Abstract
A gradient of Dorsal activity patterns the dorsoventral (DV) axis of the early Drosophila melanogaster embryo by controlling the expression of genes that delineate presumptive mesoderm, neuroectoderm, and dorsal ectoderm. The availability of the Drosophila melanogaster genome sequence has accelerated the study of embryonic DV patterning, enabling the use of systems-level approaches. As a result, our understanding of Dorsal-dependent gene regulation has expanded to encompass a collection of more than 50 genes and 30 cis-regulatory sequences. This information, which has been integrated into a spatiotemporal atlas of gene regulatory interactions, comprises one of the best-understood networks controlling any developmental process to date. In this article, we focus on how Dorsal controls differential gene expression and how recent studies have expanded our understanding of Drosophila embryonic development from the cis-regulatory level to that controlling morphogenesis of the embryo.
The transcription factor Dorsal sets up the dorsoventral axis in fly embryos. Genome-wide studies now provide a spatiotemporal atlas of the regulatory network it controls.
The classical definition of a morphogen requires that the protein be synthesized from a localized source to affect concentration-dependent outputs of gene expression. The Drosophila embryo is a specialized case because it is a syncytium at early stages, permitting the formation of transcription factor gradients, which regulate transcription in a graded, concentration-dependent manner. In the early Drosophila embryo, two maternally deposited transcription factors, Bicoid and Dorsal, specify the anteroposterior (AP) and dorsoventral (DV) axes, respectively. Different levels of these factors control the expression of distinct sets of genes. Here, we provide an overview of DV patterning of the early Drosophila embryo. We focus on the role of Dorsal and highlight the novel insights genome approaches have provided.
Initiation of the Dorsal Nuclear Gradient
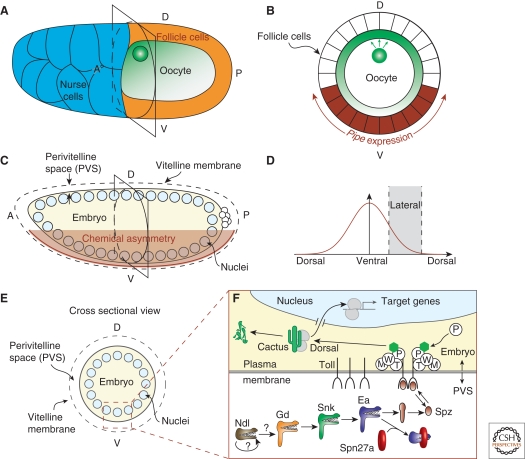
Within the nuclei of early Drosophila embryos, the Dorsal transcription factor is present in a ventral-to-dorsal gradient (Figs. 1C,D and 2A) (Moussian and Roth 2005). The maternally supplied Dorsal transcript is distributed and translated uniformly throughout the embryo; however, activation of the Toll receptor is limited to ventral and ventrolateral regions of the embryo, and this causes the Dorsal protein to be translocated into the nucleus. The positional information guiding activation of Toll is initiated by the follicle cells surrounding the developing oocyte during Stage 10 of oogenesis (Fig. 1A,B) (Anderson 1998). At this stage, epidermal growth factor receptor (EGFR) signaling through the ligand Gurken limits expression of the gene pipe to the ventral-most follicle cells (Fig. 1B) (Schupbach 1987; Sen et al. 1998). pipe encodes a protein sharing homology with vertebrate heparan sulfate 2-O-sulfotransferases, but likely functions independent of heparan (Zhu et al. 2005).
Figure 1.
Overview of the ventral signaling pathway. (A) Schematic of St. 10 egg chamber. The oocyte nucleus is located at the dorsoanterior cortex. Gurken, which is locally translated, is present in a protein gradient (green). (B) Cross section of schematic of St. 10 egg chamber. Gurken signaling (green) represses pipe expression (brown) in the follicle cells. (C) Schematic of syncytial blastoderm embryo. The ventral follicle cells, which had expressed pipe, deposit an unknown “chemical asymmetry” into the perivitelline space. (D) The chemical asymmetry results in a ventral-to-dorsal signaling gradient. (E) Cross section of schematic of syncytial blastoderm embryo. The signaling gradient is initially established within the perivitelline space, a small extracellular space between the embryo and an outer vitelline membrane. (F) Illustration of ventral signaling pathway in the early embryo. In the perivitelline space (PVS), a protease cascade (Ndl, Gd, Snk, Ea) eventually activates Spz, the ligand for the Toll receptor. The serine protease inhibitor, Spn27a, inhibits the activity of Ea. Activated Spz transduces the signal into the embryo through Toll, causing the degradation of Cactus and the nuclear translocation of the transcription factor Dorsal. The roles of Tube (T), Pelle (P), Weckle (W), and Myd88 (M) are relatively unknown, but participate in a signaling complex at the cytoplasmic tail of Toll.
