Abstract
The proteome of human salivary fluid has the potential to open new doors for disease biomarker discovery. A recent study to comprehensively identify and catalog the human ductal salivary proteome led to the compilation of 1166 proteins. The protein complexity of both saliva and plasma is large, suggesting that a comparison of these two proteomes will provide valuable insight into their physiological significance and an understanding of the unique and overlapping disease diagnostic potential that each fluid provides. To create a more comprehensive catalog of human salivary proteins, we have first compiled an extensive list of proteins from whole saliva (WS) identified through MS experiments. The WS list is thereafter combined with the proteins identified from the ductal parotid, and submandibular and sublingual (parotid/SMSL) salivas. In parallel, a core dataset of the human plasma proteome with 3020 protein identifications was recently released. A total of 1939 nonredundant salivary proteins were compiled from a total of 19 474 unique peptide sequences identified from whole and ductal salivas; 740 out of the total 1939 salivary proteins were identified in both whole and ductal saliva. A total of 597 of the salivary proteins have been observed in plasma. Gene ontology (GO) analysis showed similarities in the distributions of the saliva and plasma proteomes with regard to cellular localization, biological processes, and molecular function, but revealed differences which may be related to the different physiological functions of saliva and plasma. The comprehensive catalog of the salivary proteome and its comparison to the plasma proteome provides insights useful for future study, such as exploration of potential biomarkers for disease diagnostics.
Keywords: Biomarkers, Body fluid, MS, Plasma, Saliva
1 Introduction
Saliva is produced by the three major paired salivary glands (parotid, submandibular (SM), and sublingual (SL)) as well as by numerous minor salivary glands. Besides water, salivary fluid contains proteins, post-translationally modified proteins (e.g., glycoproteins, phosphoproteins), peptides, lipids, minerals, and other small compounds [1, 2]. Upon release of glandular secretions into the oral cavity, the fluid is mixed with a variety of exocrine, nonexocrine, cellular, and exogeneous components to ultimately form whole saliva (WS). Through its various components, saliva participates in maintenance of homeostasis in the oral cavity, lubrication of oral tissues, and facilitation of chewing, speaking, and swallowing. Furthermore, saliva protects the oral cavity from foreign invaders, such as bacteria and viruses, by digestion and inhibition of their growth [3].
Qualitative and quantitative salivary alterations in secretion or composition, induced by either systemic or oral conditions, can cause functional deficiency of the saliva [4–6]. Sjögren’s syndrome, an autoimmune disease, causes reduction in saliva volume, which leads to dry mouth, difficulties in swallowing and speaking, increased caries and periodontal diseases, and infection of the salivary gland. Saliva from Sjögren’s syndrome subjects contains increased levels of a few major salivary proteins [7–9]. We recently found 42 proteins to be significantly elevated in saliva from primary Sjögren’s syndrome subjects [8]. Oral cancers are also associated with significant changes of the salivary proteins. Using LC-MS/MS, we found five salivary proteins to be significantly elevated in oral cancer patients [10]. Also, changes in the salivary protein composition have been observed in systemic diseases. Alterations in glycosylation of salivary mucins have been associated with cystic fibrosis [11]. Increased levels of amylase and IgA are observed in diabetic patients [12, 13]. A number of salivary components, including cortisol, amylase, and lysozyme, are altered under stress conditions. These alterations suggest that analysis of saliva, especially its protein components and carbohydrate PTMs [14], may have potential for disease diagnosis and health monitoring. The relatively simple, noninvasive collection procedures and its constant availability make saliva an attractive biofluid for disease detection.
A key initial step for saliva to be of practical use for disease diagnosis and health monitoring is the cataloging of its protein components. However, because of its complexity, variation in protein abundance and PTMs, a comprehensive characterization of the protein composition of salivary fluid could not be achieved through traditional biochemical approaches until the introduction of MS-based, high-throughput proteomics technologies. Recently, several reports with the goal to comprehensive catalog the salivary proteome have been published [8, 10, 15–21], with numbers of proteins identified ranging from hundreds to over 1000. A project to catalog the proteomes from salivary gland fluids of parotid and SM/SL glands identified 1166 proteins, with 914 identified in parotid and 917 in SM/SL fluids, and 665 in common (www.hspp.ucla.edu) [22].
To appreciate the unique utility of the salivary proteome in the context of its function and potential diagnostic value, it is important to compare the saliva protein composition with other established proteomes, such as plasma. Overlap in protein content between saliva and plasma may indicate that saliva could be used as a diagnostic alternative to blood tests. Over many decades, numerous studies have uncovered how changes in the concentrations of specific plasma proteins have been associated with disease processes, leading to well-accepted clinical applications [23]. Moreover, the plasma proteome is perhaps the most extensively studied human proteome to date. The international HUPO Human Plasma Proteome Project, a collaboration of many laboratories using MS technology, compiled a core dataset of 3020 distinct proteins (with a minimum of two unique peptides per protein) [24–26]; 889 proteins were confirmed as high-confidence identifications through a rigorous statistical approach adjusting for protein length and multiple comparisons testing [27].
In this present study, we have attempted to construct a comprehensive catalog of the human salivary proteome by integrating protein identifications from both whole and ductal salivary fluids. The salivary proteome was analyzed and compared among whole and ductal saliva as well as to the human plasma proteome. These analyses should greatly facilitate the characterization of these two human body fluid proteomes and should facilitate the discovery and development of diagnostic disease biomarkers.
2 Materials and methods
The proteome of WS was contributed by datasets from four research groups: the University of Minnesota (UMN), Research Triangle Institute (RTI), Calibrant Biosystems/University of Maryland (CB/UM), and the University of California-Los Angeles (UCLA). The datasets include newly acquired data from WS as well as previously published data [17–20, 28]. The experimental methods described below are primarily for the new experiments performed to supplement the list of WS proteins. The lists of salivary protein identifications from ductal saliva, i.e., parotid and SMSL, are the result of a consortium effort by three National Institute of Dental and Craniofacial Research (NIDCR)-supported research groups (Scripps Research Institute, UCLA, and University of California-San Francisco); the methods used by each of the three groups have been described [22]. For the comparison of the salivary proteome to the plasma proteome, the published HUPO plasma proteome dataset was used [26]. The 3020 plasma protein identifications with two or more peptides were obtained from http://www.bioinformatics.med.umich.edu/hupo/ppp. The dataset is available also at the European Bioinformatics Institute (http://www.ebi.ac.uk/pride), and it has been incorporated into the Peptide Atlas at the Institute for Systems Biology (http://www.peptideatlas.org).
2.1 Saliva collection, protein fractionation, and protein identification
2.1.1 University of Minnesota (UMN)
2.1.1.1 Sample collection and processing
Whole, unstimulated saliva was collected from four healthy individuals using a previously described protocol [29]. WS (1 mL) was removed and centrifuged at 25 000 × g at 4°C for 30 min. The supernatant was collected and quantified by using the BCA protein assay with BSA as a standard control (Pierce), giving 1.05 mg of total soluble protein per mL. Equal amounts of soluble saliva (200 µL) were combined from the four individuals. The combined saliva was brought to 100 mM with HEPES, pH 8.0 and 5 mM with Tris-(2-carboxyethyl)- phosphine (TCEP) and incubated overnight with 20 µg of trypsin (Promega, Madison, WI) at 37°C. The resulting peptides were concentrated and desalted using an RP Sep-Pak cartridge (Waters, Milford, MA) and dried by vacuum centrifugation.
2.1.1.2 Protein/peptide fractionation
Preparative IEF of the tryptic peptide mixture was performed using a commercially available ProTeam free-flow electrophoresis (FFE) system (BD Biosciences, Franklin, NJ), as described previously [18]. Approximately 50% of each FFE fraction was taken from each of the microtiter plate wells containing peptides and processed as described [17], and a second step of fractionation was performed using a PolySULFOETHYL strong cation exchange (SCX) guard column (Javelin guard column, 1.0 mm id × 10 mm, 5 µm, 300 Å, PolyLC) using an automated syringe pump capable of highly accurate sub-microliter per minute flow rates (Harvard Apparatus). Each peptide fraction was re-dissolved in 200 µL of SCX loading buffer (10 mM KH2PO3 containing 20% ACN, pH 3.0) and loaded onto a preconditioned SCX column at a flow rate of 50 µL/min. Peptides were eluted with step-gradient chromatography, using steps with increasing KCl concentration, at a flow rate of 50 µL/min. Eluted fractions from salt steps of 20, 25, 50, and 200 mM KCl in loading buffer were collected (200 µL total volume); each collected fraction was concentrated by vacuum centrifugation, and reconstituted in 30 µL of HPLC loading buffer.
2.1.1.3 Protein identification
All online µLC separations were done on an automated Paradigm MS4 system (Michrom Bioresources, Auburn, CA), coupled with an LTQ linear IT mass spectrometer (ThermoFisher Scientific, San Jose, CA) as described previously [17, 18]. Acquired MS/MS spectra were searched using SEQUEST [30] (Bioworks version 3.2, Thermo Finnigan, San Jose, CA) against a nonredundant human protein sequence database from the European Bioinformatics Institute (ipi.HUMAN.v3.18.fasta, containing 62 000 entries). A reversed-sequence version of the same database was appended to the end of the forward version for the purpose of false positive rate estimation [31]. Differential amino acid mass shifts for oxidized methionine ( Da) were also included. Precursor peptide mass tolerance was ±2.0 Da with no tryptic specificity. Fragment ion tolerance was set to ±1.0 Da. To each matched peptide sequence a predicted pI using the Shimura algorithm [32] was automatically assigned using an in-house developed script developed. The search results were validated using the peptide validation program PeptideProphet [33]. The peptide sequence match results were organized and interpreted using the software tool Interact [34]. Peptide matches (regardless of assigned P score) were kept for further consideration only if their predicted pI was within ±0.5 U of the average pI value for the FFE fraction from which they were identified, and the peptide sequence was at least partially tryptic to maximize the high confidence matches [35]. The estimated false positive rate for our protein catalog was 1%.
2.1.2 Research Triangle Institute (RTI)
2.1.2.1 Sample collection and processing
Whole, unstimulated saliva was collected from a healthy individual into a 50 mL conical centrifuge tube and stored at −80°C until use. Prior to trypsin digestion, the saliva was centrifuged at 5000 × g for 5 min to remove debris. Total protein content of the supernatant was quantified using a Bradford protein assay, with BSA as a reference standard (Pierce), and a total of 1 mg of protein was digested with modified trypsin (Promega) at a ratio of 50:1 (sample/protease) at 37°C for overnight. Digests were desalted using a C18-“light” Sep-Pak (Waters).
2.1.2.2 Protein/peptide fractionation
Salivary peptides were focused on IPG-IEF strips, as previously reported [36–38]. Briefly, a 24 cm pH 3.5–4.5 IPG strip (GE Healthcare) was rehydrated overnight with 1 mg of peptides re-suspended in 8 M urea, 0.5% carrier ampholytes. The strip was subsequently focused using an IPGPhor II (GE Healthcare) according to the manufacturer’s provided protocol. The strip was manually cut into 60 fractions of ~4 mm width. Each fraction was sequentially extracted with 200 µL of 0.1% TFA, 200 µL of 0.1% TFA/50% ACN, and 200 µL of 0.1% TFA/100% ACN. The pooled peptide extracts were dried, resuspended in 0.1% TFA, and then further purified using an Oasis HLB SPE (Waters) resin in a 96-well plate format. Vacuum-dried (Speed-Vac) peptide extracts were subsequently resuspended in 40 µL of 0.1% TFA.
