Summary
Eukaryotic RNA exosomes participate in 3′-5′ processing and degradation of RNA in the nucleus and cytoplasm. RNA exosomes are multi-subunit complexes composed of at least nine distinct proteins which form the exosome core. While the eukaryotic exosome core shares structural and sequence similarity to phosphorolytic archaeal exosomes and bacterial PNPase, the eukaryotic exosome core has diverged from its archaeal and bacterial cousins and appears devoid of phosphorolytic activity. In yeast, the processive hydrolytic 3′-5′ exoribonuclease Rrp44 associates with exosomes in the nucleus and cytoplasm. While human Rrp44 appears homologous to yeast Rrp44, it has not yet been shown to associate with human exosomes. In the nucleus, eukaryotic exosomes interact with Rrp6, a distributive hydrolytic 3′-5′ exoribonuclease. To facilitate analysis of eukaryotic RNA exosomes, we will describe procedures used to clone, express, purify and reconstitute the nine-subunit human exosome and nine-, ten-, and eleven-subunit yeast exosomes. We will also discuss procedures to assess exoribonuclease activities for reconstituted exosomes.
Introduction
Classical genetics identified one of the first proteins involved in ribosome biosynthesis, the ribosomal RNA processing protein 4 (Rrp4; Mitchell et al., 1996). Protein-A tagged-Rrp4 co-purified with a 300 kDa complex including at least four additional proteins Rrp41, Rrp42, Rrp43 and Rrp44 and complementation of yeast rrp4-1 with human RRP4 facilitated purification of the human exosome and identification of additional exosome subunits in both yeast and human exosomes (Mitchell et al., 1997; Allmang et al., 1999). These and subsequent studies led to the discovery that human and yeast exosomes include a core of nine to eleven proteins, six that share similarity to bacterial RNase PH (Rrp41, Rrp45, Rrp42, Rrp43, Mtr3 and Rrp46), three that share similarity to S1/KH RNA binding domains (Rrp4, Rrp40, and Csl4), and two that share similarity to bacterial hydrolytic enzymes, RNase D (Rrp6) and RNase II/R (Rrp44). Ten of the eleven exosome proteins are essential for yeast viability, excepting Rrp6, and depletion of any elicited RNA processing defects (Allmang et al., 1999; Schneider et al., 2007).
Homologs for each exosome gene and respective protein are found in human and yeast, but their association in the cell may differ (Graham et al., 2006). In yeast, affinity-purified exosomes from both nuclear and cytoplasmic fractions contain yeast Rrp44, whereas human exosomes have not been co-purified with human Rrp44 (Allmang et al., 1999; Raijmakers et al., 2004). Consequently, it is currently believed that nine-subunit (Exo9) or ten-subunit (Exo10) exosomes comprise core cytoplasmic exosomes in human and yeast while exosomes in the nucleus associate with the additional subunit Rrp6 (Allmang et al., 1999).
The proteins involved in 3′-5′ RNA decay share evolutionary relationships amongst prokaryotic, archaeal and eukaryotic organisms. Once initiated, 3′-5′ RNA decay in bacteria is presumed to proceed via two distinct 3′-5′ exoribonuclease activities, one catalyzed by processive hydrolytic exoribonucleases (RNase II/R), and the other catalyzed by a processive phosphorylase (PNPase). Bacterial PNPase is a core constituent of an RNA decay complex, the degradosome, a complex comprised of PNPase, the endonuclease RNase E, the helicase RhlB, and enolase (Figure 1; Carpousis, 2002). Each PNPase protomer includes four domains, two C-terminal RNA binding domains (S1 and KH domains) and two core domains that bear sequence and structural homology to RNase PH (a phosphate-dependent exoribonuclease). The N-terminal RNase PH-like core domain is non-catalytic while the second RNase PH-like core domain contains the catalytic residues required to activate phosphate for nucleophilic attack at the RNA phosphodiester backbone. Both RNase PH-like core domains contribute to formation of a composite surface for substrate binding within the central pore (Figure 1; Symmons et al., 2000; Carpousis, 2002).
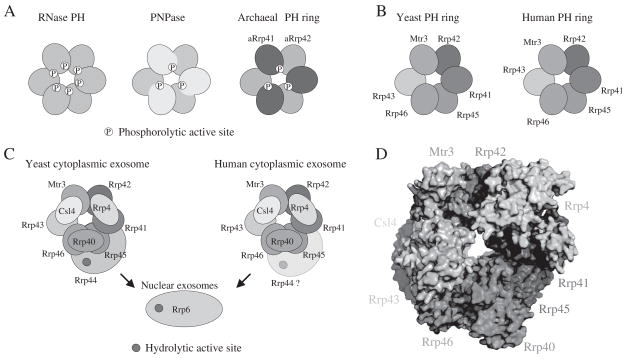
Figure 1. Schematic of bacterial, archaeal, and eukaryotic RNase PH rings and exosomes.
A) The RNase PH ring from RNase PH (left) indicating phosphorolytic active sites (circles containing a P) within the six subunits. The PNPase RNase PH-like ring (middle) indicating that PNPase encompasses a fusion between an N-terminal and C-terminal RNase PH-like domains. The C-terminal PH-like domain contains the phosphorolytic active site while the N-terminal and C-terminal PH-like domains encompass a composite substrate binding surface. Active sites are indicated. Archaeal RNase PH-like ring (right) indicating that archaeal exosomes include a RNase PH-like ring composed of a trimer of Rrp41/Rrp42 heterodimers. In this instance, the Rrp41 subunit contains the active site analogous to the active PNPase PH-like domain while Rrp42 is analogous to the PNPase N-terminal PH-like domain. B) RNase PH-like rings for yeast and human Rrp41, Rrp45, Rrp42, Mtr3, Rrp43, Rrp46. While Rrp41, Rrp46, and Mtr3 appear more similar to archaeal Rrp41 and the C-terminal PNPase PH domain, none have been shown to contain a functional phosphorolytic catalytic site. Rrp42, Rrp43, and Rrp45 appear more similar to archaeal Rrp42 and the N-terminal PNPase PH domain. The position of labels for individual subunits differences between human and yeast with respect to the sub-complexes purified. In the case of yeast, Rrp41/Rrp45, Mtr3/Rrp42, Rrp46/Rrp43 co-purified as sub-complexes whereas in human, Rrp43/Mtr3/Rrp42 and Rrp41/Rrp45 were purified as sub-complexes and Rrp46 was purified as a protomer. C) Organization of the cytoplasmic and nuclear exosomes from human and yeast. The S1/KH domain proteins Csl4, Rrp4, and Rrp40 cap the RNase PH-like ring. Rrp44 is known to associate with the yeast nine-protein core, although Rrp44 has not yet been shown to associate with the human exosome core (indicated by ‘Rrp44?’). Arrows indicate that respective cytoplasmic exosomes associate with Rrp6 in nuclear fractions. Filled circles indicate the location of hydrolytic active sites. D) The x-ray structure of the human exosome in a similar orientation to that observed in C indicating the positions of respective subunits and oriented to highlight the central pore common to RNase PH-like rings from all kingdoms of life.