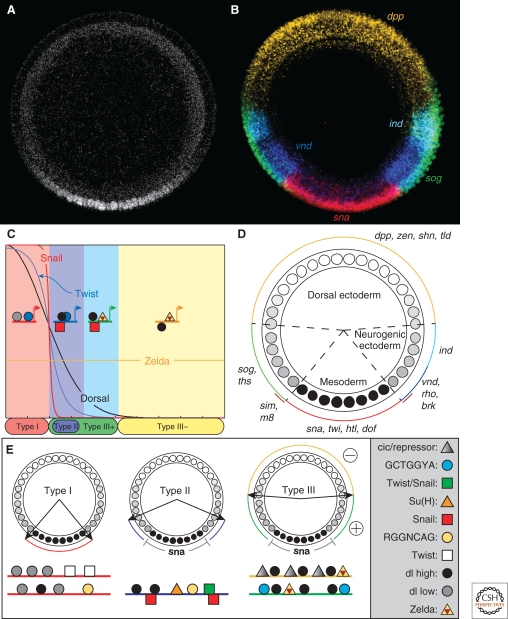
Figure 2.
Illustration of early embryonic fate map. (A) Cross section of Stage 5 Drosophila embryo, fluorescently stained with an α-Dorsal antibody. (B) Cross section of Stage 5 Drosophila embryo, fluorescently stained by in situ hybridization to detect Dorsal target gene transcripts dpp, ind, vnd, sog, and sna. (C) Dorsal and Twist cooperate to specify both Type I and Type II Dorsal target genes. Dorsal functions together with Zelda to support expression of Type III (+ and −) target genes (See legend in part E). (D) Schematic of fate map. The Dorsal nuclear gradient divides the embryo into three main subtissues: mesoderm, neurogenic ectoderm, and dorsal ectoderm. The neurogenic ectoderm can be further divided into ventral and dorsal halves. (E) Groupings of Dorsal target genes. Type I genes are expressed in the ventral-most portion of the embryo, where Dorsal nuclear levels are the highest. Type II genes have dorsal borders in the middle of the neurogenic ectoderm. These genes are also repressed by Snail. Type III genes have their dorsal (+) or ventral (−) borders at roughly 50% DV axis, and contain sites for both Dorsal binding as well as a uniformly expressed activator, such as Zelda.
As oogenesis proceeds, the pipe-expressing follicle cells deposit an unknown “chemical asymmetry” into the eggshell, which spatially regulates an extracellular protease cascade, comprised of four proteases: Nudel (Ndl), Gastrulation-defective (Gd), Snake (Snk), and Easter (Ea) (Fig. 1F) (Smith and DeLotto 1994; Sen et al. 1998; LeMosy et al. 2001; Peri et al. 2002). The protease reactions occur in the perivitelline space and culminate in the processing of Spätzle (Spz), the protein ligand for Toll (Fig. 1C–F) (Roth 1993; Morisato 2001; Zhu et al. 2005). In addition to the protease cascade that ensures localized activation of Spz in ventral regions, the gradient of active Spz is further shaped through the action of a serpin (serine-protease inhibitor), Spn27A (Hashimoto et al. 2003; Ligoxygakis et al. 2003).