2.1.2.3 Protein identification
Extracted peptide fractions were subjected to LC-MS/MS analysis on a ThermoFisher Scientific LTQ Classic quadrupole IT equipped with a New Objective (Woburn, MA) Picoview nanospray source coupled to an Eksigent (Dublin, CA) Nano-2-D LC System equipped with an integrated Valco 10-port switching valve and peltier-cooled micoautosampler. The column, which was integral with the nanospray tip, consisted of a 100 µm id × 360 µm od × 10 cm piece of fused silica packed with a monodisperse 5 µm polymeric packing material (5RPC, gift from GE Healthcare, Piscataway, NJ). Three microliter of each dried peptide fraction was loaded onto a capillary sample trap (packed with the same material as the column) and washed briefly with 0.1% aqueous formic acid (FA) (5 min) before switching in-line with the analytical column. The HPLC gradient was 80 min in length and progressed from 15 to 50% B (A: aqueous 0.1% FA, B: 70% ACN with 0.1% FA) at a flow rate of 250 nL/min.
The mass spectrometer was programmed to take sequential scans of the following mass ranges (400–600, 600–700, 700–800, 800–900, and 900–1300 m/z) followed by data-dependent MS/MS of the three most intense ions in each mass range, except in the case of 400–600 m/z where only the two most intense ions were analyzed. Dynamic exclusion was enabled with a repeat count of 2, repeat duration of 60 s, and an exclusion duration of 120 s.
The database employed was the International Protein Index (IPI), human version 3.19. A reversed version of the same database was indexed for tryptic peptides and searched against MS/MS spectra using TurboSEQUEST (ThermoFisher Scientific). Data were subjected to reverse database [31] and pI-filtering using in-house developed software (IDSieve) as previously reported [39]. Actual SEQUEST crosscorrelation score (Xcorr) cutoffs were determined for each fraction based on the Xcorr of the highest scoring reverse database hit as a function of charge state for an empirical peptide false discovery rate of ~1%.
2.1.3 Calibrant Biosystems/University of Maryland (CB/UM)
2.1.3.1 Sample collection and processing
Whole, unstimulated saliva was collected from a healthy male volunteer. One milliliter of saliva was placed in a tube containing a mixture of protease inhibitors (1 µg aprotinin, 1 µg pepstatin A, and 1 µg leupeptin) and centrifuged at 20 000 × g for 30 min. The supernatant was collected and placed in a dialysis cup (Pierce, Rockford, IL) and dialyzed overnight at 4°C against 100 mM Tris, pH 8.2. Urea and DTT were added to the sample with final concentrations of 8 M and 1 mg/mL, respectively, and incubated at 37°C for 2 h under nitrogen. Iodoacetamide was added to a concentration of 2 mg/mL and kept at room temperature for 1 h in the dark. Trypsin was added at a 1:20 w/w enzyme-to-substrate ratio and incubated overnight at 37°C. The protein digest was desalted using an RP trap column (Michrom Bioresources), eluted with a peptide concentration of 2.0 µg/µL, and lyophilized to dryness using a Speed-Vac (ThermoSavant, San Jose, CA), and then stored at −80°C.
2.1.3.2 Protein/peptide fractionation
Transient capillary isotachophoresis/CZE (CITP/CZE) was the basis of the multidimensional separations strategy employed. The CITP apparatus was constructed in-house using a CZE 1000R high-voltage power supply (Spellman High-Voltage Electronics, Plainview, NY). A 80 cm long CITP capillary was initially filled a background electrophoresis buffer of 0.1 M acetic acid at pH 2.8. The sample containing saliva protein digests was prepared in a 2% pharmalyte solution and was hydrodynamically injected into the capillary. A positive electric voltage of 24 kV was then applied to the inlet reservoir, which was filled with a 0.1 M acetic acid solution. The cathodic end of the capillary was housed inside a stainless steel needle using a coaxial liquid sheath flow configuration. A sheath liquid composed of 0.1 M acetic acid was delivered at a flow rate of 1 µL/min using a syringe pump (Harvard Apparatus 22, South Natick, MA). The stacked and resolved peptides in the CITP/CZE capillary were sequentially fractionated and loaded into individual wells on a moving microtiter plate.
2.1.3.3 Protein identification
Each peptide fraction was analyzed by nano-RP LC equipped with an Ultimate dual-quaternary pump (Dionex, Sunnyvale, CA) and a dual nano-flow splitter connected to two pulled-tip fused-silica capillaries. These two 15 cm long capillaries were packed with 3 µm Zorbax Stable Bond (Agilent, Palo Alto, CA) C18 particles. Nano-LC separations were performed in parallel in which a dual-quaternary pump delivered two identical 2 h organic solvent gradients with an offset of 1 h. Peptides were eluted at a flow rate of 200 nL/min using a 5–45% linear ACN gradient over 100 min with the remaining 20 min for column regeneration and equilibration. The peptide eluents were monitored using a linear IT mass spectrometer (LTQ, ThermoFisher Scientific) operated in a data-dependent mode.
Raw LTQ data were converted to peak list files by msn_extract.exe (Thermo Fisher Scientific). The program OMSSA was used [40] to search the peak list files against a decoyed Swiss-Prot human protein sequence database. This database was constructed by reversing all 12 484 real sequences and appending them to the end of the sequence library. Searches were performed with the following parameters: fully tryptic, 1.5 Da precursor ion mass tolerance, 0.4 Da fragment ion mass tolerance, one missed cleavage, alkylated cysteine as a fixed modification, and variable modification of Met oxidation. The false positive rate for peptide identifications was determined as 1%.
2.1.4 University of California-Los Angeles (UCLA)
2.1.4.1 Sample collection and processing
WS was obtained from healthy nonsmoking subjects in the morning prior to eating and after rinsing the mouth with water. To minimize protein degradation, protease inhibitor cocktail (Sigma Chemical, 1 µL/mL of WS) and 1 mM of sodium orthovanadate were added immediately to the saliva after sample collection. All samples were kept on ice during the entire process. Roughly 5 mL of clear WS was obtained from pooled individuals after centrifuging at 1300 × g for 5 min. A further centrifugation at 14 000 × g at 37°C for 15 min was performed to remove debris. Protein concentration was determined to be 0.4–1.0 mg/mL (BioRad Protein Assay). The samples were divided into 1 mL aliquots and stored at −80°C.
2.1.4.2 Protein/peptide fractionation
Ultracentrifugation filters (Microcon YM-10K and YM-3K, Millipore, Billerica, MA, USA) were used to prefractionate the WS into three fractions according to molecular weight: less than 3 kDa, 3–10 kDa, and greater than 10 kDa. Sample processing and trypsin digestion followed protocols described previously [16].
Additional saliva samples were prefractionated by solution IEF [22, 41, 42]. Proteins in WS were precipitated by mixing with four times the volume of 100% cold ethanol and then incubated overnight at −20°C. The mixture was centrifuged at 13 000 rpm for 15 min at 4°C. The pellet was resuspended in lysis buffer (Zoom 2D protein solubilizer, Invitrogen, Carlsbad, CA) containing Complete Protease Inhibitor (Roche Diagnostic, Indianapolis, IN), Tris base, DTT, and water and sonicated on ice. The pH of the lysate was adjusted to pH 8.5–8.7 with 1 M Tris base and then incubated for 15 min at room temperature with shaking. Sample lysate was reduced for 30 min with 99% dimethylacrylamide (DMA) at room temperature. To quench any excess of DMA, DTT was added and incubated for 5 min at room temperature. After centrifuging the sample for 30 min at 13 400 rpm at 4°C, the supernatant was collected. The protein concentration was determined by the Non-Interfering Protein Assay (Geno Technology, St. Louis, MO) to be approximately 1.5 mg/mL.
Protein lysate (1.5 mg/mL, 400 µL) was diluted to a final concentration of 0.6 mg/mL in dilution buffer consisting of Zoom IEF denaturant, Zoom focusing buffer pH 3–7 (Invitrogen), Zoom focusing buffer, pH 7–12, and 5 µL 2 M DTT. Solution IEF separation with a Zoom IEF Fractionator (Invitrogen) was performed in the standard format (pH 3.0–10). Diluted sample was loaded into each of the five chambers of the fractionator. Five fractions (pI 3–4.6, 4.6–5.4, 5.4–6.2, 6.2–7.0, and 7.0–10.0) were obtained after fractionation. Proteins from each fraction were precipitated by mixing with 70% acetone, incubating at −20°C for 3–4 h and centrifuging at 13 000 rpm for 30 min.
2.1.4.3 Protein identification
LC-MS/MS was performed on an Applied Biosystems (Foster City, CA) QSTAR Pulsar XL (QqTOF) mass spectrometer equipped with a nanoelectrospray interface (Protana, Odense, Denmark) and an LC Packings (Sunnyvale, CA) nano-LC system. The nano-LC was equipped with a homemade precolumn (75 µm × 10 mm) and an analytical column (75 µm × 150 mm) packed with Jupiter Proteo C12 resin (particle size 4 µm, Phenomenex, Torrance, CA). The released peptides were dried and dissolved in 0.1% FA solution. For each LC-MS/MS run, typically 6 µL of sample solution was loaded to the precolumn. The precolumn was washed with the loading solvent (0.1% FA) for 4 min before the sample was injected onto the LC column. The eluents used for the LC were 0.1% FA (solvent A) and 95% ACN containing 0.1% FA (solvent B). The flow was 200 nL/min, and the following gradient was used: 3% B to 35% B in 72 min, 35% B to 80% B in 18 min, and maintained at 80% B for the final 9 min. The column was equilibrated with 3% B for 15 min prior to the next run.
For online LC-MS/MS analyses, a Proxeon (Odense, Denmark) nanobore stainless steel online emitter (30 µm id) was used for spraying with the voltage set at 1900 V. Peptide product ion spectra were recorded automatically during the LC-MS/MS runs by information-dependent analysis (IDA) on the mass spectrometer. Argon was employed as the collision gas. Collision energies for maximum fragmentation efficiencies were calculated using empirical parameters based on the charge and m/z of the peptide precursor ion.
Proteins were identified by using the Mascot database search engine (Matrix Science, London, UK). All searches were performed against the EBI human IPI database (version 3.03; release date February 5, 2005). For saliva samples prefractionated by in-solution IEF, DMA modification of cysteines was added to the variable modification list. In all searches, one missed tryptic cleavage was allowed, and a mass tolerance of 0.3 Da was set for the precursor and product ions. A MASCOT score of >25 with a p-value of <0.05 was considered a significant match. False-positive rates were determined to be ~2% by using the method described by Matrix Science (www.matrixscience.com).
2.2 Data integration and reassembly of protein identifications from peptide sequences
Protein and peptide identifications collected from WS, parotid, SMSL, and plasma by the participating groups were imported into a relational database designed specially for storage of proteomics experimental data generated by the NIDCR-supported salivary proteome consortium project.