Archaeal exosomes were uncovered through genomic analysis and by affinity purification; revealing that archaeal exosomes are composed of at least four proteins (Rrp41, Rrp42, Csl4 and Rrp4), so named for their relationship to the analogous eukaryotic proteins (Koonin et al., 2001; Evguenieve-Hackenberg et al., 2003). Archaeal Rrp42 and Rrp41 share sequence and structural similarity to the first and second RNase PH-like domains in PNPase, respectively (Lorentzen et al., 2005), while Csl4 and Rrp4 share similarity to the S1 and KH domains within PNPase (Buttner et al., 2005). Structural studies have revealed the architecture of six- and nine-subunit archaeal exosomes. In the former, structures showed that archaeal exosomes form stable six-subunit RNase PH-like rings through oligomerization of three Rrp41/Rrp42 heterodimers (Figure 1; Lorentzen et al., 2005; Lorentzen and Conti, 2005; Buttner et al., 2005). In the latter, structures revealed that nine-subunit archaeal exosomes are formed by capping the PH-like ring with either three Csl4 or three Rrp4 subunits (Buttner et al., 2005; Lorentzen et al., 2007). In the cell, archaeal exosomes are presumed to form complexes with mixtures of Csl4 and Rrp4, although the stoichiometry of this complex is not known. Subsequent studies revealed that RNA substrates must penetrate the pore to gain access to the phosphorolytic active sites, although it is not currently understood whether RNA is recruited to these complexes via direct interactions with Rrp41, Rrp42, Rrp4 or Cls4 (Lorentzen et al., 2007).
The architecture of the nine-subunit human exosome core was recently determined (Liu et al., 2006), revealing structural relationships amongst the eukaryotic exosome, PNPase, and archaeal exosomes. In this case, the eukaryotic RNase PH-like ring is composed of six different gene products, and single copies of Csl4, Rrp4, and Rrp40 cap the RNase PH-like ring (Figure 1; Liu et al., 2006). While the RNase PH-like domains resemble the phosphorolytic PH-like domains in PNPase and archaeal exosomes, recent studies suggest that eukaryotic exosomes have diverged mechanistically from their bacterial and archaeal cousins (Liu et al., 2006; Liu et al., 2007; Dziembowski et al., 2007). In these studies, phosphorolytic activity was not observed for nine-subunit PH-ring complexes from either yeast or human. While phosphorolytic activities were initially reported for the human nine-subunit exosome (Liu et al., 2006), activities were later determined to be due to contamination by bacterial PNPase (Liu et al., 2007). It remains unclear why eukaryotic nine-subunit exosome cores have lost their phosphorolytic activity, although we posit that phosphorolytic activity may have been lost to facilitate evolution of binding sites within the nine-subunit core, either for interaction with RNA substrates or for interaction with protein co-factors, including the hydrolytic exoribonucleases Rrp44 and Rrp6 as well as TRAMP and Ski complexes (Houseley et al., 2006).
In this chapter, we will describe procedures used to clone and express eukaryotic exosome subunits, and protocols for reconstitution of exosomes which include the nine-subunit human exosome, and nine-, ten-, and eleven-subunit exosomes from the budding yeast Saccharomyces cerevisiae. We will also describe biochemical assays used to assess the catalytic activities of these exosome preparations.
Cloning strategies for recombinant protein expression
To prepare individual exosome proteins in the absence of potential contaminating co-factors or exosome associated subunits derived from a eukaryotic host, we embarked on expression and purification of eukaryotic exosome proteins from recombinant bacterial hosts. Some human and yeast exosome proteins could be obtained as single homogeneous polypeptides; however other exosome proteins required obligate co-expression partners to maintain solubility and to facilitate co-purification of stochiometric sub-complexes. In some instances, co-expression did not suffice and protein expression required the assistance of fusion partners.
Primer sets were designed for exosome genes from yeast and human for sub-cloning into Duet vectors (Novagen), pSMT3-Topo or pSMT3 (Mossessova and Lima; 2000; Custom-Topo adapted by Invitrogen). Duet vectors facilitate co-expression by offering the opportunity to sub-clone genes into two distinct multiple cloning sites (MCS1 and MCS2) within a single plasmid. Engineered primers for Duet or pSMT3 vectors contained sequences for unique restriction endonuclease sites to facilitate ligation into respective MCS. In contrast, the pSMT3-Topo vector utilized directional Topo cloning via flap ligation (Invitrogen). In this instance, the 5′ primer required no additional engineered sequences, while the 3′ primer required addition of a 5′-CACC-3′ to facilitate flap ligation (Cheng and Shuman, 2000). Both pSMT3 and pSMT3-Topo vectors are based on pET-28b (Novagen) and encode fusions between the respective polypeptide and an N-terminal hexa-histidine Smt3 polypeptide. Fusions to the yeast SUMO ortholog Smt3 can increase expression levels and enhance solubility (Mossessova and Lima, 2000). The His6-Smt3 fusion can be liberated from the protein of interest by incubation in the presence of the Smt3 protease Ulp1. The following protocols are the result of a number of co-expression combinations tested; the most efficient of which will be presented.
PCR and sub-cloning protocols
Coding DNA for yeast exosome genes could be amplified by PCR from S. cerevisiae genomic DNA (W303-1A) as none of the yeast genes included introns. Coding DNA for human exosome genes was amplified by PCR from human placental cDNA (Ambion, Inc.). Touch-down PCR methods were utilized in conjunction with a high-fidelity polymerase such as Pfu (Stratagene). Native stop codons were maintained in all respective coding sequences. PCR products designed for Topo ligation were gel-purified (Qiagen) and subsequently ligated into pSMT3-Topo vector using directional TOPO-ligation protocols (Invitrogen). Vectors and PCR products designated for pRSF-Duet-1, pDuet-1, or pSMT3 were digested with appropriate restriction enzymes (NEB), and the vectors were treated with calf intestinal phosphatase (NEB) to dephosphorylate 5′ ends. PCR products and vectors were gel purified (Qiagen) and ligated using T4 DNA ligase (NEB). Many RNase PH-like proteins were insoluble when expressed alone (discussed below; Liu et al., 2006; Lorentzen et al., 2005), so exosome RNase PH-like genes were cloned in pairs into pDuet-1 MCS1 and MCS2 in sequential fashion. Cloning into MCS1 encodes an N-terminal fusion to a hexahistidine polypeptide (His6), whereas an untagged polypeptide was encoded through utilization of MCS2. Ligated products were transformed into chemically competent One Shot E. coli TOP10 cells (Invitrogen). Plasmid DNA was obtained from a single colony, sequence-verified, and used to transform E. coli expression strains.
PCR and sub-cloning protocols for yeast cDNA
Yeast Rrp41/Rrp45
DNA encoding yeast RRP41 and RRP45 was inserted into MCS1 and MCS2 of plasmid pRSF-Duet-1 (Novagen), respectively, utilizing restriction sites engineered into positions flanking the coding sequence. The plasmid pRSF-RRP41(MCS1)/RRP45(MCS2) encodes two polypeptides, an N-terminal His-tagged Rrp41 fusion and untagged Rrp45 (Table I). This plasmid was used to transform E. coli BL21 (DE3) Codon Plus RIL (Stratagene).
Table I.