Downstream of the Spz-mediated activation of Toll, intracellular signaling occurs through the action of several maternal factors. These proteins, which include Weckle, Myd88, Tube, and Pelle, function together to facilitate the degradation of Cactus, a cytoplasmic tethering protein, thereby releasing Dorsal from cytoplasmic retention (Fig. 1F) (Hecht and Anderson 1993; Belvin et al. 1995; Grosshans et al. 1999; Sun et al. 2004; Chen et al. 2006). With the exception of Weckle, this intracellular signaling module is well conserved in vertebrates in which the homologs are involved in regulation of the immune response (Belvin and Anderson 1996). In addition to freeing Dorsal from Cactus, Toll signaling likely potentiates Dorsal nuclear translocation in a Cactus-independent manner, as even in cactus mutants, Dorsal is not fully nuclear on the dorsal side of the embryo (Bergmann et al. 1996). However, despite the lack of Toll-mediated signal on the dorsal side of the embryo, recent in vivo imaging studies using a Dorsal-GFP fusion protein show a constant shuttling of the protein between the cytoplasm and the nuclei, including the dorsal-most nuclei (DeLotto et al. 2007). Therefore, it appears that exclusion of Dorsal from nuclei is not achieved by simply preventing Dorsal nuclear import, but by a balance between slow import and rapid export.
Differential Expression of Dorsal Target Genes
The gradient of nuclear-localized Dorsal regulates a number of genes in a concentration-dependent manner (Fig. 2B,D) (reviewed in Stathopoulos and Levine 2004). High levels of nuclear Dorsal present in ventral regions activate genes such as twist (twi) and snail (sna), which are required for specification of the mesoderm (Simpson 1983; Thisse et al. 1987; Jiang et al. 1991; Pan et al. 1991; Ray et al. 1991; Ip et al. 1992a). Intermediate levels of nuclear Dorsal, present in ventrolateral regions, induce the expression of genes such as rhomboid (rho) and ventral neuroblasts defective (vnd), which are important for specification of the neurogenic ectoderm (Bier et al. 1990; Ip et al. 1992a; Jimenez et al. 1995; Stathopoulos et al. 2002). Low levels of nuclear Dorsal activate genes such as short-gastrulation (sog) and thisbe (ths) in broad lateral domains across the embryo (Markstein et al. 2002; Stathopoulos et al. 2002). These genes are required for patterning the dorsal ectoderm, amnioserosa, and dorsal mesoderm (Francois et al. 1994; Stathopoulos et al. 2004).
Dorsal functions as both an activator of transcription to induce gene expression and a repressor to keep genes silenced (Jiang et al. 1992; Dubnicoff et al. 1997). The same low levels of Dorsal that activate genes in lateral regions of the embryo also mediate repression of certain targets, such as decapentaplegic (dpp), tolloid (tld), and zerknüllt (zen), thereby limiting their expression to regions where nuclear Dorsal is absent (Ip et al. 1991; Huang et al. 1993; Kirov et al. 1994). These genes are required for proper patterning of the dorsal ectoderm and amnioserosa, which develops in the dorsal-most regions of the embryo (Rushlow and Levine 1990; Ferguson and Anderson 1992b; Ferguson and Anderson 1992a).
Different levels of activated Toll direct graded Dorsal-dependent gene expression outputs. Characterization of several Toll alleles (Anderson et al. 1985) supports this view (e.g., Stathopoulos et al. 2002). Dominant mutations in the Toll gene (Toll10B), which presumably constitutively activate the receptor (Schneider et al. 1991), result in ubiquitous activation of genes such as twi and sna that are normally only expressed in ventral regions of the embryo. Concomitantly, genes such as rho, sog, or dpp are repressed. Specific recessive alleles (Tollrm9 and Tollrm10), which are presumed to result in a partially active receptor (Schneider et al. 1991), direct ubiquitous activation of lower-response genes such as rho and sog, whereas genes such as sna and dpp are absent. In the absence of Toll-mediated signaling, genes such as dpp and zen are ubiquitously expressed, as no nuclear Dorsal is present and therefore genes activated by Dorsal fail to be expressed (e.g., sna, vnd, etc.).
Perhaps the most compelling evidence that the levels of activated Toll can instruct distinct target gene expression profiles stems from ectopic expression experiments. These studies demonstrated that the DV axis can be reoriented such that genes normally expressed along the DV axis are instead expressed along the AP axis (Huang et al. 1997). This reorientation of axes was accomplished through the ectopic expression of an anteriorHIGH-posteriorLOW gradient of constitutively activated Toll receptor. This gradient of activated Toll receptor was sufficient to support differential Dorsal-target gene expression. Thus, like activin-dependent receptor signaling in Xenopus (Dyson and Gurdon 1998), it appears that cells in the early embryo are responsive to different levels of activated Toll receptor. Although the levels of nuclear Dorsal that result from differential activation of the Toll receptor have not been quantified, taken together, these results suggest that differential activation of the Toll receptor regulates the level of nuclear Dorsal along the DV circumference, which in turn determines gene expression output.