The list of protein and peptide identifications from WS was derived from several protein database sources. To create a consensus list of protein identifications for each biological sample source (i.e., WS, parotid, SM/SL, or plasma) and to make an effective comparison among the sample sources, the mandatory first step is to standardize the protein identifications in reference to the same protein database through a reproducible algorithm. Therefore, we reassembled the protein identifications based on peptide sequences and chose protein database IPI v3.32 (released in August 2007) as the reference database. The strategy of reassembly (inference) of protein identifications from the peptide level was used previously in both plasma and brain proteome studies [26, 43] and also in the integration of the human peptide sequences with the human genome [44]. The algorithm we used seeks to find the minimum protein identification in a given sample source by the following steps:
Construct a unique protein list that includes all proteins from which each peptide identified from a sample source might be derived. The unique peptide sequence list for each sample source was extracted from the database. Each unique peptide in the list was subsequently searched against the reference protein database IPI v3.32. All protein entries containing exact matches to the peptide sequence were recorded into the database. The unique peptide sequences that could not find an exact match in IPI v3.32, which is usually caused by protein sequence changes during the periodic database updates or sequence differences between protein databases, were discarded from further analysis. During this step, all proteins from which each unique peptide could be derived were identified. The unique protein list with all these potential protein candidates involved represents the maximum number of detected proteins for that sample source.
Construct a unique peptide list for each protein inferred from step (i). During this step, peptides that matched to the same protein are combined. In some cases, a peptide can be combined with other peptides and matched to more than one protein.
Cluster proteins that were identified by the same set of peptides. The proteins inferred by the same set of peptides were defined as equivalent protein identifications and were clustered together. These equivalent proteins are usually paralogs, isoforms, or proteins sharing the same functional domains. In some cases, the peptide list used for protein identifications can be the subset of the peptide list for other protein identifications. These proteins were also clustered together. A minimum protein list for a sample source was created after a representative protein was chosen from each cluster. The representative protein was chosen by applying the following procedures sequentially: (i) Select the protein that contains the highest number of peptides. (ii) Select the protein that is crossreferenced to the UniProt/Swiss-Prot database. (iii) Select the protein with detailed descriptions rather than proteins described as “hypothetical,” “putative,” “fragment,” “similar to,” or “cDNA.” (iv) Select the protein that has the lowest IPI number.
2.3 Sequence feature prediction
Protein identifications were classified based on whether they contained sequence features of secreted signal sequence, transit sequence, or transmembrane domain. The sequence features of the protein identifications were either extracted from the protein annotation file obtained from UniProt/Swiss-Prot database or predicted using the sequence feature prediction programs, SignalP for secretion signal sequences [45], TargetP for organelle presequences [46], and TMHMM for transmembrane helix sequences [47]. These programs were obtained from the Center for Biological Sequence Analysis, Technical University of Denmark DTU (http://www.cbs.dtu.dk/services).
2.4 Data sources for protein annotation, gene ontology (GO) analysis, and disease association
IPI protein sequence database and its crossreferences file released in August, 2007 were obtained from ftp://ftp.ebi.ac.uk/pub/databases/IPI/. A flat file format of GO for biological process, molecular function, and cellular component were obtained from the GO database website http://www.geneontology.org/Go.downloads.shtml. A gene map of the online Mendelian inheritance in man (OMIM) was obtained from ftp://ftp.ncbi.nih.gov/repository/OMIM/genemap. Biological pathway information was obtained from the KEGG database (http://www.genome.jp/kegg/).
2.5 Statistical analysis
The significance of comparisons of GO distributions was estimated using the χ2 test. The χ2 test was performed using the statistical package SAS. The adjustments for protein length and multiple comparisons testing reported for the Plasma Proteome Project [24] were not applied to the salivary proteome results.
2.6 Web interface and database
The WS peptide and protein identifications and its comparison to the human plasma proteome were stored in a relational database. The details of the relational database can be accessed through the http://www.hspp.ucla.edu/. Briefly, the database was implemented using the open source relational database package MySQL. The database has web interface features that allow users to search and query the database through a variety of parameters including saliva source, protein accession numbers, and keywords.
3 Results
3.1 Human whole saliva proteome
In parallel to the analysis of ductal salivary proteomes recently reported [22], the present study reports the characterization of the human WS proteome. The WS protein and peptide identifications include those derived from the high-throughput MS-based experiments performed independently by four research groups reported here, as well as results from previous efforts [16, 18–20, 28]. In total, the four groups submitted 12 679 distinct peptide identifications with a false positive rate of less than 2% and 3196 distinct protein identifications. The four groups implemented diversified protocols for protein fractionation, peptide separation, MS, and database searching algorithms and databases (Table 1).
Table 1.
Summary of experimental approaches for WS protein identifications
| Laboratory | Prefractionation | Peptide Separation | MS | Search Program | Sequence Database | FPRa) | No. of peptides |
|---|---|---|---|---|---|---|---|
| CB/UM | None | CIEF | LTQ | OMSSA | Swiss-Prot | ≤1% | 7582 |
| CITP/CZE | |||||||
| UMN | None | FFE-SCX-capLC | LTQ | SEQUEST | IPI v3.18 | ≤1% | 5468 |
| RTI | None | IPG-IEF | LTQ | SEQUEST/IdSieve | IPI v3.19 | ≤1% | 537 |
| UCLA | Zoom-IEF Ultracentrifuge | RPLC | QSTAR | MASCOT | IPI v3.03 | ≤2% | 1571 |
FPR (peptide false positive rate).
To create a consensus comprehensive list of WS protein components, we integrated and standardized the heterogeneous protein identifications to the IPI database (IPI v3.32, August 2007 release). The integration process started at the peptide level and resolved a nonredundant minimal set of protein identifications, defined such that within a group of proteins that contain the sequences with 100% identity to a set of peptides, one of them was selected to represent the group of proteins and reported. The computational approach for the integration and standardization was similar to the method introduced previously [48], and the selection of a representative protein from a group of proteins was similar to that used for the HUPO Plasma Proteome Project [26]. A total of 12 602 of the 12 679 original submitted peptides were found to exactly match that found in IPI v3.32; these peptides were used to infer 2158 distinct proteins. Within the 2158 WS proteins, 702 resulted from single-peptide-based identifications, which were subsequently excluded. We utilized the remaining 1456 identifications, derived from 2 or more peptides, as high confidence identifications for further analyses and comparisons.
Besides proteins from human sources, proteins derived from bacterial sources found in the oral cavity were observed within WS. To exclude these bacterial contaminant proteins, the peptides used to derive the 1456 WS identifications were searched against bacterial protein databases. Only 12 out of the 1456 WS identifications contained peptides that matched also to bacterial proteins; these proteins were excluded from the WS identifications, reducing the number of WS protein identifications to 1444.
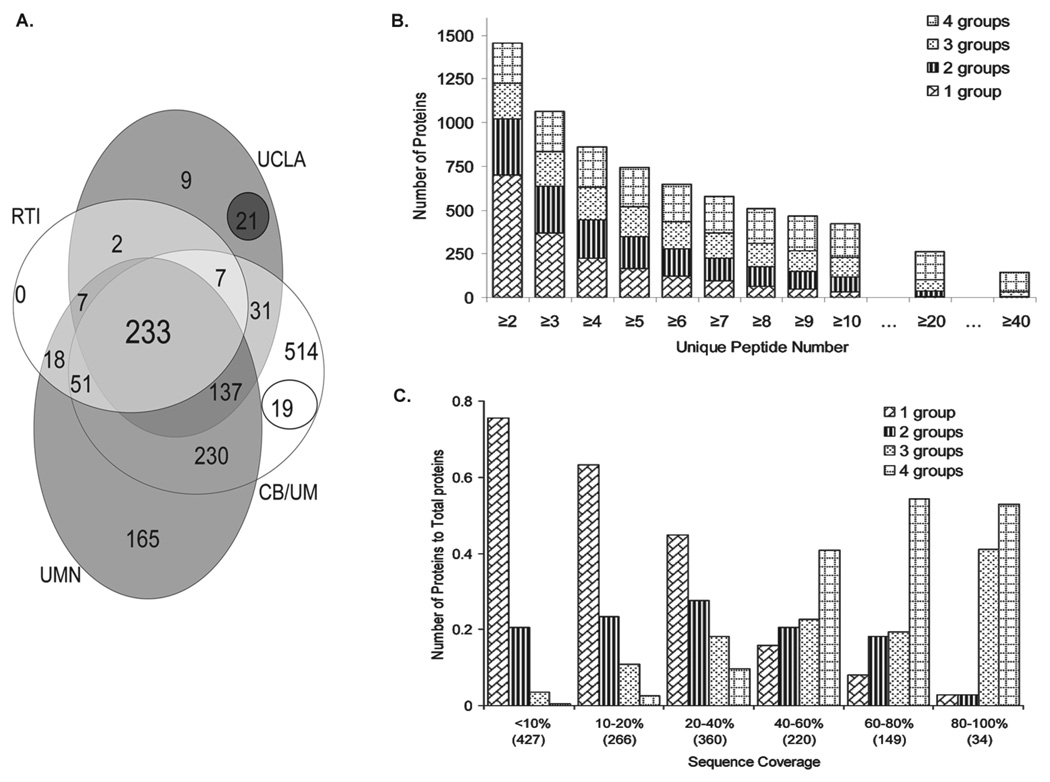
A total of 233 out of the 1444 WS proteins were confirmed by all four collaborating laboratories and approximately one half of the proteins (756) were supported by at least two laboratories (Fig. 1A). An approximate relative abundance of the WS proteins was estimated by the number of unique peptides used to derive the identifications and by sequence coverage (Fig. 1). The concordance among the groups increased with proteins having increased number of unique peptides per protein identification (Fig. 1B). Similarly, protein identifications by multiple groups were related to the sequence coverage of the protein (Fig. 1C). The number of identifications confirmed by any two groups reached a maximum when the coverage was 40–50%, while three and four group matches dominated at higher sequence coverage (Fig. 1C).
Figure 1.
WS protein identifications based on the overlap of the identifications, unique peptide number, and sequence coverage. (A) Venn diagram showing the overlap of the WS proteins between laboratories with total identifications from each group as 1222 CB/UM (CB/UM), 337 RTI (Research Triangle Institute), 447 UCLA, 862 UMN. (B) Number of WS proteins identified as a function of number of unique peptide detected; each bar is demarcated by the number of labs making the identifications. (C) WS protein identifications classified based on protein sequence coverage. The number in parentheses represents the total number of proteins within the sequence coverage range.
