signal sequence has a strong preference for specific amino acids at particular positions. For example, the residue at the −1 position must be small, whereas the residue at the −3 position must not be aromatic (eg, Tyr, Trp), charged (eg, Asp), or large and polar (eg, Asn). Accordingly, missense mutations at the −1 and −3 positions of signal sequences have now been identified as the causative lesion in several serious forms of genetic diseases.45–47 By analogy, a tryptophan residue so close to the cleavage site of the apoAV signal sequence could reduce the processing of this preprotein. In transgenic mice,11 apoAV deficiency is associated with the development of high serum triglyceride levels, suggesting that the rare sequence variant at the APOA5c.56C>G locus might represent the lesion that confers susceptibility to FCHL. However, we have not formally proven that this sequence variant is the actual lesion that contributes to the transmission of FCHL, because this would require functional studies and induced mutations in mice. In addition, we note that our results do not exclude the possibility that the rare APOA5c.56G allele might be in LD with an allele that increases serum triglyceride levels through an effect on APOC3 or APOA5 mRNA levels. This would provide a plausible explanation for our results, because previous studies have established that mice expressing altered levels of these mRNAs develop elevated serum triglyceride levels.11–13 In addition, we have excluded the possibility that the APOA5c.56G allele is in LD with a sequence variant in the APOA1/C3/A4/A5 genomic interval that alters an amino acid codon (other than codon 19 of APOA5) of APOA1, C3, or A5. We have also shown that the frequency of the minor allele at the APOA4Val-8Met locus, which is in LD with the APOA5c.56G allele, is too low to account for the increased transmission of the APOA5c.56G allele in FCHL and that the APOA5c.56G allele is not in LD with an allele that alters the Kozak consensus sequence or a consensus intron/exon splice site sequences of APOA1, C3, A4, or A5.
In summary, we have used a novel combination of linkage and association analyses to establish the importance of 2 distinct alleles at the APOA1/C3/A4/A5 genomic interval in the transmission of FCHL. The results provide new insights into the early genetic studies performed in this condition and additionally indicate that the transmission of this condition is likely to occur through sequence variation at multiple loci7,8 in a substantial proportion of families and that some of these loci may have a small to moderate impact on lipid or apoB levels in the general population.
Acknowledgments
The work was supported by the British Heart Foundation (grants PG/98159 and PG/2001015) and Medical Research Council, the Hammersmith Hospital NHS Trust, London UK, the United States Department of Energy (contract No. DE-AC03-76SF00098 to E.M.R.), and the NIH-NHLBI program for Genomic Applications (grant HL66681to E.M.R.). The authors are indebted to all family participants and to Saro Nithyananathan, Stephanie A Bonney, Regina Cole, Sister Clare K.Y. Neuwirth, Daniel Savic, and Linda Smith for key technical support. They also thank Dr David Perkins (Imperial College London), Dr Martin Farrall (Wellcome Trust Centre for Human Genetics, University of Oxford, UK), Dr Goncalo Abecasis (Center for Statistical Genetics, University of Michigan, Ann Arbor, Mich), Ms Meredyth Bass and Dr Eden R. Martin (Center for Human Genetics, Duke University Medical Center, Durham, NC) and Dr Janet Sinsheimer (Department of Human Genetics and Biomathematics, UCLA, Los Angeles, Calif) for guidance on the statistical software packages.