cis-Regulatory Control of DV Axis Patterning
Classical genetic screens identified 10 genes that function in the early embryo to control DV patterning (sna, twi, single-minded [sim], m8, brinker [brk], rho, sog, tld, zen, and dpp) (reviewed in Stathopoulos and Levine, 2004; Klambt et al. 1989; Bier et al. 1990; Leptin and Grunewald 1990; Nambu et al. 1990; Rushlow and Levine 1990; Kosman et al. 1991; Ferguson and Anderson 1992b; Ferguson and Anderson 1992a; Jimenez et al. 1995; Jazwinska et al. 1999). Early experiments to dissect the logic of how Dorsal controls the expression of these genes involved the use of highly laborious methods to identify the genomic sequence controlling target-gene expression. These cis-regulatory sequences are commonly defined as noncoding genomic sequences able to drive expression of a lacZ reporter in a manner similar to the endogenous patterns of genes. This requires that the reporter mimics both spatial and temporal aspects of the endogenous gene. To identify the respective cis-regulatory sequences for five of these Dorsal target genes (i.e., sna, rho, tld, dpp, and zen) “pre-genome sequencing,” genomic walks and associated promoter analysis using reporter genes were necessary (Ip et al. 1991; Jiang et al. 1991; Ip et al. 1992b; Huang et al. 1993; Kirov et al. 1994; Stathopoulos and Levine 2002a). Nevertheless, this small sampling offered the first insights into the general mechanisms used to control patterning of the DV axis by Dorsal, and these still stand today.
First, classical studies based on the analysis of cis-regulatory sequences showed that Dorsal can function as either an activator or a repressor to control gene expression along the DV axis (Jiang et al. 1993). Context-dependent interactions were proposed as the mechanism controlling whether Dorsal functions as a repressor; specifically, those resulting from cooperative interactions with DNA-binding proteins occupying associated AT-rich sequences (Kirov et al. 1993). For example, the Torso receptor tyrosine kinase signaling pathway, which is activated at the poles, selectively masks the ability of Dorsal to function as a transcriptional repressor at these positions (Rusch and Levine 1994). Torso signaling modulates the ability of Capicua (Cic), Cut, and Dri to function, proteins that influence the ability of Dorsal to function as a transcriptional repressor (Valentine et al. 1998; Jimenez et al. 2000). More recently, the activation and repression activities of Dorsal have been uncoupled, opening the way for a detailed assessment of these functions in vivo (Ratnaparkhi et al. 2006).
Combinatorial interactions between Dorsal and other transcription factors were found to be important in particular regions of the embryo. For instance, synergistic DNA binding between Dorsal and Twist, a bHLH transcription factor, permits gene expression in more lateral regions of the embryo where neither Dorsal nor Twist is capable of inducing gene expression independently (Gonzalez-Crespo and Levine 1993; Jiang and Levine 1993; Stathopoulos and Levine 2002b). Furthermore, transcriptional repressors function to refine the expression domains produced by activators. For instance, to restrict rho expression to ventrolateral stripes in the embryo, Dorsal and Twist activation is antagonized in ventral regions by the Snail repressor, resulting in the lateral stripe pattern of gene expression exhibited by these genes (Kosman et al. 1991; Ip et al. 1992a).
Lastly, the affinity of Dorsal binding sites within cis-regulatory sequences was found to influence the domain of target-gene expression (Jiang and Levine 1993). A twi cis-regulatory sequence, which normally directs expression to ventral regions of the embryo, exhibited a dorsally expanded expression domain (i.e., the ventrolateral domain) when the low affinity Dorsal binding sites were mutated to high-affinity ones.
In summary, despite the limited nature of this initial set of cis-regulatory sequences, Dorsal-mediated patterning was observed to follow three principles: (1) Dorsal functions as a context-dependent activator or repressor; (2) combinatorial regulation between Dorsal and other transcription factors affects transcriptional outputs; and (3) the binding affinity of Dorsal binding sites can influence the spatial extent of gene regulation. Following publication of the Drosophila genome, bioinformatic studies revealed these principles to be general.