3.2 Comparison of proteomes from WS, ductal saliva,and plasma
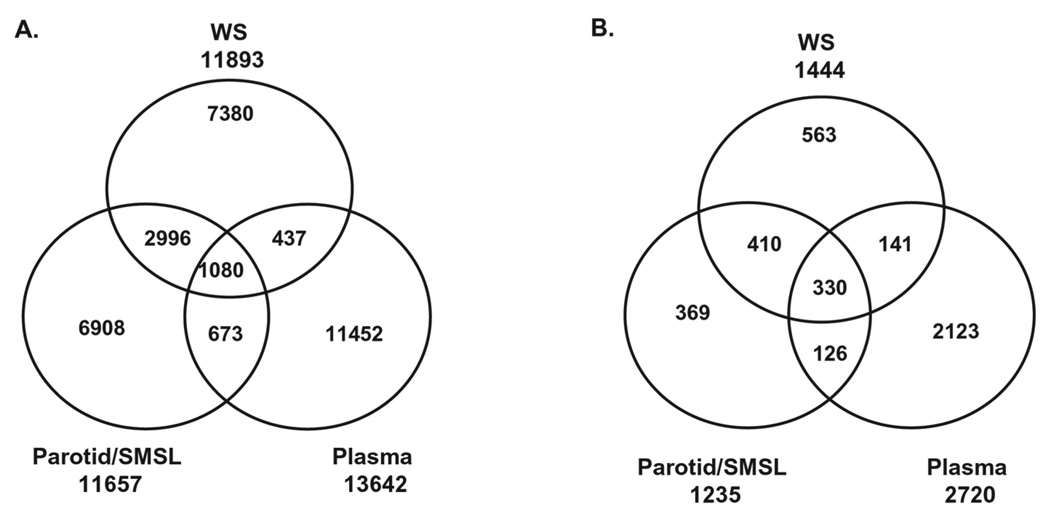
To study the origin of the salivary proteins, we compared the WS proteome to the ductal parotid/SMSL saliva proteome [22]. Similarly, to examine the common nature of saliva and blood, we compared the saliva proteins to the plasma proteome. To make the comparison effective, the parotid/SMSL proteomes derived from IPI v3.24, and the plasma proteome from IPI v2.23 were integrated and standardized to the reference database IPI v3.32 following the same procedures as implemented for WS. As shown in Fig. 2, 34 and 10% of the distinct peptides identified in WS overlap with the peptides identified in the parotid/SMSL proteome and plasma proteome, respectively. At the protein level, 51% of the 1444 WS proteins overlap with the 1235 parotid/SMSL proteins and 33% overlap with the plasma proteins. The higher overlap observed at the protein level indicates that the same proteins found in the two proteomes do not necessarily depend on the same overlapped peptides. A similar phenomenon was noted in a comparison of brain, plasma, and platelet proteomes [43].
Figure 2.
Venn diagrams showing the overlapping peptide and protein identifications between WS, parotid/SMSL, and plasma. (A) Peptide identifications; (B) protein identifications.
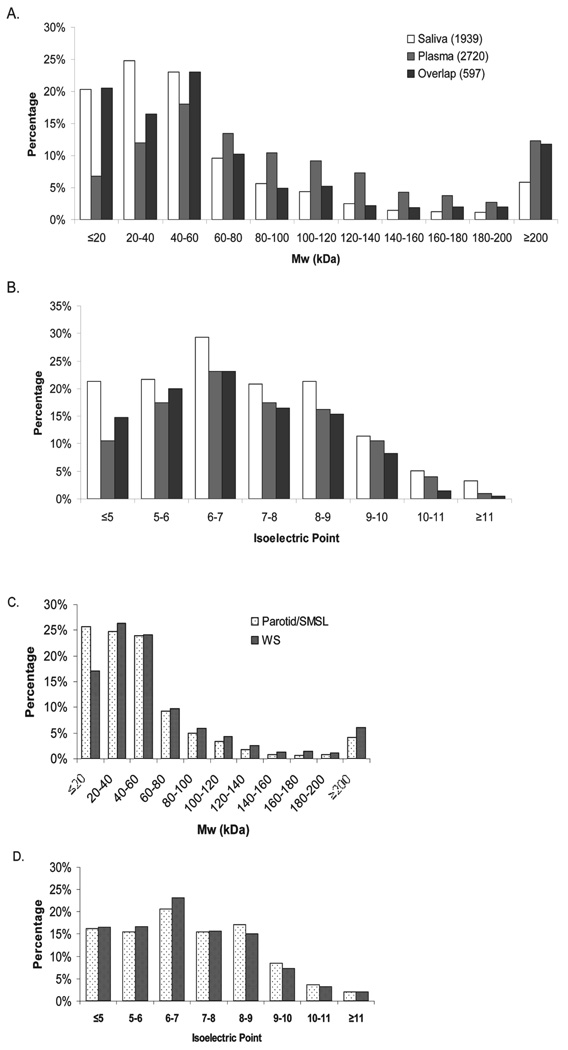
To create a comprehensive catalog of the human salivary proteome, the proteins found in WS and ductal saliva were combined, resulting in a total of 1939 proteins. This combined WS/ductal salivary proteome was compared to the plasma proteome with regard to their theoretical molecular weight and pI (Fig. 3). The salivary proteome contains a large proportion (20%) of low molecular weight proteins (<20 kDa) in contrast to only 7% for the plasma proteome. In total, 68% of the saliva proteins have molecular weight less than 60 kDa compared to the 37% of the plasma proteins.With regard to the proteins found in common between saliva and plasma, the molecular weight distributions show similarity to the distributions of the salivary proteome with a tendency toward the low molecular weight end, except in the highest MW range (≥200 kDa). A pI comparison of the saliva and plasma proteomes revealed that saliva contains more proteins in the lower and (≤5) higher end (≥11) of the pI scale (Fig. 3B), with an average protein pI of 7.03 and 7.13 for saliva and plasma, respectively. The trend toward a higher proportion of proteins with MW less than 20 kDa observed in the saliva proteome is further manifested in the ductal parotid/SMSL proteome. Compared to 17% in WS, 26% of the parotid/SMSL proteins are less than 20 kDa in size (Fig. 3C). In contrast to the difference in the pI distribution of saliva and plasma, parotid/SMSL, and WS proteomes show very similar pI distributions (Fig. 3D).
Figure 3.
Comparison of molecular weight and isoelectric point of saliva proteome to plasma proteome and ductal parotid/SMSL saliva proteome to WS proteome.
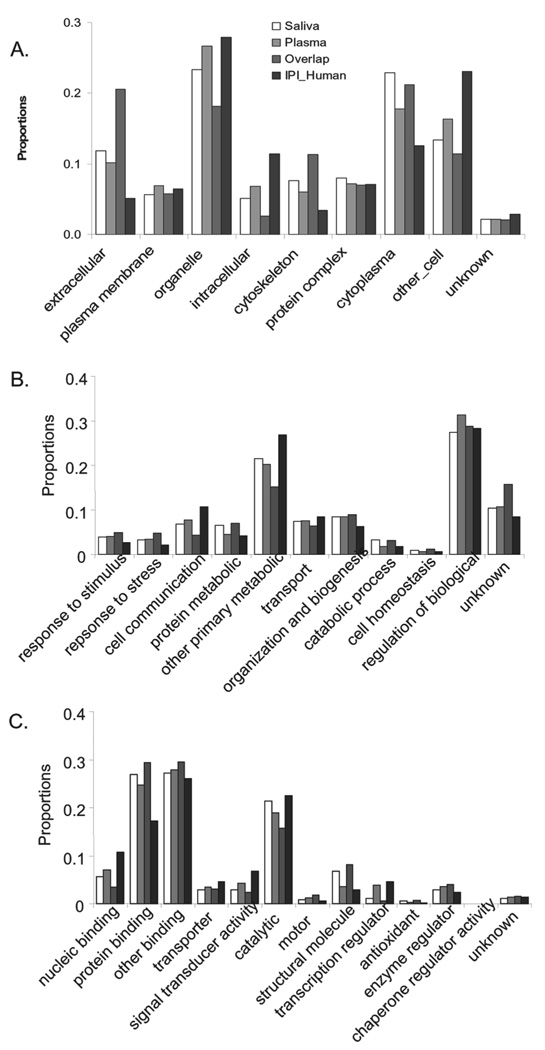
The salivary and plasma proteomes were further compared based on their annotation in GO terms of cellular component, molecular process, and biological function (Fig. 4). As expected, compared to the total human proteome, the salivary and plasma proteomes are over-represented in the extracellular component, an indication of secretion (p<0.001). The level of over-representation in the extra-cellular component is further enhanced in the proteins that coexist in saliva and plasma. The salivary and plasma proteins are also over-represented in the cytoplasmic and cytoskeleton components (p<0.001). In contrast, intracellular components are under-represented in saliva and plasma. With regard to biological processes (Fig. 4B), compared to the human proteome, saliva, and plasma are over-represented in the categories of response-to-stimulus, responseto- stress, and cell organization and biogenesis, but are underrepresented in cell communication and other primarily metabolic processes. Interestingly, the distributions of the salivary proteins are significantly enhanced in protein metabolic and catabolic processes compared to plasma (p<0.001). In the GO molecular functional categories, the salivary and plasma proteomes are significantly over-represented in protein binding but are under-represented in nucleic acid binding, transporter activity, and signal transducer activity (p<0.001) (Fig. 4C). In general, the salivary and plasma proteomes showed similar distributions in the GO molecular functional categories. However, exceptions were found in the structural, transcription regulator, and antioxidant functions. Compared to plasma, saliva is significantly over-represented in structural molecule and antioxidant functions but under-represented in the transcription regulator function (p<0.001). The proteins common to saliva and plasma generally show an enhanced tendency in the over-represented and under-represented categories of the salivary and plasma proteins. The distributions of the overlapping proteins are significantly enhanced in the extracellular and cytoplasm of the cellular component, response-to-stimulus, response-to-stress, protein metabolic and catabolic processes, and protein binding, motor, structural molecule, antioxidant, and enzyme regulator of molecular function, but are under-represented in organelle and intracellular of the cellular component, cell communication, and other primary metabolic of the biological process, and nucleic binding, signal transducer, catalytic, and transcription regulator of molecular function.
Figure 4.
GO SLIM distributions of saliva proteome, plasma proteome, overlapping proteins of saliva and plasma, and IPI human proteins. (A) Cellular component; (B) biologic process; (C) molecular function.
To test our hypothesis that the body fluids are enriched with proteins that contain secretion sequence signals, we examined the sequence features present in the salivary and plasma proteomes, based on the sequence categories of signal sequence (prepeptide), transit peptide, glycosylation site, and transmembrane region. The sequence annotations were obtained either from the UniProt/Swiss-Prot protein knowledgebase or through the sequence feature prediction programs, signalp, targetp, and TMHMM. As shown in Table 2, 1436 out of 1939 salivary proteins and 1966 out of 2720 plasma proteins have their corresponding entries in the UniProt/Swiss-Prot database. A large portion of the salivary proteins (27%) and plasma proteins (23%) are annotated with a signal sequence at the N-terminus. Consistent with the observations that many salivary and plasma proteins can be glycosylated [49], the sequence feature annotation shows that 24% of the salivary proteins and 26% of the plasma proteins contain N-linked glycosylation site(s). Both saliva and plasma contain putative transmembrane proteins (11% in saliva and 18% in plasma). In contrast to the high percentage of proteins with a signal sequence, the proportion of proteins with a transit peptide sequence required for protein transport across organelle membranes are low in both saliva and plasma (3.3% in WS and 1.3% in plasma). Considering that the part of WS is from the secretion of the ductal fluids, we also compared the sequence feature of WS to parotid/SMSL saliva. The result shows that the ductal saliva proteome contains 37% proteins with secreted signal peptide in contrast to 21% in WS.