References
- 1.Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG. Hyperlipidemia in coronary heart disease, II: genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest. 1973;52:1544–1568. doi: 10.1172/JCI107332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikkila EA, Aro A. Family study of serum lipids and lipoproteins in coronary heart-disease. Lancet. 1973;1:954–959. doi: 10.1016/s0140-6736(73)91598-5. [DOI] [PubMed] [Google Scholar]
- 3.Grundy SM, Greenland P, Herd A, Huebsch JA, Jones RJ, Mitchell JH, Schlant RC. Cardiovascular and risk factor evaluation of healthy American adults: a statement for physicians by an ad hoc committee appointed by the Steering Committee, American Heart Association. Circulation. 1987;75:1340A–1362A. [PubMed] [Google Scholar]
- 4.Rose HG, Kranz P, Weinstock M, Juliano J, Haft JI. Inheritance of combined hyperlipoproteinemia: evidence for a new lipoprotein phenotype. Am J Med. 1973;54:148–160. doi: 10.1016/0002-9343(73)90218-0. [DOI] [PubMed] [Google Scholar]
- 5.Austin MA, Horowitz H, Wijsman E, Krauss RM, Brunzell J. Bimodality of plasma apolipoprotein B levels in familial combined hyperlipidemia. Atherosclerosis. 1992;92:67–77. doi: 10.1016/0021-9150(92)90011-5. [DOI] [PubMed] [Google Scholar]
- 6.Cullen P, Farren B, Scott J, Farrall M. Complex segregation analysis provides evidence for a major gene acting on serum triglyceride levels in 55 British families with familial combined hyperlipidemia. Arterioscler Thromb. 1994;14:1233–1249. doi: 10.1161/01.atv.14.8.1233. [DOI] [PubMed] [Google Scholar]
- 7.Aouizerat BE, Allayee H, Cantor RM, Davis RC, Lanning CD, Wen PZ, Dallinga-Thie GM, de Bruin TW, Rotter JI, Lusis AJ. A genome scan for familial combined hyperlipidemia reveals evidence of linkage with a locus on chromosome 11. Am J Hum Genet. 1999;65:397–412. doi: 10.1086/302490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pajukanta P, Terwilliger JD, Perola M, Hiekkalinna T, Nuotio I, Ellonen P, Parkkonen M, Hartiala J, Ylitalo K, Pihlajamaki J, Porkka K, Laakso M, Viikari J, Ehnholm C, Taskinen MR, Peltonen L. Genomewide scan for familial combined hyperlipidemia genes in Finnish families, suggesting multiple susceptibility loci influencing triglyceride, cholesterol, and apolipoprotein B levels. Am J Hum Genet. 1999;64:1453–1463. doi: 10.1086/302365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porkka KV, Nuotio I, Pajukanta P, Ehnholm C, Suurinkeroinen L, Syvanne M, Lehtimaki T, Lahdenkari AT, Lahdenpera S, Ylitalo K, Antikainen M, Perola M, Raitakari OT, Kovanen P, Viikari JS, Peltonen L, Taskinen MR. Phenotype expression in familial combined hyperlipidemia. Atherosclerosis. 1997;133:245–253. doi: 10.1016/s0021-9150(97)00134-2. [DOI] [PubMed] [Google Scholar]
- 10.Moller DE. New drug targets for type 2 diabetes and the metabolic syndrome. Nature. 2001;414:821–827. doi: 10.1038/414821a. [DOI] [PubMed] [Google Scholar]
- 11.Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 12.Ito Y, Azrolan N, O’Connell A, Walsh A, Breslow JL. Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science. 1990;249:790–793. doi: 10.1126/science.2167514. [DOI] [PubMed] [Google Scholar]
- 13.Masucci-Magoulas L, Goldberg IJ, Bisgaier CL, Serajuddin H, Francone OL, Breslow JL, Tall AR. A mouse model with features of familial combined hyperlipidemia. Science. 1997;275:391–394. doi: 10.1126/science.275.5298.391. [DOI] [PubMed] [Google Scholar]
- 14.Hegele RA, Brunt JH, Connelly PW. Multiple genetic determinants of variation of plasma lipoproteins in Alberta Hutterites. Arterioscler Thromb Vasc Biol. 1995;15:861–871. doi: 10.1161/01.atv.15.7.861. [DOI] [PubMed] [Google Scholar]