Cloning and expression
| Human | Vector | MCS | 5′ site | 3′ site | Non-native N-terminal amino acids |
|---|---|---|---|---|---|
| Rrp45 | pDuet-1 | MCS I | EcoRI | SalI | MGSSHHHHHHSQDPNSH |
| Rrp41 | MCS II | BglII/BamHI* | SalI/XhoI* | MADP | |
| Rrp42 | pRSFDuet-1 | MCS I | BamHI | XhoI/SalI* | MGSSHHHHHHSQDP |
| Mtr3 | MCS II | NdeI | SalI/XhoI* | - | |
| Rrp43 | pSMT3 | MCS | BamHI | SalI/XhoI* | DP |
| Rrp46 | pSMT3-Topo | - | Blunt | Flap | SL |
| Csl4 | pRSFDuet-1 | MCS I | BamHI | HindIII | MGSSHHHHHHSQDP |
| Rrp4 | pRSFDuet-1 | MCS I | BamHI | SacI | MGSSHHHHHHSQDPH |
| Rrp40 | pRSFDuet-1 | MCS I | BamHI | PstI | MGSSHHHHHHSQDP |
| Yeast | Vector | MCS | 5′ site | 3′ site | Non-native N-terminal amino acids |
|---|---|---|---|---|---|
| Rrp41 | pRSFDuet-1 | MCSI | BamHI | SalI | MGSSHHHHHHSQDPH |
| Rrp45 | MCSII | BglII/BamHI* | SalI/XhoI* | MADPH | |
| Mtr3 | pRSFDuet-1 | MCSI | EcoRI | XhoI/SalI* | MGSSHHHHHHSQDPNSH |
| Rrp42 | MCSII | BglII/BamHI* | SalI/XhoI* | MADPH | |
| Rrp46 | pRSFDuet-1 | MCSI | BamHI | SalI | MGSSHHHHHHSQDPH |
| Rrp43 | MCSII | BglII/BamHI* | SalI/XhoI* | MADP | |
| Csl4 | pRSFDuet-1 | MCSI | BamHI | SalI | MGSSHHHHHHSQDP |
| Rrp4 | pRSFDuet-1 | MCSI | BamHI | SalI | MGSSHHHHHHSQDP |
| Rrp40 | pRSFDuet-1 | MCSI | BamHI | SalI | MGSSHHHHHHSQDPH |
| Rrp44 | pRSFDuet-1 | MCSI | EcoRI | XhoI/SalI * | MGSSHHHHHHSQDPNS |
| Rrp6 | pRSFDuet-1 | MCSI | EcoRI | SalI | MGSSHHHHHHSQDPNS |
| Rrp6 | pSMT3-Topo | - | Blunt | Flap | SL |
denotes loss of restriction site after ligation
Yeast Mtr3/Rrp42
DNA encoding yeast MTR3 and RRP42 was inserted into MCS1 and MCS2 of plasmid pRSF-Duet-1, respectively, utilizing restriction sites engineered into positions flanking the coding sequence. The plasmid pRSF-MTR3(MCS1)/RRP42(MCS2) encodes two polypeptides, an N-terminal His-tagged Mtr3 fusion and untagged Rrp42 (Table I). This plasmid was used to transform E. coli BL21 (DE3) Codon Plus RIL.
Yeast Rrp46/Rrp43
DNA encoding yeast RRP46 and RRP43 was inserted into MCS1 and MCS2 of plasmid pRSF-Duet-1, respectively, utilizing restriction sites engineered into positions flanking the coding sequence. The plasmid pRSF-RRP46(MCS1)/RRP43(MCS2) encodes two polypeptides, an N-terminal His-tagged Rrp46 fusion and untagged Rrp43 (Table I). This plasmid was used to transform E. coli BL21 (DE3) Codon Plus RIL.
Yeast Rrp4, Rrp40, Csl4, Rrp44, Rrp6
DNA encoding yeast RRP4, RRP40, CSL4, RRP44, or RRP6 were individually inserted into MCS1 within plasmid pRSF-Duet-1 to generate plasmids pRSF-RRP4, pRSF-RRP40, pRSF-CSL4, pRSF-RRP44, pRSF-RRP6, respectively. These constructs encoded in-frame fusions of the respective exosome polypeptides to N-terminal hexahistidine polypeptides (Table I). These plasmids were used to transform E. coli BL21 (DE3) Codon Plus RIL. To facilitate cloning within pRSF-Duet-1 MCS1, deliberate point mutations were inserted within the 5′ primers to encode proteins with the following amino acid substitutions: Rrp4 (S2A), Rrp6 (T2A) and Rrp44 (S2A). The gene for RRP40 contained a point mutation that resulted in substitution of leucine for phenylalanine at position 160. This mutation was inherent to the parental yeast strain (W303-1A) that was used to prepare genomic DNA.
Yeast Rrp6 fused to Smt3
DNA encoding RRP6 was also ligated into pSMT3-Topo (Mossessova and Lima, 2000; custom plasmid; Invitrogen) resulting in plasmid pSMT3-RRP6 which encodes a fusion between an N-terminal His6-Smt3 polypeptide and Rrp6. The His6-Smt3 polypeptide can be liberated from Rrp6 by digestion with the Smt3 protease Ulp1 to generate Rrp6 with a non-native N-terminal Ser-Leu polypeptide (Table I). This plasmid was used to transform E. coli BL21 (DE3) Codon Plus RIL.
PCR and sub-cloning protocols for human cDNA
Human Rrp45/Rrp41
DNA encoding human RRP45 and RRP41 was inserted into MCS1 and MCS2 of plasmid pDuet-1 (Novagen), respectively (Table I). Plasmid pETDuet-RRP45(MCS1)/RRP41(MCS2) encodes two polypeptides, an N-terminal His-tagged Rrp45 fusion and untagged Rrp41. This plasmid was used to transform E. coli BL21(DE3) Codon Plus RIL.
Human Rrp42/Mtr3
DNA encoding human RRP42 and MTR3 were inserted into pRSF-Duet-1. RRP42 was placed into MCS1 whereas MTR3 was integrated into MCS2 (Table I). Plasmid pRSF-RRP42(MCS1)/MTR3(MCS2) encodes two polypeptides, an N-terminal His-tagged Rrp42 fusion and untagged Mtr3. This plasmid was used to transform E. coli BL21 (DE3) STAR (Invitrogen).
Human Rrp43 and Rrp46 fused to Smt3
DNA encoding human RRP43 or RRP46 were sub-cloned into pSMT3-Topo (Mossessova and Lima, 2000; custom plasmid; Invitrogen) resulting in plasmids pSMT3-RRP43 and pSMT3-RRP46, respectively (Table I). These plasmids generate fusions between an N-terminal His6-tagged Smt3 polypeptide and the respective exosome polypeptide. These plasmids were used to transform E. coli BL21 (DE3) Codon Plus RIL.
Human Rrp4, Rrp40, and Csl4
DNA encoding human RRP4, RRP40, or CSL4 was inserted into MCS1 of plasmid pRSF-Duet-1 to generate plasmids pRSF-RRP4, pRSF-RRP40, and pRSF-CSL4, respectively (Table I). These expression constructs encode in-frame polypeptide fusions that contain an N-terminal hexahistidine-tag followed by the respective exosome polypeptides. These plasmids were used to transform E. coli BL21 (DE3) Codon Plus RIL.