Table 2.
Sequence features of saliva and plasma proteomes and overlapping proteins between saliva and plasma
| Source | Swiss_Prot_Annotationa) (saliva: 1436; plasma: 1966; overlapped: 395) | Sequence feature predicationb) (saliva: 1939; plasma: 2720; overlapped: 597) | ||||||
|---|---|---|---|---|---|---|---|---|
| Signal | Target | GlycSite | Transmem | Signalp | Targetp RC ≤2 | TMHMM | ||
| p ≥0.99 | p ≥0.75 | |||||||
| Saliva | 380 (27%) | 47 (3.3%) | 348 (24%) | 153 (10.7%) | 553 (29%) | 667 (34%) | 211 (11%) | 260 (13%) |
| Plasma | 457 (23%) | 25 (1.3%) | 513 (26%) | 350 (18%) | 530 (20%) | 702 (26%) | 291 (115%) | 457 (17%) |
| Overlap | 135 (34%) | 4 (1.0%) | 129 (33%) | 38 (9.6%) | 233 (39%) | 259 (43%) | 73 (12%) | 58 (9.7%) |
Sequence feature information extracted from the UniProt knowledgebase. The number represents the proteins with the entry in the UniProt. Signal: protein with a signal sequence (prepeptide); Target: protein with a transit peptide (mitochondrion, chloroplast, thylakoid,cyanelle, or microbody); Transmem: protein with transmembrane domain; GlycSite: protein with glycosylation site.
Sequence feature information obtained from signalp, targetp, and TMHMM sequence feature prediction programs. The number represents the total proteins used for the predictions. p: HMM signal peptide probability; RC: reliability classes.
We examined the distinct salivary proteins that are observed in saliva but not in plasma. Because it can be expected that some of the plasma proteins can be present in very low abundance in saliva, we compared the salivary proteome composed from WS, parotid, and SMSL to the plasma protein list including one peptide-based identifications (9555 total proteins). The proteins unique to saliva include those with well known salivary functions, such as proline-rich protein isoforms, amylase, cystatin isoforms, lactoperoxidase, and statherin. Antioxidant proteins, peroxiredoxin-4 and 6, proteinase kallikrein-1, and myeloperoxidase are also identified as unique proteins to saliva (Table 3). The abundance of these distinct salivary proteins are ranked based on the number of unique peptides used to derive their identifications from WS (Table 3).
Table 3.
Distinct human salivary proteins
| AC_V332 | Description | Num_Pep | Num_Group |
|---|---|---|---|
| IPI00745705 | LOC730924 similar to pancreatic α-amylase | 89 | 4 |
| IPI00374315 | C6orf58 uncharacterized protein C6orf58 | 68 | 3 |
| IPI00032294 | CST4 cystatin-S | 67 | 4 |
| IPI00305477 | CST1 cystatin-SN | 59 | 4 |
| IPI00013382 | CST2 Cystatin-SA | 58 | 4 |
| IPI00304557 | C20orf70 short palate, lung and nasal epithelium carcinoma-associated protein 2 | 49 | 4 |
| IPI00007244 | MPO isoform H17 of myeloperoxidase | 45 | 3 |
| IPI00060800 | LOC124220 protein UNQ773/PRO1567 | 42 | 4 |
| IPI00009650 | LCN1 lipocalin-1 | 39 | 4 |
| IPI00291410 | C20orf114 isoform 1 of long palate, lung and nasal epithelium carcinoma-associated protein 1 | 39 | 3 |
| IPI00025023 | LPO lactoperoxidase | 38 | 4 |
| IPI00010796 | P4HB protein disulfide-isomerase | 38 | 3 |
| IPI00026256 | FLG filaggrin | 37 | 3 |
| IPI00031547 | DSG3 desmoglein-3 | 35 | 4 |
| IPI00002851 | CST5 cystatin-D | 34 | 4 |
| IPI00025846 | DSC2 isoform 2A of desmocollin-2 | 34 | 3 |
| IPI00219525 | PGD 6-phosphogluconate dehydrogenase, decarboxylating | 32 | 3 |
| IPI00186290 | EEF2 elongation factor 2 | 32 | 2 |
| IPI00008274 | CAP1 adenylyl cyclase-associated protein 1 | 31 | 4 |
| IPI00386755 | ERO1L ERO1-like protein alpha | 31 | 3 |
| IPI00305622 | TGM1 protein-glutamine γ-glutamyltransferase K | 29 | 2 |
| IPI00784295 | HSP90AA1 isoform 1 of heat shock protein HSP 90-α | 29 | 2 |
| IPI00297056 | CRNN cornulin | 28 | 3 |
| IPI00082931 | SPRR3 small proline-rich protein 3 | 26 | 4 |
| IPI00011285 | CAPN1 calpain-1 catalytic subunit | 26 | 2 |
| IPI00027444 | SERPINB1 leukocyte elastase inhibitor | 25 | 4 |
| IPI00291006 | MDH2 malate dehydrogenase, mitochondrial | 25 | 3 |
| IPI00169383 | PGK1 phosphoglycerate kinase 1 | 23 | 4 |
| IPI00219077 | LTA4H isoform 1 of leukotriene A-4 hydrolase | 23 | 3 |
| IPI00744692 | TALDO1 transaldolase | 23 | 3 |
| IPI00031461 | GDI2 Rab GDP dissociation inhibitor β | 21 | 4 |
| IPI00299729 | TCN1 transcobalamin-1 | 21 | 3 |
| IPI00303476 | ATP5B ATP synthase subunitβ, mitochondrial | 21 | 2 |
| IPI00006560 | SERPINB13 isoform 1 of serpin B13 | 19 | 3 |
| IPI00007797 | FABP5;LOC728641 fatty acid-binding protein, epidermal | 19 | 3 |
| IPI00025512 | HSPB1 heat shock protein β-1 | 19 | 3 |
| IPI00003817 | ARHGDIB Rho GDP-dissociation inhibitor 2 | 18 | 4 |
| IPI00453476 | Uncharacterized protein ENSP00000348237 | 18 | 3 |
| IPI00010133 | CORO1A coronin-1A | 16 | 4 |
| IPI00012503 | PSAP isoform Sap-mu-0 of proactivator polypeptide | 16 | 3 |
| IPI00550363 | TAGLN2 transgelin-2 | 16 | 2 |
| IPI00295741 | CTSB cathepsin B | 15 | 4 |
| IPI00152154 | MUC7 mucin-7 | 15 | 3 |
| IPI00103242 | POF1B isoform 1 of protein POF1B | 15 | 1 |
| IPI00004656 | B2M β-2-microglobulin | 14 | 4 |
| IPI00220301 | PRDX6 peroxiredoxin-6 | 14 | 3 |
| IPI00000875 | EEF1G elongation factor 1-γ | 14 | 2 |
| IPI00440493 | ATP5A1 ATP synthase subunitα, mitochondrial | 14 | 1 |
| IPI00019038 | LYZ lysozyme C | 13 | 4 |
| IPI00304808 | KLK1 kallikrein-1 | 13 | 4 |
| IPI00646304 | PPIB peptidylprolyl isomerase B | 13 | 4 |
| IPI00291175 | VCL isoform 1 of vinculin | 13 | 3 |
| IPI00296777 | SPARCL1 SPARC-like protein 1 | 13 | 3 |
| IPI00216984 | CALML3 calmodulin-like protein 3 | 12 | 3 |
| IPI00219446 | PEBP1 phosphatidylethanolamine-binding protein 1 | 12 | 3 |
| IPI00414320 | ANXA11 annexin A11 | 12 | 1 |
| IPI00013895 | S100A11 protein S100-A11 | 11 | 4 |
| IPI00009123 | NUCB2 nucleobindin-2 | 11 | 3 |
| IPI00335168 | MYL6 isoform nonmuscle of myosin light polypeptide 6 | 11 | 2 |
| IPI00010896 | CLIC1;DDAH2 chloride intracellular channel protein 1 | 11 | 1 |
| IPI00011937 | PRDX4 peroxiredoxin-4 | 10 | 3 |
| IPI00026185 | CAPZB isoform 1 of F-actin-capping protein subunit β | 10 | 3 |
| IPI00060143 | FAM3D protein FAM3D | 10 | 3 |
| IPI00006995 | P11 placental protein 11 | 10 | 2 |
| IPI00024145 | VDAC2 isoform 1 of voltage-dependent anion-selective channel protein 2 | 10 | 1 |
| IPI00296526 | NAGK N-acetylglucosamine kinase | 9 | 3 |
| IPI00296713 | GRN isoform 1 of granulins | 9 | 3 |
| IPI00008580 | SLPI antileukoproteinase | 9 | 2 |
| IPI00026260 | NME1;NME2 nucleoside diphosphate kinase B | 9 | 2 |
| IPI00299571 | PDIA6 isoform 2 of protein disulfide-isomerase A6 | 9 | 2 |
| IPI00332828 | CES2 carboxylesterase 2 isoform 1 | 9 | 2 |
| IPI00377025 | PRH1; PRH2 PRH1 protein (fragment) | 9 | 1 |
| IPI00220828 | TMSB4X thymosin β-4 | 8 | 4 |
| IPI00012011 | CFL1 cofilin-1 | 8 | 3 |
| IPI00827847 | BPI bactericidal permeability-increasing protein | 8 | 3 |
| IPI00008529 | RPLP2 60S acidic ribosomal protein P2 | 8 | 2 |
| IPI00009856 | PLUNC protein Plunc | 8 | 2 |
| IPI00024915 | PRDX5 isoform mitochondrial of peroxiredoxin-5, mitochondrial | 8 | 2 |
| IPI00304171 | H2AFY isoform 2 of core histone macro-H2A.1 | 8 | 2 |
| IPI00216308 | VDAC1 voltage-dependent anion-selective channel protein 1 | 8 | 1 |
| IPI00023011 | SMR3B submaxillary gland androgen-regulated protein 3 homolog B | 7 | 4 |
| IPI00027463 | S100A6 protein S100-A6 | 7 | 3 |
| IPI00216088 | CRABP2 cellular retinoic acid-binding protein 2 | 7 | 3 |