THE DORSAL GENE REGULATORY NETWORK: INITIAL INSIGHTS ACQUIRED FROM THE GENOMIC SEQUENCE
The availability of the sequenced genome of Drosophila melanogaster in 2000 facilitated the analysis of Dorsal-dependent gene expression on a whole-genome scale (Adams et al. 2000). Newly developed bioinformatic approaches permitted scanning the entire genome for clusters of transcription-factor binding sites in a matter of seconds (Berman et al. 2002; Markstein et al. 2002). Furthermore, microarrays containing probes for predicted open reading frames were made commercially available and included many previously uncharacterized genes. Using a microarray-based approach, many additional Dorsal-dependent genes were identified (Stathopoulos et al. 2002), bringing the estimated total number of genes regulated by Dorsal to about 50. Surprisingly, all genes differentially expressed along the DV axis could be categorized into six basic patterns of gene expression (Fig. 2D). Before such a large-scale analysis, it was unclear, with only 10 known target genes, whether common boundaries of gene expression existed or if genes might be expressed in many different domains along the DV axis. Bioinformatic approaches rapidly identified an additional 20 cis-regulatory sequences for genes expressed along the DV axis. This increased the total fivefold from the five found by the highly laborious methods in the preceding 10-year period, and this number is constantly growing as more and more cis-regulatory sequences are characterized (reviewed in Markstein et al. 2002; Stathopoulos et al. 2002; Markstein et al. 2004; Stathopoulos and Levine 2004; Stathopoulos et al. 2004; Stathopoulos and Levine 2005b).
Classification of DV Patterns
Based on their expression patterns along the DV axis, this first set of 25 cis-regulatory sequences were categorized into four classes: ventral (Type I), ventrolateral (Type II), broad lateral (Type III+), and dorsal (Type III−) domains (Fig. 2E) (Stathopoulos and Levine 2004). Analysis of multiple Dorsal-dependent cis-regulatory sequences for coexpressed genes led to generalizations and “combinatorial codes” for which transcription factor binding sites control particular expression patterns. For instance, in addition to the presence of Dorsal sites in all the classes, the motif GCTGGYA was identified in enhancers that direct expression in a broad lateral stripe. In contrast, CACATGT and RGGNCAG motifs were identified in enhancers supporting expression in ventrolateral stripes (Stathopoulos et al. 2002; Markstein et al. 2004). The factors that bind to the GCTGGYA and RGGNCAG sites remain unidentified, whereas both Twist and Snail have been shown to bind the CACATGT site (Ip et al. 1992a).
A genome-wide computational search also identified an additional set of sequences, denoted the “TAGteam” (CAGGTAG and slight variations of this sequence), which is present in cis-regulatory elements of many genes expressed at the onset of zygotic transcription (ten Bosch et al. 2006; De Renzis et al. 2007). This sequence is recognized by a ubiquitous, maternally provided transcription factor, Zelda, and is present in several Dorsal-dependent enhancers, most especially genes of the Type III+/− classes, such as sog, dpp, and zen (Liang et al. 2008; Liberman and Stathopoulos 2009). Moreover it appears that Zelda is required for correct spatial patterning of these genes: In Zelda mutants, expression of the Type III− target genes dpp and zen disappear, whereas the Type III+ gene sog diminishes and acquires the characteristics of a Type II pattern (Liang et al. 2008).
It is interesting to note that three of the known cis-regulatory sequences—those directing expression of sim, m8, and intermediate neuroblasts defective (ind) genes—drive expression in domains that are somewhat different from any of the others (Kasai et al. 1998; Stathopoulos and Levine 2005b; Zinzen et al. 2006a). However, the enhancers for sim and m8 contain many transcription factor binding sites also found in Type II enhancers. This suggests that these responses should be similar, yet sim and m8 are only expressed in stripes a single cell wide, compared with vnd, brk, and rho, which are expressed in stripes 5–7 cells wide (Fig. 2D,E). It is likely that Notch signaling is required for activation of sim and m8, which would explain their more refined expression domain (Morel and Schweisguth 2000; Cowden and Levine 2002). Furthermore, recent analysis of the ind enhancer has suggested that this pattern is actually similar to the Type III+ responses, such as sog and ths, in terms of activation potential. However, the ind gene is refined by the action of localized activators (i.e., downstream of EGFR signaling) and repressors. Thus, to refine ind, Vnd functions in ventrolateral regions of the embryo, and an unknown repressor presumably functions in dorsal and/or dorsolateral regions of the embryo. Together, these repressors restrict ind to its lateral expression domain (Fig. 2B,D,E) (Cowden and Levine 2003; Stathopoulos and Levine 2005b).