Expression and Purification of Yeast Exosome Proteins
Large-scale expression cultures were obtained by fermentation in a BioFlo 3000 bioreactor (New Brunswick). 2 ml of LB culture was inoculated with 50 μl glycerol stock and incubated for 5–6 hours at 37°C. This starter culture was used to inoculate 500 ml Superbroth (SB) media (Teknova) which was then incubated at 37°C overnight, but for no more than 12 hours. The overnight culture was used to inoculate the 10 L of SB in the BioFlo 3000. Cultures were maintained at 37°C and infused with air by mixing at 800 rpm. When the OD600 reached 1.5, cultures were cooled to 30°C and expression was induced by addition of IPTG to a final concentration of 0.75 mM before the culture reached OD600 of 2. Cultures were fermented for an additional 3–4 hours at 30°C. Cell cultures for expression of recombinant yeast exosome proteins were harvested by centrifugation at 4°C at 9000 × g (Beckman JLA-8.1). Supernatant was discarded and cell pellets were suspended in 20% sucrose and 50 mM Tris-HCl pH 8.0 at a concentration of 0.5 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. Unless otherwise noted, cell suspensions from 3.5 L of the 10 L fermentor culture were thawed and prepared for sonication in buffer containing 20% sucrose, 50 mM Tris-HCl pH 8.0, 0.5 M NaCl, 20 mM imidazole, 0.1% IGEPAL, 1 mM β-mercaptoethanol (BME), 1 mM phenylmethanesulphonyl fluoride (PMSF) and 10 μg/mL DNase (Sigma). Over 20 minutes, cells were disrupted by sonication using 8 × 30 second pulses at 80% power (Branson Digital Sonifier). Insoluble cell debris was removed by centrifugation at 39000 × g (Beckman JA-20).
All protein purification was conducted at 4°C. Metal-affinity chromatography (Ni-NTA SuperFlow; Qiagen) was utilized to affinity purify each recombinant yeast protein that contained a fusion to an N-terminal hexahistidine polypeptide. Lysate was applied to a chromatography column that contained 5 to 10 ml Ni-NTA resin equilibrated in Ni-wash buffer (20 mM imidazole, 350 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM BME) and washed with an additional 5 column volumes of Ni-wash buffer. Proteins were eluted from the resin in batch by application of Ni-elution buffer (250 mM imidazole, 350 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME). Fractions containing the protein or proteins of interest were pooled and passed through a 0.2 μm filter prior to application of the sample to a HiLoad Superdex 200 26/60 (GE Healthcare) gel filtration column equilibrated with gel filtration buffer (350 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME). Fractions were analyzed by SDS-PAGE (4–12% Bis-Tris polyacrylamide, MES buffer; Invitrogen), and those containing the protein or proteins of interest were pooled and concentrated using Centriplus or Centricon filtration devices (Amicon). DTT was added to the concentrated sample to a final concentration of 1 mM. Samples were snap-frozen in liquid nitrogen and stored at −80°C.
Most of the yeast RNase PH-like proteins did not behave well when expressed as individual polypeptides. Yeast Rrp46 and Rrp43 were insoluble when expressed individually. When co-expressed and co-purified, both were obtained in the soluble fraction. Yeast Rrp41 was obtained as a soluble protein after small scale expression (40 ml), but it was insoluble when expressed at preparative scales (10 L). Yeast Mtr3 was soluble, but this protein was prone to aggregation during size exclusion chromatography. With the exception that Rrp45 expression was not attempted at large scale, Rrp42 was the only yeast RNase PH-like protein that we could express, purify, and obtain as a monodisperse sample after size exclusion chromatography. As a result of these studies, the six yeast RNase PH-like proteins were expressed in pair-wise manner as described below. In each case, one of the respective RNase PH-like proteins contained an N-terminal His6-tag for affinity purification by Ni-NTA chromatography while the other untagged PH-like protein co-purified via interactions with its His6-PH-like protein partner.
Yeast Rrp41/Rrp45
A 40 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and cleared by centrifugation. Yeast Rrp41/Rrp45 was purified by metal-affinity (Ni-NTA) chromatography. Fractions containing Rrp41/Rrp45 were pooled and applied to a gel filtration column (Superdex 200). Fractions containing Rrp41/Rrp45 were pooled, concentrated to 10 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 30 mg of Rrp41/Rrp45 from 40 g cell pellet.
Yeast Mtr3/Rrp42
A 50 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and cleared by centrifugation. Yeast Mtr3/Rrp42 was purified by metal-affinity chromatography. Fractions containing Mtr3/Rrp42 were pooled and applied to a gel filtration column (Superdex 200). This sample contained the Mtr3/Rrp42 heterodimer and excess Mtr3. While the heterodimer appeared monodisperse during gel filtration, Mtr3 was prone to aggregation and eluted in the void volume of the column. Fractions containing Mtr3/Rrp42 were pooled, concentrated to 7 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 15 mg of Mtr3/Rrp42 from 50 g cell pellet.
Yeast Rrp46/Rrp43
Co-expression and purification of Rrp46/Rrp43 initially failed using SB media and large-scale fermentation insomuch as Rrp46 underwent limited degradation. While Rrp46 degradation products maintained the ability to interact with Rrp43, these subcomplexes were not competent for reconstitution into exosomes. As such, we sought alternative methods to obtain a more homogenous sample. Strains were cultured in ten 2 L baffled flasks with each flask containing 1 L of SB media. Cultures were inoculated with 5 mL overnight culture (SB) and grown at 37°C to an OD600 of 2. Cultures were cooled on ice for 30 minutes. Ethanol was added to a final concentration of 2% and IPTG was added to a final concentration of 0.25 mM. Cultures were then incubated at 18°C for 18 hours. The 10 L culture was harvested, and the cell pellet (80 g; wet weight) was suspended in lysis buffer, sonicated, and cleared by centrifugation. Yeast Rrp46/Rrp43 was purified by metal-affinity chromatography. Fractions containing Rrp46/Rrp43 were pooled, concentrated to 8 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 10 mg of Rrp46/Rrp43 from 80 g cell pellet. We attempted additional purification steps to improve the quality of these preparations, including ion exchange chromatography and fusion to His6-Smt3, but none have resulted in improved yields or purity for this heterodimer.
Yeast Rrp4, Rrp40, and Csl4
These three proteins were purified independently, but as their purification schemes are nearly identical, we will discuss them within a single paragraph. A 40 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and cleared by centrifugation. Yeast Rrp4, Rrp40, and Csl4 were purified by metal-affinity chromatography. Fractions containing the respective protein were pooled and applied to a gel filtration column (Superdex 200). Yeast Rrp4, Rrp40, and Csl4 could each be isolated as monodisperse proteins by gel filtration. Proteins were concentrated to 10 mg/ml, snap frozen in liquid nitrogen, and stored at −80°C. We typically obtain 40 mg, 15 mg, and 30 mg of Rrp4, Rrp40, and Csl4, respectively, from 40 g cell pellet.
Yeast Rrp44
A 40 g cell pellet (wet weight) was obtained, suspended in lysis buffer, sonicated, and cleared by centrifugation. Yeast Rrp44 was purified by metal-affinity (Ni-NTA) chromatography and fractions containing Rrp44 were pooled and applied to a gel filtration column (Superdex 200). Fractions containing Rrp44 were pooled, concentrated to 10 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 40 mg of Rrp44 from 40 g cell pellet.