| IPI00002818 | KLK11 isoform 1 of kallikrein-11 | 7 | 2 |
| IPI00010270 | RAC2 Ras-related C3 botulinum toxin substrate 2 | 7 | 2 |
| IPI00017987 | SPRR1A cornifin-A | 7 | 2 |
| IPI00465431 | LGALS3 galectin-3 | 7 | 2 |
| IPI00010214 | S100A14 Protein S100-A14 | 7 | 1 |
| IPI00019533 | CHI3L2 chitinase-3-like protein 2 | 7 | 1 |
| IPI00028064 | CTSG cathepsin G | 7 | 1 |
| IPI00299078 | PRH1; PRH2 salivary acidic proline-rich phosphoprotein 1/2 | 7 | 1 |
| IPI00075248 | CALM3; CALM1; CALM2 calmodulin | 6 | 3 |
| IPI00456429 | UBA52 ubiquitin and ribosomal protein L40 | 6 | 3 |
| IPI00017992 | SPRR2B small proline-rich protein 2B | 6 | 2 |
| IPI00022810 | CTSC dipeptidyl-peptidase 1 | 6 | 2 |
| IPI00025366 | CS citrate synthase, mitochondrial | 6 | 2 |
| IPI00298237 | TPP1 isoform 1 of tripeptidyl-peptidase 1 | 6 | 2 |
| IPI00304903 | SPRR1B cornifin-B | 6 | 2 |
| IPI00783680 | SOD1 superoxide dismutase | 6 | 2 |
| IPI00062120 | S100A16 protein S100-A16 | 6 | 1 |
| IPI00295542 | NUCB1 nucleobindin-1 | 5 | 3 |
| IPI00414896 | RNASET2 isoform 1 of ribonuclease T2 | 5 | 2 |
| IPI00441498 | FOLR1 folate receptor alpha | 5 | 2 |
| IPI00013881 | HNRPH1 heterogeneous nuclear ribonucleoprotein H | 5 | 1 |
| IPI00478198 | PRB1 proline-rich protein BstNI subfamily 1 isoform 1 | 5 | 1 |
| IPI00034319 | CUTA isoform A of protein CutA | 4 | 3 |
| IPI00003919 | QPCT glutaminyl-peptide cyclotransferase | 4 | 2 |
| IPI00011302 | CD59 CD59 glycoprotein | 4 | 2 |
| IPI00016513 | RAB10 Ras-related protein Rab-10 | 4 | 2 |
| IPI00329538 | PRSS8 prostasin | 4 | 2 |
| IPI00786921 | SPRR1B similar to cornifin B | 4 | 2 |
| IPI00023038 | PRB1 basic salivary proline-rich protein 1 | 4 | 1 |
| IPI00028066 | ADH7 class IV alcohol dehydrogenase 7 mu or sigma subunit | 4 | 1 |
| IPI00029699 | RNASE4 ribonuclease 4 | 4 | 1 |
| IPI00177543 | PAM uncharacterized protein PAM | 4 | 1 |
| IPI00219029 | GOT1 aspartate aminotransferase, cytoplasmic | 4 | 1 |
| IPI00301579 | NPC2 epididymal secretory protein E1 | 4 | 1 |
| IPI00376005 | EIF5A isoform 2 of eukaryotic translation initiation factor 5A-1 | 4 | 1 |
| IPI00382404 | PRB4 PRB4 protein | 4 | 1 |
| IPI00010182 | DBI isoform a 1 of acyl-CoA-binding protein | 3 | 3 |
| IPI00012024 | HTN1 histatin-1 | 3 | 3 |
| IPI00329801 | ANXA5 annexin A5 | 3 | 3 |
| IPI00215997 | CD9 CD9 antigen | 3 | 2 |
| IPI00291488 | WFDC2 isoform 1 of WAP four-disulfide core domain protein 2 | 3 | 2 |
| IPI00000877 | HYOU1 hypoxia upregulated protein 1 | 3 | 1 |
| IPI00003881 | HNRPF heterogeneous nuclear ribonucleoprotein F | 3 | 1 |
| IPI00012585 | HEXB β-hexosaminidase beta chain | 3 | 1 |
| IPI00299086 | SDCBP syntenin-1 | 3 | 1 |
| IPI00465315 | CYCS cytochrome c | 3 | 1 |
| IPI00022990 | STATH statherin | 2 | 3 |
| IPI00027019 | PRR4; PRH1; PRH2 proline-rich protein 4 | 2 | 2 |
| IPI00027851 | HEXA β-hexosaminidase alpha chain | 2 | 2 |
| IPI00028714 | MGP matrix Gla protein | 2 | 2 |
| IPI00067738 | FAM3B isoform B of protein FAM3B | 2 | 2 |
| IPI00293276 | MIF macrophage migration inhibitory factor | 2 | 2 |
| IPI00328960 | LOC147645 similar to carcinoembryonic antigen-related cell adhesion molecule 1 | 2 | 2 |
| IPI00003176 | HTRA1 serine protease HTRA1 | 2 | 1 |
| IPI00003802 | MAN2A1 α-mannosidase 2 | 2 | 1 |
| IPI00006713 | DNAJC3 isoform 1 of DnaJ homolog subfamily C member 3 | 2 | 1 |
| IPI00012540 | PROM1 prominin-1 | 2 | 1 |
| IPI00018236 | GM2A ganglioside GM2 activator | 2 | 1 |
| IPI00018387 | FURIN furin | 2 | 1 |
| IPI00168884 | ATP6AP2 renin receptor | 2 | 1 |
| IPI00183695 | S100A10 protein S100-A10 | 2 | 1 |
| IPI00329482 | LAMA4 isoform 1 of laminin subunit α-4 | 2 | 1 |
| IPI00333140 | DNER delta and Notch-like epidermal growth factor-related receptor | 2 | 1 |
The list contains the proteins that are not found in plasma but found in Whole, Parotid, and SMSL salivas. The numbers for the Num_Pep and Num_Group columns are represented by the data from WS dataset.
To further examine the biological roles of the salivary and plasma proteins, we examined and classified the proteins based on their biological pathways extracted from the KEGG pathway database (Table 4). Saliva and plasma proteins contain highest pathway activities in cell communication, carbohydrate and amino acid metabolism, immune system, and signal transduction. Exceptions were found in the signaling molecule and interaction pathway, to which the 108 out of 1415 total entries of the plasma proteins in KEGG were matched, in contrast to only 47 of 1527 total entries for the saliva proteins.
Table 4.
Involvement of salivary and plasma proteomes in KEGG biological pathways
| KEGG pathways | Saliva (entries = 1527) | Plasma (entries = 1415) | Overlap (entries = 368) |
|---|---|---|---|
| Cell communication | 205 | 195 | 85 |
| Carbohydrate metabolism | 160 | 89 | 40 |
| Amino acid metabolism | 164 | 95 | 35 |
| Immune system | 127 | 180 | 44 |
| Signal transduction | 99 | 167 | 20 |
| Lipid metabolism | 67 | 42 | 9 |
| Energy metabolism | 63 | 18 | 11 |
| Translation/transcription | 57 | 17 | 2 |
| Endocrine system | 58 | 62 | 12 |
| Cell motility | 50 | 53 | 18 |
| Nucleotide metabolism | 51 | 34 | 7 |
| Infectious diseases | 52 | 25 | 12 |
| Xenobiotics biodegradation and metabolism | 38 | 33 | 6 |
| Neurodegenerative disorders | 38 | 37 | 16 |
| Others | 52 | 38 | 8 |
| Folding, sorting, and degradation | 27 | 20 | 6 |
| Metabolism of cofactors and vitamins | 28 | 28 | 6 |
| Nervous system | 27 | 28 | 4 |
| Signaling molecules and interaction | 47 | 108 | 16 |
| Cancers | 21 | 25 | 0 |
| Development | 23 | 35 | 1 |
| Glycan biosynthesis and metabolism | 47 | 25 | 5 |
| Cell growth and death | 13 | 40 | 4 |
| Metabolic disorder | 14 | 21 | 1 |
The number of entries represents the total pathways containing saliva proteins, plasma proteins, or the overlapping proteins of saliva and plasma. A protein can be involved in multiple pathways.
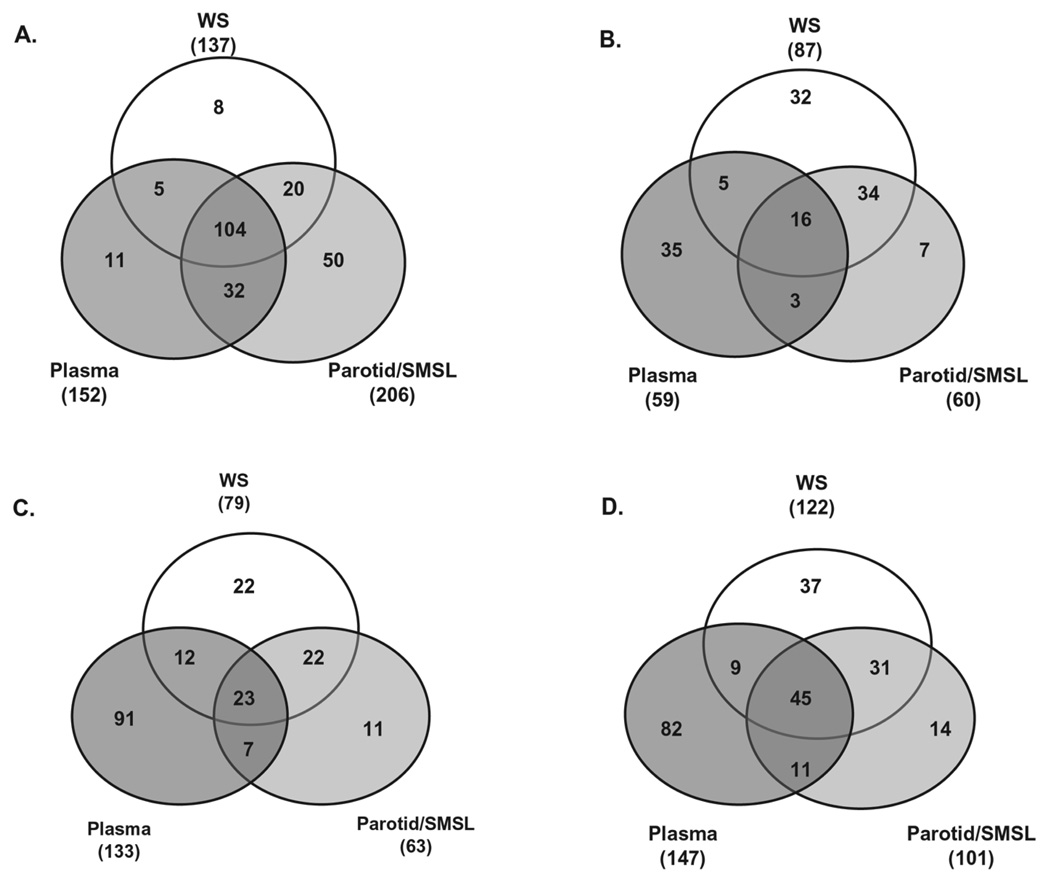
3.3 Enrichment of Igs in salivary and plasma proteomes
Besides the KEGG pathways shared by many salivary and plasma proteins, a detailed examination revealed that these two proteomes are enriched with Igs. Consistent with the previous report that Igs make up about 5–15% of the total number of salivary proteins [50], the saliva proteome from the integrated results of WS and parotid/SMSL show that 219 (11.3%) of the 1939 salivary protein components are Igs. Interestingly, a majority of these Igs (141 out of 219) were shown to overlap with the plasma Igs, even though the specimens of saliva and plasma are not from the same individuals (Fig. 5). We also compared the salivary and plasma proteins participating in the KEGG carbohydrate metabolism, immune system, and cell communication pathways. In contrast to the striking high overlap (61%, 141 out of 230) of Igs found in saliva and plasma, only 18% (24 out of 132) overlap was found in the carbohydrate metabolism pathway, 22% (42 out of 188) in immune system, and 27% (65 out of 239) in cell communication. When the comparisons are performed between WS and ductal parotid/SMSL proteomes, the higher overlaps are observed in these KEGG pathways with 57% in carbohydrate metabolism, 46% in immune system, and 43% in cell communication (Fig. 5). This higher overlap between WS and ductal parotid/SMSL is consistent with the closer biological and physiological similarity of these two fluids than between saliva and plasma.