In other words, although there are apparently six different gene-expression patterns, it seems that only three distinct activation “thresholds” exist (Types I, II, and III), and the diversity of patterns is generated by repressors or additional requirements for activation acting downstream of Dorsal (Fig. 2E). Together, these three activation thresholds delineate distinct gene-expression boundaries and thus subdivide the embryo into four domains (Types I, II, and III+/−). From this analysis and the previous studies, the following generalizations can be made (Fig. 2C,E): First, Twist and Dorsal function in a synergistic manner to regulate expression of both Type I and Type II target genes. Type II genes are distinguished from Type I genes because, generally, they are repressed by Sna and require higher-affinity Dorsal sites or stronger Dorsal/Twist synergy (Jiang and Levine 1993; Papatsenko and Levine 2005). Second, gene expression in dorsolateral regions of the embryos (Type III+) requires the function of both Dorsal and Zelda, while in dorsal regions (Type III−), Dorsal activity must be essentially absent but Zelda is still required (Liang et al. 2008; Liberman and Stathopoulos 2009). Further analysis of cis-regulatory region architecture suggested that the organization of sites relative to each other, in particular of the relationship between Dorsal and Twist transcription factors, may be important to support expression in given domains (Jiang and Levine 1993; Erives and Levine 2004; Papatsenko and Levine 2005; Zinzen et al. 2006b).
Insights into the cis-Regulatory Mechanisms Controlling Dorsal Target Gene Expression: Tiling Arrays and ChIP Techniques
Tiling arrays, in which probes are designed to span the entire genomic sequence, typically at ∼100 base-pair intervals, facilitated an assay of all sequences including the noncoding regions that were absent from previous open-reading-frame restricted arrays. In addition to presenting an unbiased platform useful for gene expression studies, the tiling arrays also allow whole-genome chromatin immunoprecipitation (ChIP) experiments (i.e., ChIP-chip) that assay in vivo occupancy of transcription factor binding.
Tiling arrays were used in expression-based screening to identify transcripts that are differentially expressed along the DV axis. Besides increasing the number of genes exhibiting Dorsal-dependent regulation from approximately 50 predicted by standard chip hybridizations (Stathopoulos et al. 2002) to approximately 100 (Biemar et al. 2006), the tiling arrays also identified specific, differentially expressed splice products (e.g., bunched) and miRNAs (mir1 and mir9a), that could not be distinguished with standard arrays containing probes designed only to assay predicted gene products.
Experiments to identify cis-regulatory regions using ChIP-chip methodology were able to identify in vivo binding site occupancy for Dorsal, Twist, and Snail transcription factors (Sandmann et al. 2007; Zeitlinger et al. 2007). Hundreds of newly predicted cis-regulatory regions were found throughout the genome, based on the assumption that occupancy of a factor on the DNA is associated with changes in gene expression observed for nearby genes. Five novel Dorsal-binding (Zeitlinger et al. 2007) and six novel Twist-binding cis-regulatory sequences (Sandmann et al. 2007) were confirmed. These studies also suggested extensive cross talk between DV and AP patterning (Fig. 3C) (Zeitlinger et al. 2007). Furthermore, other ChIP-chip analyses conducted in the early Drosophila embryo showed that the occupancy of genomic sites by most transcription factors is extensive (Li et al. 2008), the suggestion being that much of the binding identified by ChIP analyses is not necessarily required for the spatial patterns of gene expression. Future studies will be required to confirm how many occupied sites produce a functional output for each of the transcription factors in question. Nevertheless, ChIP-chip analyses represent a significant advance over pure in silico bioinformatic approaches in the identification of cis-regulatory sequences.