Yeast Rrp6
A 25 g cell pellet (wet weight) was obtained, suspended in lysis buffer, sonicated, and cleared by centrifugation. Rrp6 was purified by metal-affinity chromatography. Fractions containing Rrp6 were pooled and applied to a gel filtration column (Superdex 200). Fractions containing Rrp6 were pooled, concentrated to 3 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 2 mg of His6-Rrp6 from 25 g cell pellet. The yield for Rrp6 was approximately one order of magnitude below that achieved for other yeast exosome proteins, however large-scale expression of His6-Smt3-Rrp6 improved the purity of this protein. Cultures were grown and processed as previously described. After gel filtration, His6-Smt3-Rrp6 was incubated with the Ulp1 SUMO protease at a ratio of 1000:1 at 4°C for 6–8 hours to liberate Rrp6 from His6-Smt3 (Mossessova and Lima, 2000). Rrp6 was separated from Smt3 by a second gel filtration step (Superdex 200). Fractions containing Rrp6 were pooled, concentrated to 3 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 2 mg of Rrp6 from a 40 g cell pellet. The overall yield was lower when compared to His6-Rrp6, but the purity was superior.
Expression and purification of recombinant human exosome proteins
For strains containing plasmids encoding human exosome proteins, 500 ml LB cultures containing appropriate antibiotics were inoculated with 200 μl from the respective glycerol stock and grown overnight at 37°C. The 500 ml overnight culture was used to inoculate 10 L of SuperBroth (SB). Cultures were grown by fermentation using a BioFlo 3000 reactor (New Brunswick) at 37°C to an OD600 of 2–3, cooled to 30°C, induced for expression by addition of 0.75 mM IPTG, and grown for 4 hours at 30°C. Strains containing Rrp42/Mtr3 were induced for 6 hours. Cells were harvested by centrifugation at 6000 × g (Beckman JLA-8.1) for 15 minutes at 4°C. The supernatant was discarded and cell pellets were suspended in 50 mM Tris-HCl pH 8.0 and 20% sucrose at a concentration of 0.5 g cell wet weight per ml. Suspended pellets were distributed into 50 ml conical tubes followed by snap-freezing in liquid nitrogen prior to storage at −80°C. Cell suspensions were equilibrated in lysis buffer (50 mM Tris-HCl pH 8.0, 20 % sucrose, 350 mM NaCl, 10 mM imidazole, 1 mM BME, 0.1 % IGEPAL, 10 μg/ml DNase, and 1 mM PMSF). Cells were disrupted by sonication over 20 minutes using 8 × 30 second pulses at 80% power (Branson Digital Sonifier). Insoluble cell debris was removed by centrifugation at 44000 × g (Beckman JA-20) for 1 hour at 4°C.
All protein purification was conducted at 4°C. Metal-affinity chromatography (Ni-NTA SuperFlow; Qiagen) was utilized to affinity purify each recombinant human protein that contained N-terminal hexahistidine or His6-Smt3 polypeptide fusions. Human proteins were isolated from the soluble fraction of E. coli lysate by mixing the supernatant with 10 ml of Ni-NTA Superflow resin with stirring for 1–2 hours in Ni-wash buffer (20 mM Tris-HCl pH 8.0, 350 mM NaCl, 10 mM imidazole, and 1 mM BME). The slurry was loaded into a chromatography column and washed with 100–200 ml Ni-wash buffer. Proteins were eluted by batch via application of Ni-elution buffer (20 mM Tris-HCl pH 8.0, 350 mM NaCl, 250 mM imidazole, and 1 mM BME). Fractions containing the protein or proteins of interest were pooled and the protein concentration determined by Bradford method using Bio-Rad Protein Assay reagent. Where noted, samples were applied to a gel filtration column (Superdex 200 26/60 or Superdex 75 26/60) equilibrated with gel filtration buffer (20 mM Tris-HCl pH 8.0, 350 mM NaCl, and 1 mM BME). Prior to any steps involving FPLC chromatography columns, samples were passed through a 0.2 μm filter. After each purification step, fractions were analyzed by SDS-PAGE using 4–12% polyacrylamide gradient gels (NuPAGE, MES buffer; Invitrogen), and fractions containing the protein or proteins of interest were pooled and concentrated using Centriplus or Centricon filtration devices (Amicon). DTT was added to the concentrated samples to a final concentration of 1 mM. Samples were snap-frozen in liquid nitrogen and stored at −80°C.
As observed for the yeast system, several of the human RNase PH-like proteins did not behave well when expressed as individual polypeptides, with notable exception. Unlike yeast Rrp46/Rrp43, human Rrp46 did not co-purify with human Rrp43, so Rrp43 and Rrp46 were expressed as individual polypeptides. As a result of these studies, we expressed four of the human RNase PH-like proteins in a pair-wise manner as described below. In these two cases, one RNase PH-like protein contained an N-terminal His6-tag for affinity purification by Ni-NTA chromatography while the other untagged PH-like protein co-purified via interactions with its His6-PH-like protein partner.
Human Rrp45/Rrp41
A 20 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and the lysate cleared by centrifugation. Rrp45/Rrp41 was purified by metal-affinity chromatography. Fractions containing Rrp45/Rrp41 were pooled and applied to a gel filtration column (Superdex 200), and fractions containing Rrp45/Rrp41 were pooled, concentrated to 5–15 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 30 mg of Rrp45/Rrp41 from 20 g cell pellet.
Human Rrp42/Mtr3
A 40 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and cleared by centrifugation. Rrp42/Mtr3 was purified by metal-affinity chromatography and fractions containing Rrp42/Mtr3 were pooled and concentrated. Typical yields were 10–15 ml at 5–10 mg/ml, although this is an overestimate of Rrp42/Mtr3 yield due to contaminating proteins after elution from Ni-NTA. Samples were snap-frozen in liquid nitrogen, and stored at −80°C. This sample will be used in reconstitution of the Rrp42/Mtr3/Rrp43 complex (see below).
Human Rrp43
A 20 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and cleared by centrifugation. His6-Smt3-Rrp43 was purified by metal-affinity chromatography. Fractions containing Smt3-Rrp43 were pooled and concentrated. Typical yields were 10–15 ml at 10 mg/ml, although this is an overestimate of Rrp43 yield due to contaminating proteins after elution from Ni-NTA, and because the Smt3 tag was not liberated from Rrp43 until it was mixed with Rrp42/Mtr3 (see below). Samples containing Smt3-Rrp43 were snap-frozen in liquid nitrogen and stored at −80°C.
Purification of human Rrp42/Mtr3/Rrp43
This ternary complex was obtained by reconstituting Rrp42/Mtr3 with Smt3-Rrp43 (see above) by mixing in equimolar ratio in the presence of Ulp1 at a ratio of 1000:1 Smt3-Rrp43:Ulp1 (Mossessova and Lima, 2000). This mixture was incubated at 4°C overnight or until Smt3 cleavage was complete. The sample was applied to a gel filtration column (Superdex 200). Fractions containing Rrp42/Mtr3/Rrp43 were pooled, concentrated, and buffer-exchanged into 100 mM NaCl, Tris-HCl pH 8.0, 1 mM BME. The sample was then applied to anion exchange resin (Mono Q 10/10) and eluted with a NaCl gradient from 100 mM to 450 mM over 15 column volumes. Fractions containing Rrp42/Mtr3/Rrp43 were pooled and concentrated to 5 mg/ml. Typical yields for Rrp42/Mtr3/Rrp43 were 10–20 mg from 40 g cell pellet for Rrp42/Mtr3 and 20 g cell pellet for Rrp43. Samples were snap-frozen in liquid nitrogen and stored at −80°C.