Figure 5.
Venn diagram showing the salivary and plasma overlap in Igs and proteins participating in a KEGG pathway. (A) Igs; (B) carbohydrate metabolism; (C) immune system. (D) cell communication. The number inside the parentheses represents the total number of proteins participating in the pathway from the fluid.
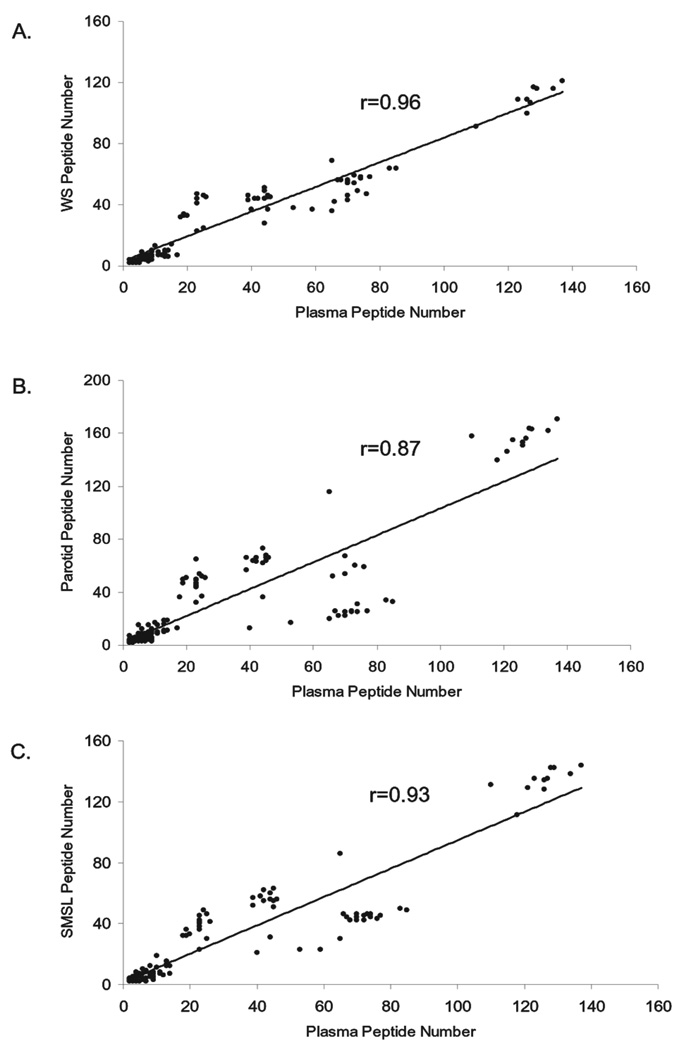
The relative abundance of these Igs varied greatly. The Igs in saliva are identified with 2 to 171 unique peptides and with sequence coverages from below 0.1% to as high as 91%. In plasma, the Igs are derived from 2 to 137 unique peptides. Figure 6 demonstrates that linear correlation of the number of unique peptides observed for the Igs exists between WS and plasma, parotid and plasma, and SMSL and plasma.
Figure 6.
Linear correlation of the unique peptide numbers for the Ig proteins coexisting in WS, parotid, SMSL, and plasma. Each point represents an overlapping Ig protein. (A) Correlation of Ig proteins in WS and plasma; (B) correlation of Ig proteins in parotid and plasma; (C) correlation of Ig proteins in SMSL and plasma. The straight line shows that the number of unique peptides used for the Ig identifications in WS, parotide, or SMSL has a linear correlation with the number of the peptides used for the plasma Ig identifications.
4 Discussion
4.1 WS and ductal saliva proteomes
To achieve a comprehensive human salivary proteome, we began with construction of the WS proteome. Similar to other large-scale proteome projects such as the HUPO Plasma and Brain Proteome Projects and most recent ductal parotid and SMSL proteome, the intrinsic complexity of the WS proteome made its characterization challenging and is influenced by sample source and collection process, sample preparation, and the protein identification process. The power of combining the datasets from different experimental approaches results in a more comprehensive proteome than any single approach can achieve. A core dataset with 1444 WS proteins was assembled from the integration process. Similar to the reports from the plasma proteome, brain proteome, and parotid/SMSL proteome studies, a large portion (52%) of the WS proteins are identifications measured by only one laboratory. Besides the various sample preparation and experimental measurement approaches employed, variations in saliva sample source and protein concentration also may induce differences in protein identification. The WS proteins confirmed by the four groups (233 proteins) should represent the essential salivary components that are least susceptible to differences in methodology and sample source. These protein identifications include such wellknown salivary proteins as Igs, amylases, cystatins (D, S, C, SA, and SN), proline-rich protein 3, keratins, and mucin-5B.
With our previous characterization of the ductal parotid and SMSL proteomes, our present study showed that the salivary protein components vary with the source and origin of the fluid, i.e., WS or ductal salivas. Although 740 out of 1939 salivary proteins coexist in WS and ductal saliva, 563 are specific to WS and 369 specific to parotid/SMSL. It is known that proteins in WS originate from not only the secretion of salivary glands (i.e., SM, SL, parotid, and minor glands) but also from leakage of plasma, secretion of bronchial and nasal sources, gingival crevicular fluid, bacteria, food debris, and epithelial or other cell debris.
The functions of saliva include lubrication, antimicrobial, protection of mucosal integrity, and digestion. Proteins that participate in one or more of these salivary functions include mucins, amylases, defensins, cystatins, histatins, proline-rich proteins, statherin, lactoperoxidase, lysozyme, lactoferrin, and Igs. The functions of these proteins can be redundant and overlapping. Our study indicates that all of these protein family/isoforms were shared between WS and parotid and SM/SL fluids, although one or more proteins in the family can be specific to the WS, parotid or SM/SL proteome. These observations support the previous hypothesis that a specific protein may not be critical for a specific salivary function because other protein families can maintain its function [50, 51].
4.2 Similarities and differences between saliva and plasma proteomes
Except for Igs, proteins with known salivary functions were commonly, but not always, absent in the plasma proteome. For example, statherin and histatin protein families are specific to saliva. The number of isoforms and abundance of mucin, cystatin, and proline-rich protein families in plasma were significantly lower in plasma than in the WS and parotid/SMSL proteomes.
Similarity and distinction of the salivary and plasma proteomes were revealed also through analysis of their cellular components, molecular functions, biological processes, sequence features, and biological pathways. As expected for body fluids, the GO study of cellular components displayed that both saliva and plasma are over-represented with extra-cellular proteins when compared with the overall human proteome. Surprisingly, saliva and plasma are also enriched with the cytoplasmic proteins, which could result from cell death.However, specific transport pathways may also exist. A recent study of the tear proteome revealed that cytoplasmic proteins are enriched [52]; a few intracellular proteins were demonstrated as originating from cellular shedding of the epithelium [53]. Tears are produced from the lacrimal gland with a structure similar to serous acini of the salivary gland. Whether the cytoplasmic proteins in saliva and plasma also originate from cellular shedding of the epithelium, as in tear fluid, remains to be determined. The GO analysis demonstrated that saliva and plasma are over-represented in response-to-stimulus and response-to-stress processes, presumably reflecting the functions of these two body fluids in the body’s defense system. Saliva is over-represented in catabolic and protein metabolic processes, which may reflect its major physiologic function in food digestion. As expected, the sequence feature analysis indicated that saliva and plasma contain high proportions of proteins with a signal peptide sequence required for targeting proteins to the ER for subsequent transport through the secretory pathway.
Glycosylation of salivary proteins is believed to play a role in the salivary protective functions. Characterizations of the glycosylated proteins in saliva and plasma have reported 45 proteins in saliva and 303 in plasma as glycosylated proteins [20, 49]. The result of the annotation information extracted from the UniProt knowledge indicates that potentially more glycosylated proteins exist in saliva and plasma.
Several sources can contribute to the overlap of protein identifications of saliva and plasma: (i) leakage of plasma into saliva through intracellular or extracellular routes, including outflow of gingival crevicular fluid; (ii) plasma and saliva may share essential proteins needed to maintain their physiological functions as body fluids; (iii) proteins derived from cell debris may be in close contact with either fluid. We expected that the overlapping proteins from different sources would show different abundance patterns. Classification of the salivary and plasma proteins based on their function in the KEGG pathways revealed that the abundance correlations of the overlapping proteins of saliva and plasma vary with their biological functions. Previous estimates established that Igs contribute 5–15% of total salivary proteins. In the present study, 11% of total salivary proteins identified were Igs, and 64% of these were found in plasma. The source of the Igs in saliva was previously proposed as either from salivary gland secretions or from crevicular fluid [50, 54]. Our study reveals that there is a high correlation between the abundance of the overlapping Igs in saliva and plasma, suggesting that these overlapping Igs could result from leakage from plasma.
4.3 Clinical value of saliva and plasma
The ultimate goal of cataloging the proteins found in body fluids is to use the information for health screening and disease detection. To that end, plasma proteins have proved their value as clinical analytes. Saliva has attracted increased attention in that it provides advantages over other body fluids in its noninvasive collection, constant availability, little need for special equipment, and cost-effectiveness. Diseases such as Sjögren’s syndrome, bacterial and viral infectious diseases, and oral cancer cause alterations of salivary protein expression. Comparison of the salivary proteome with the plasma proteome helps to identify the salivary specific biomarkers as well as plasma-derived biomarkers that have been used in the diagnostics of a variety of human diseases.
Our search of the OMIM database indicated that salivary and plasma proteomes contain a large number of proteins associated with genetic disorders, some of which have known phenotypes. Table 5 shows the gene entries of the salivary and plasma proteomes in OMIM. The saliva proteins were matched to 1183 entries in OMIM; 1089 are disease genes with known sequences and 91 are related with diseases with known phenotypes. Similar distributions are observed for plasma proteins. Proteins present in both saliva and plasma were matched to 310 entries in the diseases with known gene sequence and 47 entries in the diseases with phenotype. Interestingly, a few plasma proteins that are used in clinical diagnostics [55, 56] are also identified in the saliva, including creatine kinase B-type, fibrinogen, hemoglobin, rheumatoid factor, and Igs. These results enhance the potential value of salivary proteins as biomarkers for diagnostics. However, it remains to be determined qualitatively and quantitatively whether these proteins carrying genetic disorders or in combination with the diagnostic plasma proteins can be used as disease biomarkers.
Table 5.
Entries of the salivary and plasma proteins in OMIM
| OMIM category | Saliva | Plasma | Overlap |
|---|---|---|---|
| Genes with known sequence | 1089 | 1288 | 310 |
| Genes of known sequence and a phenotype | 91 | 115 | 47 |
| Confirmed phenotype with molecular basis unknown | 2 | 0 | 0 |
| Descriptive entry | 1 | 2 | 1 |
Acknowledgments
This work was supported by the National Institutes of Health (U01 DE16275 to D.T.W. and J.A.L., U01DE016267 to J.R.Y. and J.E.M., U54DA021519 to G.O., RO1DE17734 to T.J.G.), MEDC grant GR687 (to G.O.), and SAIC/NCI contract SAIC/ NCI 23X110A. J.L.B., J.R.S., B.J.C. and J.L.S. acknowledge funding from the National Institute of Allergy and Infectious Disease, National Institutes of Health under contract No. HHSN266200400067C. J.A.L. acknowledges also support from the W. M. Keck Foundation for the establishment of the UCLA Functional Proteomics Center.