- 15.Vu-Dac N, Gervois P, Torra IP, Fruchart JC, Kosykh V, Kooistra T, Princen HM, Dallongeville J, Staels B. Retinoids increase human apo C-III expression at the transcriptional level via the retinoid X receptor: contribution to the hypertriglyceridemic action of retinoids. J Clin Invest. 1998;102:625–632. doi: 10.1172/JCI1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterworth DM, Ribalta J, Nicaud V, Dallongeville J, Humphries SE, Talmud P. ApoCIII gene variants modulate postprandial response to both glucose and fat tolerance tests. Circulation. 1999;99:1872–1877. doi: 10.1161/01.cir.99.14.1872. [DOI] [PubMed] [Google Scholar]
- 17.Hayden MR, Kirk H, Clark C, Frohlich J, Rabkin S, McLeod R, Hewitt J. DNA polymorphisms in and around the Apo-A1-CIII genes and genetic hyperlipidemias. Am J Hum Genet. 1987;40:421–430. [PMC free article] [PubMed] [Google Scholar]
- 18.Monsalve MV, Young R, Wiseman SA, Dhamu S, Powell JT, Greenhalgh RM, Humphries SE. Study of DNA polymorphisms of the apolipoprotein AI-CIII-AIV gene cluster in patients with peripheral arterial disease. Clin Sci (Lond) 1989;76:221–228. doi: 10.1042/cs0760221. [DOI] [PubMed] [Google Scholar]
- 19.Wojciechowski AP, Farrall M, Cullen P, Wilson TM, Bayliss JD, Farren B, Griffin BA, Caslake MJ, Packard CJ, Shepherd J. Familial combined hyperlipidaemia linked to the apolipoprotein AI-CII-AIV gene cluster on chromosome 11q23-q24. Nature. 1991;349:161–164. doi: 10.1038/349161a0. [DOI] [PubMed] [Google Scholar]
- 20.Tybjaerg-Hansen A, Nordestgaard BG, Gerdes LU, Faergeman O, Humphries SE. Genetic markers in the apo AI-CIII-AIV gene cluster for combined hyperlipidemia, hypertriglyceridemia, and predisposition to atherosclerosis. Atherosclerosis. 1993;100:157–169. doi: 10.1016/0021-9150(93)90202-6. [DOI] [PubMed] [Google Scholar]
- 21.Dallinga-Thie GM, Bu XD, Linde-Sibenius TM, Rotter JI, Lusis AJ, de Bruin TW. Apolipoprotein A-I/C-III/A-IV gene cluster in familial combined hyperlipidemia: effects on LDL-cholesterol and apolipoproteins B and C-III. J Lipid Res. 1996;37:136–147. [PubMed] [Google Scholar]
- 22.Wijsman EM, Brunzell JD, Jarvik GP, Austin MA, Motulsky AG, Deeb SS. Evidence against linkage of familial combined hyperlipidemia to the apolipoprotein AI-CIII-AIV gene complex. Arterioscler Thromb Vasc Biol. 1998;18:215–226. doi: 10.1161/01.atv.18.2.215. [DOI] [PubMed] [Google Scholar]
- 23.Tahvanainen E, Pajukanta P, Porkka K, Nieminen S, Ikavalko L, Nuotio I, Taskinen MR, Peltonen L, Ehnholm C. Haplotypes of the ApoA-I/C-III/A-IV gene cluster and familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 1998;18:1810–1817. doi: 10.1161/01.atv.18.11.1810. [DOI] [PubMed] [Google Scholar]
- 24.Assmann G, Schulte H. Relation of high-density lipoprotein cholesterol and triglycerides to incidence of atherosclerotic coronary artery disease (the PROCAM experience): Prospective Cardiovascular Munster study. Am J Cardiol. 1992;70:733–737. doi: 10.1016/0002-9149(92)90550-i. [DOI] [PubMed] [Google Scholar]
- 25.Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet. 2002;11:3031–3038. doi: 10.1093/hmg/11.24.3031. [DOI] [PubMed] [Google Scholar]
- 26.Shoulders CC, Grantham TT, North JD, Gaspardone A, Tomai F, de Fazio A, Versaci F, Gioffre PA, Cox NJ. Hypertriglyceridemia and the apolipoprotein CIII gene locus: lack of association with the variant insulin response element in Italian school children. Hum Genet. 1996;98:557–566. doi: 10.1007/s004390050259. [DOI] [PubMed] [Google Scholar]
- 27.Talmud PJ, Hawe E, Martin S, Olivier M, Miller GJ, Rubin EM, Pennacchio LA, Humphries SE. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum Mol Genet. 2002;11:3039–3046. doi: 10.1093/hmg/11.24.3039. [DOI] [PubMed] [Google Scholar]