Human Rrp46
A 20 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and the lysate cleared by centrifugation. His6-Smt3-Rrp46 was purified by metal-affinity chromatography. Smt3 was liberated from Rrp46 by incubating the mixture overnight at 4°C at a ratio of 1000:1 for Smt3-Rrp46:Ulp1. The sample was applied to a gel filtration column (Superdex 75). Fractions containing Rrp46 were pooled, concentrated to 10 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 20–30 mg of Rrp46 from 20 g cell pellet.
Human Rrp4, Rrp40, and Csl4
For each strain, a 20 g cell pellet (wet weight) was suspended in lysis buffer, sonicated, and the lysate cleared by centrifugation. Human Rrp4, Rrp40, or Csl4 were purified by metal-affinity chromatography and fractions containing the respective protein were applied to a gel filtration column (Superdex 75). Fractions containing the respective protein were pooled, concentrated to 10–15 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C. We typically obtain 20–30 mg for Csl4, Rrp4, or Rrp40 from a 20 g cell pellet.
Reconstitution and purification of human and yeast exosomes
Eukaryotic exosomes are reconstituted by combining individual proteins or protein sub-complexes (see above; Liu et al., 2006). Unlike archaeal exosomes, six-subunit RNase PH-like rings do not assemble spontaneously using the six human or yeast RNase PH-like proteins, although nine-subunit complexes could be reconstituted by combining the eukaryotic RNase PH-like proteins (Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3) along with the S1/KH-domain proteins (Rrp4, Csl4 and Rrp40). To form the yeast ten- or eleven-subunit exosomes, Rrp44 and Rrp6 were included in the reconstitution. Equimolar ratios for respective subunits were ensured prior to reconstitution by determining protein concentrations on the day of reconstitution using the Bradford method. We conducted reconstitution experiments at two scales. Small-scale reconstitutions utilized ~2.5 mg of total protein in 1 ml (7 nmol per subunit) while large-scale reconstitutions have utilized as much as 30 mg of total protein in 10 ml (100 nmol per subunit).
In each instance, reconstitution began by mixing the proteins in high salt buffer (350 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME). The samples are placed into dialysis tubing with a molecular cut-off no greater than 15 kDa (Spectra-Por). Reconstitution takes place in two steps. The samples are first dialyzed against reconstitution buffer containing 100 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME and then transferred for dialysis against low salt reconstitution buffer (50 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME).
The yeast nine-subunit exosome
An equimolar ratio of subunits was combined in high salt reconstitution buffer and placed into dialysis tubing. The sample was dialyzed against 1 L reconstitution buffer containing 100 mM NaCl for 2–4 hours at 4°C. The sample was transferred and dialyzed against 1 L low salt reconstitution buffer for 6–8 hours at 4°C. After dialysis, the sample was filtered and applied to a gel filtration column (Superdex 200) equilibrated with low salt reconstitution buffer. If reconstitution was successful, the resulting complex eluted as a monodisperse peak with an apparent molecular weight of 400 kDa. Fractions containing the complex were pooled, concentrated to 5–6 mg/ml in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM TCEP (Tris(2-Carboxyethyl) phosphine), snap-frozen in liquid nitrogen, and stored at −80°C.
Incorrect estimation of concentrations for the respective subunits often led to unsuccessful reconstitution experiments. In these instances, exosome complexes eluted as two distinct but overlapping peaks during gel filtration. The observed peaks differed from peak positions observed for individual protomers or sub-complexes, suggesting the presence of partially assembled complexes. In most cases, poor reconstitution profiles for the yeast exosome core were due to sub-stoichiometric quantities of Rrp46, presumably due to Rrp46 degradation products acquired during co-expression with Rrp43 (see above). While Rrp46 degradation products interact with Rrp43, these products do not integrate into nine-subunit complexes. By adding 2.5-fold higher molar ratios of Rrp46/Rrp43 to the reconstitution mixture, reconstitutions resulted in a single monodisperse peak on gel filtration which corresponded to a nine-subunit complex as assessed by SDS-PAGE and Coomassie staining (10% polyacrylamide, MOPS buffer, Invitrogen).
The yeast ten-subunit exosome
As with the yeast nine-subunit exosome, stoichiometric quantities of the ten yeast exosome proteins were combined in high salt reconstitution buffer, including Rrp44 at 1.5 x molar excess while maintaining Rrp46/Rrp43 at an 2.5 x molar excess. Once mixed, the sample was placed into dialysis tubing and dialyzed in two steps against reconstitution buffer containing 100 mM NaCl and 50 mM NaCl, respectively (see above). After dialysis, the sample was filtered and applied to a gel filtration column (Superdex 200) equilibrated with low salt reconstitution buffer. If reconstitution was successful, the complex eluted from gel filtration as a monodisperse peak with an apparent molecular weight of 500 kDa. Fractions containing the complex were pooled, concentrated to 6 mg/ml in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM TCEP, snap-frozen in liquid nitrogen, and stored at −80°C.
The yeast eleven-subunit exosome
The eleven yeast proteins were mixed in a molar ratio (Rrp46/Rrp43 at 2.5 X, Rrp44 at 1 X, Rrp6 at 1.5 X) in high salt reconstitution buffer. The sample was dialyzed as above in two steps against reconstitution buffer containing 100 mM NaCl and 50 mM NaCl, respectively. The mixture was filtered and applied to a gel filtration column (Superdex 200). The complex eluted from gel filtration as a monodisperse peak with an apparent molecular weight of 600 kDa. SDS-PAGE and Coomassie staining revealed that fractions from this peak contained each of the eleven proteins. Fractions containing the complex were pooled, concentrated to 6 mg/ml in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM TCEP, snap-frozen in liquid nitrogen, and stored at −80°C.