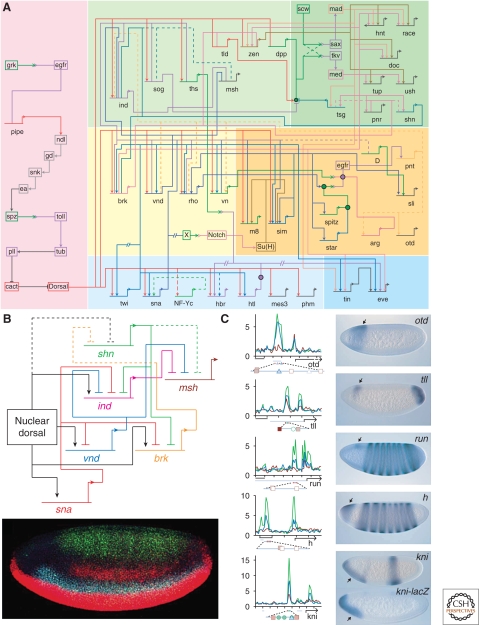
Figure 3.
Fruits of genomic approaches. (A) Dorsal gene regulatory network. The number of known Dorsal target genes increased roughly fivefold, from 10 to 50, through the use of genomic approaches. Careful study of the interactions among these genes allows for the construction of a network diagram. (Reprinted, with permission, from Levine and Davidson 2005, ©National Academy of Sciences.) (B) Network of repressors. Dorsal activity along the dorsoventral (DV) axis initiates a cascade of repressors (upper panel). Whole-mount embryo (lower panel) depicts the spatial organization of such genes by in situ hybridization using riboprobes to detect transcripts, with sna (red) on the ventral side, vnd (cyan) in the ventral neurogenic ectoderm (brk not shown), ind (dim, red) just dorsal of vnd, and schnurri (shn, green) in the dorsal ectoderm. msh (not shown) is expressed in a narrow stripe just dorsal to ind. Dashed connections are only hypothesized. It remains to be determined whether shn or another repressor functions in dorsal regions. (Image of embryo modified, with permission, from Stathopoulos and Levine 2005a, ©Elsevier.) (C) Cross talk between the Dorsal and AP patterning networks. ChIP-chip analyses with Dorsal, Snail, and Twist antibodies reveal strong binding peaks (left) for one or more of these proteins in several AP patterning genes. In situ hybridizations of reporter gene expression in whole-mount embryos (right) reveal DV asymmetries in these genes. (Reprinted, with permission, from Zeitlinger et al. 2007.)
Biological Insights into Patterning: The Gene Regulatory Network
The regulatory interactions responsible for patterning the early Drosophila embryo are summarized in a gene-regulatory network, a circuit diagram that describes all the genetic and cis-regulatory interactions uncovered to date (Fig. 3A) (Levine and Davidson 2005; Stathopoulos and Levine 2005a). The information depicted includes genetic interactions based on mutant analysis, cis-regulatory information, and ectopic expression experiments. The cis-regulatory sequences serve as a platform by which information is processed in particular cells and a decision made to either express or silence a gene. Arrows at the end of lines symbolize binding to cis-regulatory regions by activators, whereas blunt ends symbolize repressors binding to cis-regulatory regions.
Analysis of this network reveals that the Drosophila embryo uses different mechanisms to establish domains of expression. Feed-forward loops, such as the requirement of the Dorsal-target gene, Twist, to turn on the repressor Snail, function as timing mechanisms that ensure that genes are expressed in proper sequence. In addition, patterning is initiated when the embryo itself is a syncytium, an unusual aspect of Drosophila embryogenesis. The common cytoplasm of the Drosophila syncytium permits the formation of transcription-factor gradients, which can directly act on gene-expression outputs. This stands in contrast to patterning in a cellularized environment, which relies heavily on cell–cell communication coupled to signal transduction (Fig. 3B). Clearly, some signaling is active within the syncytium as Toll signaling controls nuclear import of Dorsal, but it is unclear how many other signaling pathways are also functioning. In contrast, it has been shown that several signaling pathways become active immediately following cellularization.