- 28.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin ER, Bass MP, Kaplan NL. Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet. 2001;68:1065–1067. doi: 10.1086/319525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinsheimer JS, Blangero J, Lange K. Gamete-competition models. Am J Hum Genet. 2000;66:1168–1172. doi: 10.1086/302826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, Martin R, Lewitzky S. Estimation of haplotype frequencies from diploid data. Am J Hum Genet. 2001;69:114. Abstract. [Google Scholar]
- 33.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 34.Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Todorov AA, Borecki IB, Rao DC. Linkage analysis of complex traits using affected sibpairs: effects of single-locus approximations on estimates of the required sample size. Gen Epidemiol. 1997;14:389–401. doi: 10.1002/(SICI)1098-2272(1997)14:4<389::AID-GEPI4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 37.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–46. [PubMed] [Google Scholar]
- 38.Dallinga-Thie GM, Linde-Sibenius TM, Rotter JI, Cantor RM, Bu X, Lusis AJ, de Bruin TW. Complex genetic contribution of the Apo AI-CIII-AIV gene cluster to familial combined hyperlipidemia: identification of different susceptibility haplotypes. J Clin Invest. 1997;99:953–961. doi: 10.1172/JCI119260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL. An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 33 untranslated region polymorphisms. Proc Natl Acad Sci U S A. 1993;90:4562–4566. doi: 10.1073/pnas.90.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sijbrands EJ, Hoffer MJ, Meinders AE, Havekes LM, Frants RR, Smelt AH, De Knijff P. Severe hyperlipidemia in apolipoprotein E2 homozygotes due to a combined effect of hyperinsulinemia and an SstI polymorphism. Arterioscler Thromb Vasc Biol. 1999;19:2722–2729. doi: 10.1161/01.atv.19.11.2722. [DOI] [PubMed] [Google Scholar]
- 41.Endo K, Yanagi H, Araki J, Hirano C, Yamakawa-Kobayashi K, Tomura S. Association found between the promoter region polymorphism in the apolipoprotein A-V gene and the serum triglyceride level in Japanese schoolchildren. Hum Genet. 2002;111:570–572. doi: 10.1007/s00439-002-0825-0. [DOI] [PubMed] [Google Scholar]
- 42.Nabika T, Nasreen S, Kobayashi S, Masuda J. The genetic effect of the apoprotein AV gene on the serum triglyceride level in Japanese. Atherosclerosis. 2002;165:201–204. doi: 10.1016/s0021-9150(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 43.Ribalta J, Figuera L, Fernandez-Ballart J, Vilella E, Castro CM, Masana L, Joven J. Newly identified apolipoprotein AV gene predisposes to high plasma triglycerides in familial combined hyperlipidemia. Clin Chem. 2002;48:1597–1600. [PubMed] [Google Scholar]
- 44.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Chan D, Ho MS, Cheah KS. Aberrant signal peptide cleavage of collagen X in Schmid metaphyseal chondrodysplasia: implications for the molecular basis of the disease. J Biol Chem. 2001;276:7992–7997. doi: 10.1074/jbc.M003361200. [DOI] [PubMed] [Google Scholar]
- 46.Siggaard C, Rittig S, Corydon TJ, Andreasen PH, Jensen TG, Andresen BS, Robertson GL, Gregersen N, Bolund L, Pedersen EB. Clinical and molecular evidence of abnormal processing and trafficking of the vaso-pressin preprohormone in a large kindred with familial neurohypophyseal diabetes insipidus due to a signal peptide mutation. J Clin Endocrinol Metab. 1999;84:2933–2941. doi: 10.1210/jcem.84.8.5869. [DOI] [PubMed] [Google Scholar]
- 47.Sunthornthepvarakul T, Churesigaew S, Ngowngarmratana S. A novel mutation of the signal peptide of the preproparathyroid hormone gene associated with autosomal recessive familial isolated hypoparathyroidism. J Clin Endocrinol Metab. 1999;84:3792–3796. doi: 10.1210/jcem.84.10.6070. [DOI] [PubMed] [Google Scholar]