Further purification of yeast exosomes
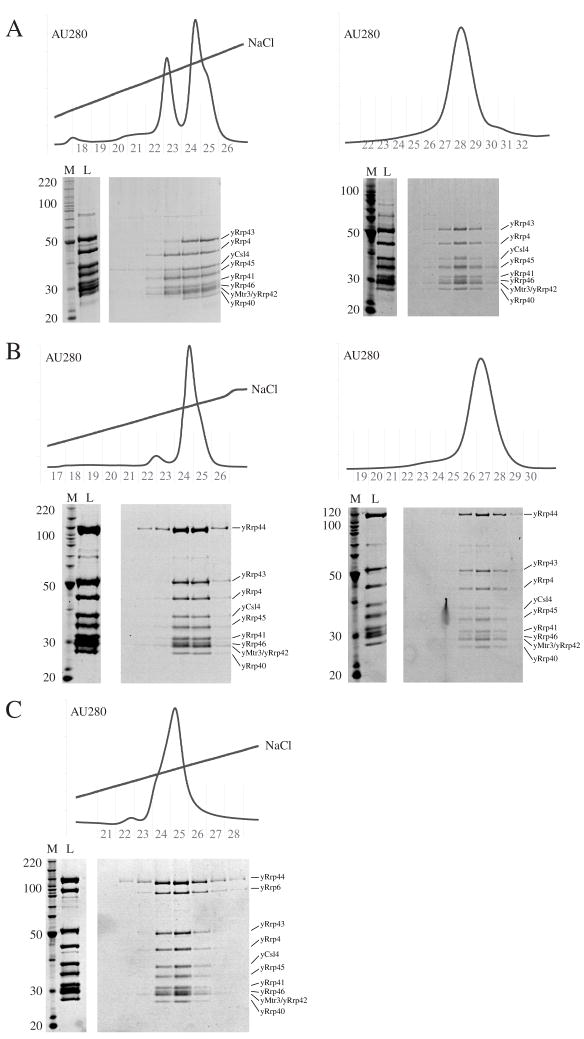
The quality of exosome preparations can be significantly improved through purification of the assembled complexes by ion exchange chromatography. The yeast nine-subunit exosome does not bind to cation exchange resin, but it does interact with anion exchange resin (Mono Q, GE Healthcare). After gel filtration, the complex was applied to a Mono Q 5/5 column equilibrated with 50 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME and eluted by a NaCl gradient from 50 mM NaCl to 1 M NaCl over 20 column volumes (Figure 2A, left). The nine-subunit complex eluted in two peaks and SDS-PAGE analysis revealed that the minor peak lacked Rrp40 and Rrp43 while the major peak contained each of the nine subunits. The major peak eluted at approximately 300 mM NaCl. To ensure that the complex did not disassemble after anion exchange, fractions containing the peak were subjected to analytical gel filtration (Superose 6 10/30 GL; GE Healthcare; equilibrated in low salt reconstitution buffer). The sample eluted as a monodisperse peak which contained all nine proteins (Figure 2A, right).
Figure 2. Ion exchange and gel filtration for yeast exosomes.
A) The nine-subunit yeast exosome. The left panel indicates the chromatogram and SDS-PAGE gel for purification of the yeast nine-subunit complex by anion exchange (Mono Q 5/5). The right panel depicts the gel filtration profile (Superose 6 10/30) and SDS-PAGE analysis for the yeast nine-subunit exosome after purification by anion exchange. B) The ten-subunit yeast exosome. The left panel indicates the chromatogram and SDS-PAGE gel for purification of the yeast ten-subunit complex by anion exchange (Mono Q 5/5). The right panel depicts the gel filtration profile (Superose 6 10/30) and SDS-PAGE analysis for the yeast ten-subunit exosome after purification by anion exchange. C) The yeast eleven-subunit exosome. The panel indicates the chromatogram and SDS-PAGE gel for purification of the yeast eleven-subunit complex by anion exchange (Mono Q 5/5). We have not yet obtained material after anion exchange for purification by gel filtration. Subunit positions are labeled adjacent to the respective gels. Fractions and lanes are aligned to assist in relating chromatograms to respective SDS-PAGE gels. Fraction numbers in chromatograms indicate 0.5 ml volumes post-injection. SDS-PAGE gels are stained with Sypro-Ruby (Bio-Rad).
To improve purification, the ten-subunit complex was applied to a Mono Q 5/5 column and eluted as described for the nine-subunit complex. As before, the complex eluted in two peaks, one which included a sub-stochiometric complex (apparent absence of Rrp40 and Rrp43) and one that included a stochiometric complex. To further improve reconstitution of the ten-subunit exosome, we mixed nine-subunit yeast exosomes purified by gel filtration and anion exchange with Rrp44 in high salt reconstitution buffer. The sample was placed into dialysis tubing and dialyzed in two steps into low salt reconstitution buffer (see above). When the mixture was applied to gel filtration (Superdex 200), the resulting peak contained all ten proteins as observed previously. This ten-subunit exosome was then applied to Mono Q 5/5 and eluted using a NaCl gradient, resulting in one major peak that eluted at approximately 300 mM NaCl and contained all ten proteins in apparent molar ratios (Figure 2B, left). To ensure that the complex did not disassemble after anion exchange, fractions containing the peak were subjected to analytical gel filtration (Superose 6 10/30 GL; equilibrated in low salt reconstitution buffer). The sample eluted as a mono-disperse peak which contained all ten proteins (Figure 2B, right).
This procedure was also employed to improve the quality of the eleven-subunit exosome. The eleven-subunit complex was applied to anion exchange resin (Mono Q 5/5) and eluted as described for the nine- and ten-subunit complexes. In this instance, the eleven-subunit exosome eluted in a major peak at approximately 300 mM NaCl, although a shoulder is apparent on the left side of the peak. SDS-PAGE analysis of the resulting fractions revealed a complex which contained each of the eleven subunits (Figure 2C).
The human nine-subunit exosome
Reconstitution of the human nine-subunit exosome was performed by mixing stochiometric quantities of Rrp41/Rrp45 with Rrp42/Mtr3/Rrp43, Rrp46, Csl4, Rrp4, and Rrp40 in high salt reconstitution buffer. This mixture (10 ml) was placed into dialysis tubing and dialyzed for 4–6 hours at 4°C against 1 L 20 mM Tris-HCl pH 8.0, 100 mM NaCl, and 1 mM BME. The sample was then dialyzed overnight at 4°C against 1 L low salt reconstitution buffer containing 10 mM Tris-HCl pH 8.0, 50 mM NaCl, and 1 mM BME. Insoluble material was removed by centrifugation and by passing the mixture through a 0.2 μm filter. The sample was then loaded onto a gel filtration column (Superdex 200) equilibrated with low salt reconstitution buffer. The complex eluted as a monodisperse peak and fractions containing the desired complex were analyzed by SDS-PAGE (4–12% Nu-PAGE gels and Coomassie-staining). Fractions containing the complex were pooled, concentrated to 6–9 mg/ml in 10 mM Tris-HCl, 50 mM NaCl, 1 mM DTT, snap-frozen in liquid nitrogen, and stored at −80°C.
To improve the quality and purity of the nine-subunit exosome, the sample was applied to either cation- or anion exchange resin (Mono S or Mono Q, respectively). Unlike the yeast complex, the human exosome core interacts with both cation- and anion exchange resins. In each instance, the complex was eluted from the ion exchange column with a gradient of NaCl from 50 mM NaCl to 400 mM NaCl, eluting at approximately 300 mM NaCl from either Mono Q or Mono S (Liu et al., 2007). As discussed previously, this step was critical to purify the human exosome away from contaminating exoribonucleases which are carried over from the recombinant E. coli host. For the human exosome core, separation was best achieved by cation exchange chromatography.
Exoribonuclease assays
Archaeal exosomes and bacterial PNPase catalyze phosphate-dependent (phosphorolytic) exoribonuclease activity (Lorentzen et al. 2005 and Symmons et al., 2000). Due to similarities observed amongst PNPase, archaeal Rrp41/Rrp42, and eukaryotic PH-like subunits, it was hypothesized that the eukaryotic exosome might possess phosphate-dependent activity. The eukaryotic exosome also possesses hydrolytic exoribonuclease activities, namely through association with Rrp44 and Rrp6, two subunits that share sequence similarity to bacterial RNase II/R and RNase D, respectively. Previous biochemical analyses on a few exosome subunits supported the hypothesis that the majority of eukaryotic exosome proteins possessed 3′-5′ exoribonuclease activity (reviewed in Raijmakers et al. 2004); however more recent analysis suggests this is not true (Liu et al., 2006, 2007; Dziembowski et al. 2007). To determine exoribonuclease activities of the exosome, we tested each reconstituted exosome as well as each individual exosome protein or sub-complex in assays for hydrolytic and phosphorolytic 3′-5′ exoribonuclease activity (Liu et al., 2006).