Abbreviations
- CITP
Transient capillary isotachophoresis
- DMS
dimethylacrylamide
- FA
formic acid
- FFE
free-flow electrophoresis
- GO
gene ontology
- IPI
International Protein Index
- OMIM
online Mendelian inheritance in man
- SCX
strong cation exchange
- SL
sublingual
- SM
submandibular
- WS
whole saliva
Footnotes
The authors have declared no conflict of interest.
References
- 1.Humphrey SP, Williamson RT. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001;85:162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 2.Turner RJ, Sugiya H. Understanding salivary fluid and protein secretion. Oral Dis. 2002;8:3–11. doi: 10.1034/j.1601-0825.2002.10815.x. [DOI] [PubMed] [Google Scholar]
- 3.Defabianis P, Re F. The role of saliva in maintaining oral health. Minerva Stomatol. 2003;52:301–308. [PubMed] [Google Scholar]
- 4.Streckfus CF, Bigler LR. Saliva as a diagnostic fluid. Oral Dis. 2002;8:69–76. doi: 10.1034/j.1601-0825.2002.1o834.x. [DOI] [PubMed] [Google Scholar]
- 5.Drake RR, Cazare LH, Semmes OJ, Wadsworth JT. Serum, salivary and tissue proteomics for discovery of biomarkers for head and neck cancers. Expert Rev. Mol. Diagn. 2005;5:93–100. doi: 10.1586/14737159.5.1.93. [DOI] [PubMed] [Google Scholar]
- 6.Amado FM, Vitorino RM, Domingues PM, Lobo MJ, et al. Analysis of the human saliva proteome. Expert Rev.Proteomics. 2005;2:521–539. doi: 10.1586/14789450.2.4.521. [DOI] [PubMed] [Google Scholar]
- 7.Rujner J, Socha J, Barra E, Gregorek H, et al. Serum and salivary antigliadin antibodies and serum IgA anti-endomysium antibodies as a screening test for coeliac disease. Acta Paediatr. 1996;85:814–817. doi: 10.1111/j.1651-2227.1996.tb14157.x. [DOI] [PubMed] [Google Scholar]
- 8.Hu S, Wang J, Meijer J, Ieong S, et al. Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum. 2007;56:3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen AM, Reibel J, Nordgarden H, Bergem HO, et al. Primary Sjogren’s syndrome: Salivary gland function and clinical oral findings. Oral Dis. 1999;5:128–138. doi: 10.1111/j.1601-0825.1999.tb00077.x. [DOI] [PubMed] [Google Scholar]
- 10.Hu S, Yu T, Xie Y, Yang Y, et al. Discovery of oral fluid biomarkers for human oral cancer by mass spectrometry. Cancer Genomics Proteomics. 2007;4:55–64. [PubMed] [Google Scholar]
- 11.Shori DK, Kariyawasam HH, Knight RA, Hodson ME, et al. Sulphation of the salivary mucin MG1 (MUC-5B) is not correlated to the degree of its sialylation and is unaffected by cystic fibrosis. Pflugers Arch. 2001;443:S50–S54. doi: 10.1007/s004240100644. [DOI] [PubMed] [Google Scholar]
- 12.Aydin S. A comparison of ghrelin, glucose, alpha-amylase and protein levels in saliva from diabetics. J. Biochem. Mol.Biol. 2007;40:29–35. doi: 10.5483/bmbrep.2007.40.1.029. [DOI] [PubMed] [Google Scholar]
- 13.Hagewald SJ, Fishel DL, Christan CE, Bernimoulin JP, et al. Salivary IgA in response to periodontal treatment. Eur. J. Oral Sci. 2003;111:203–208. doi: 10.1034/j.1600-0722.2003.00040.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, LaRoche T, Hamelinck D, Bergsma D, et al. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat. Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 15.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, et al. Large-scale identification of proteins in human salivary pro-teome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 17.Xie H, Bandhakavi S, Griffin TJ. Evaluating preparative isoelectric focusing of complex peptide mixtures for tandem mass spectrometry-based proteomics: A case study in profiling chromatin-enriched subcellular fractions in Saccharomyces cerevisiae. Anal. Chem. 2005;77:3198–3207. doi: 10.1021/ac0482256. [DOI] [PubMed] [Google Scholar]
- 18.Xie H, Rhodus NL, Griffin RJ, Carlis JV, et al. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol. Cell. Proteomics. 2005;4:1826–1830. doi: 10.1074/mcp.D500008-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Guo T, Rudnick PA, Wang W, Lee CS, et al. Characterization of the human salivary proteome by capillary isoelectric focusing/nanoreversed-phase liquid chromatography coupled with ESI-tandem MS. J. Proteome Res. 2006;5:1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandran P, Boontheung P, Xie Y, Sondej M, et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 21.Wilmarth PA, Riviere MA, Rustvold DL, Lauten JD, et al. Two-dimensional liquid chromatography study of the human whole saliva proteome. J. Proteome Res. 2004;3:1017–1023. doi: 10.1021/pr049911o. [DOI] [PubMed] [Google Scholar]
- 22.Denny P, Hagen FK, Hardt M, Liao L, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasper DL, Braunwald E, Hauser S, Longo D, et al. Harrison’s Principles of Internal Medicine. New York: McGraw-Hill; 2004. [Google Scholar]
- 24.Omenn GS. Exploring the human plasma proteome. Proteomics. 2005;5:3223–3225. doi: 10.1002/pmic.200590056. [DOI] [PubMed] [Google Scholar]
- 25.Omenn GS, States DJ, Adamski M, Blackwell TW, et al. Overview of the HUPO Plasma Proteome Project Results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 26.Adamski M, Blackwell T, Menon R, Martens L, et al. Data management and preliminary data analysis in the pilot phase of the HUPO Plasma Proteome Project. Proteomics. 2005;5:3246–3261. doi: 10.1002/pmic.200500186. [DOI] [PubMed] [Google Scholar]
- 27.States DJ, Omenn GS, Blackwell TW, Fermin D, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat. Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 28.Fang X, Yang L, Wang W, Song T, et al. Comparison of electrokinetics-based multidimensional separations coupled with electrospray ionization-tandem mass spectrometry for characterization of human salivary proteins. Anal.Chem. 2007;79:5785–5792. doi: 10.1021/ac070611a. [DOI] [PubMed] [Google Scholar]
- 29.Rhodus NL, Cheng B, Myersq S, Bowles W, et al. A comparison of the pro-inflammatory, NF-kappaB-dependent cytokines: TNF-alpha, IL-1-alpha, IL-6, and IL-8 in different oral fluids from oral lichen planus patients. Clin. Immunol. 2005;114:278–283. doi: 10.1016/j.clim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Eng J, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 31.Peng J, Elias JE, Thoreen CC, Licklider LJ, et al. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 32.Shimura K, Kamiya K, Matsumoto H, Kasai K. Fluorescence-labeled peptide pI markers for capillary isoelectric focusing. Anal. Chem. 2002;74:1046–1053. doi: 10.1021/ac0108010. [DOI] [PubMed] [Google Scholar]
- 33.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal.Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 34.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie H, Griffin TJ. Trade-off between high sensitivity and increased potential for false positive peptide sequence matches using a two-dimensional linear ion trap for tandem mass spectrometry-based proteomics. J. Proteome Res. 2006;5:1003–1009. doi: 10.1021/pr050472i. [DOI] [PubMed] [Google Scholar]
- 36.Cargile BJ, Bundy JL, Freeman TW, Stephenson JL., Jr Gel based isoelectric focusing of peptides and the utility of isoelectric point in protein identification. J. Proteome Res. 2004;3:112–119. doi: 10.1021/pr0340431. [DOI] [PubMed] [Google Scholar]
- 37.Cargile BJ, Stephenson JL., Jr An alternative to tandem mass spectrometry: Isoelectric point and accurate mass for the identification of peptides. Anal. Chem. 2004;76:267–275. doi: 10.1021/ac0352070. [DOI] [PubMed] [Google Scholar]
- 38.Cargile BJ, Talley DL, Stephenson JL., Jr Immobilized pH gradients as a first dimension in shotgun proteomics and analysis of the accuracy of pI predictability of peptides. Electrophoresis. 2004;25:936–945. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 39.Essader AS, Cargile BJ, Bundy JL, Stephenson JL., Jr A comparison of immobilized pH gradient isoelectric focusing and strong-cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 2005;5:24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 40.Geer LY, Markey SP, Kowalak JA, Wagner L, et al. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 41.Zuo X, Speicher DW. Comprehensive analysis of complex proteomes using microscale solution isoelectrofocusing prior to narrow pH range two-dimensional electrophoresis. Proteomics. 2002;2:58–68. [PubMed] [Google Scholar]
- 42.Ramachandran P, Boontheung P, Xie Y, Sondej M, et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 43.Martens L, Muller M, Stephan C, Hamacher M, et al. A comparison of the HUPO Brain Proteome Project pilot with other proteomics studies. Proteomics. 2006;6:5076–5086. doi: 10.1002/pmic.200600291. [DOI] [PubMed] [Google Scholar]
- 44.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bendtsen JD, Nielsen H, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol.Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 46.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 47.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol.Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 48.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Liu T, Qian WJ, Gritsenko MA, Camp DG, II, et al. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Nieuw Amerongen A, Bolscher JGM, Veerman ECI. Salivary proteins: Protective and diagnostic value in cariology . Caries Res. 2004;38:247–253. doi: 10.1159/000077762. [DOI] [PubMed] [Google Scholar]
- 51.Rudney JD, Hickey KL, Ji Z. Cumulative correlations of lysozyme, lactoferrin, peroxidase, S-IgA, amylase, and total protein concentrations with adherence of oral viridans streptococci to microplates coated with human saliva. J.Dent. Res. 1999;78:759–768. doi: 10.1177/00220345990780030801. [DOI] [PubMed] [Google Scholar]
- 52.de Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Haeringen NJ, Glasius E. Enzymes of energy-producing metabolism in human tear fluid. Exp. Eye Res. 1974;18:407–409. doi: 10.1016/0014-4835(74)90118-3. [DOI] [PubMed] [Google Scholar]
- 54.Huq NL, Cross KJ, Ung M, Myroforidis H, et al. A review of the salivary proteome and peptidome and saliva-derived peptide therapeutics. Int. J. Pept. Res. Ther. 2007;13:547–564. [Google Scholar]
- 55.Burtis CA, Ashwood EA, editors. Tietz Fundamentals of Clinical Chemistry. 5th Edn. Philadelphia, PA: W. B. Saunders; 2001. [Google Scholar]
- 56.Wallach JB, editor. Interpretation of Diagnostic Tests. Philadelphia PA: Lippincott Williams & Wilkins; 2006. [Google Scholar]