All RNA substrates were synthetically derived (Invitrogen), and each included a fluorescein attached to the 5′ end of the substrate to enable detection of reaction products by exciting fluorescein at 473 nm and detecting fluorescence at 520 nm. To detect reaction products, samples were separated by denaturing polyacrylamide gel electrophoresis (PAGE) and detected by scanning gels on a flat transparent surface using a Fuji FLA-5000 equipped with a FITC filter.
Preliminary biochemical analysis employed end point assays in combination with protein titration from 100 pM to 1 μM. Each protein or sub-complex was prepared as a 10 X stock (1 nM to 10 μM) by diluting proteins in protein dilution buffer (10 mM Tris-HCl pH 8.0, 10 mM DTT, 50 mM KCl, 5 mM MgCl2, 1 U/μL RNase Inhibitor (NEB) with and without 10 mM sodium phosphate buffer (pH 8.0)). The reaction buffer was analogous to protein dilution buffer, but included 11.12 nM RNA. Reactions were initiated by mixing 2 μL 10 X protein stock and 18μL reaction buffer to yield samples that contained 1 X protein and 10 nM RNA. Reactions were incubated at 37°C and quenched after 45 minutes by addition of 20 μL TBE loading buffer (Invitrogen). Reaction products were separated using 15% TBE-Urea polyacrylamide gels (Invitrogen). RNA products were detected by excitation of fluorescein at 473 nM using a Fuji FLA-5000 equipped with a FITC filter. Images were processed using a linear scale with Fuji Film Multi-Gauge V2.02.
Two RNA substrates were initially assessed, an AU-rich RNA (ARE) substrate that contained two tandem repeats of the AU-rich domain derived from the 3′-UTR of the TNFα gene and a generic RNA sequence derived from previous studies (Wang and Kiledjian, 2001; Liu et al., 2006). These assays revealed that none of the yeast exosome proteins contain phosphate-dependent exoribonuclease activity, and that only two of the subunits were capable of catalyzing hydrolytic exoribonuclease activity, Rrp44 and Rrp6 (Liu et al., 2006; Mitchell et al. 1997; Burkard and Butler, 2000). For the human exosome, we initially reported phosphorolytic activities for the Rrp41/Rrp45 heterodimer and for the nine-subunit human exosome (Liu et al., 2006), but these activities were later determined to result from contamination by E. coli polynucleotide phosphorylase (PNPase; Liu et al., 2007). This discovery necessitated further purification of reconstituted exosome complexes from yeast and human by ion exchange chromatography (described above) to remove PNPase from our samples. Our results now suggest that neither human nor yeast nine-subunit exosomes are capable of catalyzing phosphorolytic activity, distinguishing eukaryotic exosomes from their archaeal and bacterial counterparts.
Comparative exoribonuclease assays with different RNA substrates
We have assessed ten different 49 nucleotide RNA substrates in comparative biochemical assays with our exosome preparations, including the aforementioned AU-rich RNA and generic RNA. In addition, we utilized a poly-adenylate RNA (Poly(A)) and three RNA chimeras that included generic RNA sequences followed by Poly(A), generic RNA sequences followed by AU-rich RNA, and a substrate containing AU-rich RNA followed by Poly(A) (Liu et al., 2006). Four 49 nucleotide AU-rich RNA substrates were also synthesized which contained a 20 nucleotide GNRA stem loop (eight GC base pairs and GCAA tetraloop; Heus and Pardi, 1991). The GNRA substrates differed from each other with respect to the position of the stem loop within the 49 nucleotide substrate such that the AU-rich sequence elements 3′ to the GNRA stem loop varied from 5, 10, 20 to 29 nucleotides in length with commensurate shortening of AU-rich sequences 5′ to the GNRA stem loop. These substrates were used to assess the effect of stable RNA secondary structure on exosome activities, and to test the hypothesis that RNA substrates must pass through the pore to gain access to exosome catalytic sites as suggested previously for archaeal exosomes (Lorentzen and Conti, 2005, Lorentzen et al. 2007).
Exosome proteins and complexes were analyzed for activity over time using fixed concentrations of substrate and protein. For these experiments, 100 μL reactions contained 10 mM Tris-HCl pH 8.0, 10 mM DTT, 50 mM KCl, 5 mM MgCl2, 1 U/μL RNase Inhibitor (NEB), 10 nM RNA and 10 nM protein. Reactions were initiated by addition of the 10 X protein stock (as above). Reactions were incubated at 37°C and 10 μL were removed at discrete time points, usually after 1, 2, 4, 8 or 16 minutes. Samples were quenched by addition of 10 μL TBE loading buffer followed by snap-freezing in liquid nitrogen. Reaction products were separated by electrophoresis using 15% TBE-Urea gels (Invitrogen) and detected by fluorescence using the FLA-5000. Images were processed using with a linear scale using Fuji Film Multi-Gauge V2.02.
Conclusions
In this Chapter, we described methods to clone, express, purify, and reconstitute eukaryotic exosomes from human and yeast. To date, biochemical characterization of exosomes has been mainly accomplished by purifying exosomes from their endogenous source by affinity techniques. While it appears possible to purify exosomes from yeast to near homogeneity, these preparations suffer from potential contamination by endogenous co-factors, and there are still problems associated with purification of stochiometric complexes (Dziembowski et al., 2007; Wang et al., 2007). Purification of human exosomes from tissue or cell culture appears even more daunting. The protocols presented herein should enable analysis of exosome function through reconstitution of mutant subunit isoforms, and by analysis of substrate dependencies on pure components obtained prior to and after exosome reconstitution. These methods, combined with biochemical and genetic analysis, should facilitate functional studies of RNA exosomes during RNA processing and decay.
Acknowledgments
We thank Quansheng Liu for his contributions to this chapter, particularly methods used to purify and reconstitute human exosomes. J.C.G. is a trainee in the Tri-Institutional Program in Chemical Biology. J.C.G. and C.D.L. are supported in part by a grant from the National Institutes of Health (GM079196). C.D.L. acknowledges additional support from the Rita Allen Foundation.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard KT, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of Archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 30:150–155. [PubMed] [Google Scholar]
- Cheng C, Shuman S. Recombinogenic flap ligation pathway for intrinsic repair of topoisomerase IB-induced double-strand breaks. Mol Cell Biol. 2000;21:8059–8068. doi: 10.1128/mcb.20.21.8059-8068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struc Molec Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Solfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell. 2006;17:1399–409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Bio. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Gen Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, Activities, and Structure of the Eukaryotic RNA Exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, Activities, and Structure of the Eukaryotic RNA Exosome. Cell. 2007;131:188–189. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Molec Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→ 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Raijmakers R, Schilders G, Pruijn GJ. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur J Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Nat Acad Sci. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]