Differential Expression of Signaling-pathway Components Along the DV Axis Controls Patterning and Cell Movement
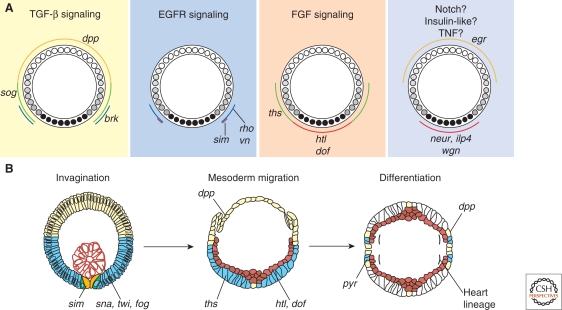
Roughly half of the estimated Dorsal target genes encode signaling molecules. These genes are essential for the localized activation of FGF (fibroblast growth factor), EGFR, and TGF (transforming growth factor)-β signaling pathways in the mesoderm, neurogenic ectoderm, and dorsal ectoderm of pregastrula embryos, respectively (Fig. 4A) (reviewed in Stathopoulos and Levine 2004). For example, localized TGF-β signaling in the dorsal ectoderm depends on Dorsal-regulated silencer elements that are associated with three target genes, dpp, tld, and zen, as well as restricted expression of sog and brk in ventrolateral regions of the embryo. Through spatial restriction of multiple signaling pathway components, Dorsal ultimately controls not only further patterning, as in the case of TGF-β, but also cell movements and subsequent differentiation events, as in the case of FGF signaling.
Figure 4.
Dorsal target genes are integral components of signaling pathways that function to control gastrulation and differentiation. (A) Dorsal regulates the activation of the TGF-β, EGFR, and FGF signaling pathways by spatial regulation of pathway components. Dorsal target genes also include components of Notch, Insulin-like, TNF, and Wnt (not shown) pathways. Whether this results in regulated activation remains to be determined. (Modified, with permission, from Stathopoulos and Levine 2004, ©Elsevier.) (B) Shown are cross sections through an embryo with the indicated expression patterns of specific genes involved in the respective processes: invagination (the invaginated mesoderm has formed a tube [red] and ectodermal cells are at the surface [blue and yellow]), mesoderm migration (the ectoderm forms a surface on which the mesoderm migrates), and differentiation of the cardiac mesoderm (dorsal somatic lineages including heart precursors are induced when the mesoderm contacts Dpp-expressing cells). Expression of ths (Blue), htl and downstream of Fgf (dof) (Red), and dpp (Yellow). (Reprinted, with permission, from Stathopoulos and Levine 2004, ©Elsevier.)
The FGF receptor (FGFR), Heartless (Htl), and its two ligands—Pyramus (Pyr) and Thisbe (Ths)—are all Dorsal targets (Stathopoulos et al. 2004). Htl is one of only two FGFRs present in Drosophila (reviewed in Szebenyi and Fallon 1999), and controls mesoderm migration during gastrulation (Beiman et al. 1996; Gisselbrecht et al. 1996). htl expression requires peak levels of nuclear Dorsal and/or Twist (Figs. 2D and 4A) (Stathopoulos et al. 2004). pyr and ths were first discovered in microarray screens designed to identify genes expressed along DV axis (Stathopoulos et al. 2002). The pyr and ths genes, which are linked on the genome, encode related FGF ligands and show dynamic patterns of gene expression in the embryo (Stathopoulos et al. 2004; Gryzik and Müller 2004). The early expression of the ths gene is Dorsal-dependent and it is induced by even the lowest levels of nuclear Dorsal present in dorsolateral regions of the embryo (Figs. 2D and 4A) (Stathopoulos et al. 2004). After internalization of presumptive mesoderm cells (i.e., invagination), cells containing the FGFR are free to contact cells expressing the ligands, enabling activation of the FGF signaling pathway.
In general, cell movements are orchestrated during gastrulation through the action of signaling pathways, which become active after cellularization. Twist and Snail transcription factors play an instrumental role in controlling invagination and ventral furrow formation by regulated expression of the genes folded-gastrulation, concertina, and T48 (Parks and Wieschaus 1991; Dawes-Hoang et al. 2005; Sandmann et al. 2007). This results in an epithelial-to-mesenchymal transition (EMT) of the presumptive mesodermal cells located in the interior of the embryo. These cells then proceed to migrate along the ectodermal cells toward dorsal regions of the embryo (Fig. 4B). This migration is required for subsequent differentiation of the mesoderm into distinct cell types such as cardiac, dorsal somatic, and visceral mesoderm cell lineages. FGF signaling likely functions at multiple steps during the migration to control the directed movement of cells (McMahon et al. 2008; Kadam et al. 2009). As a result of this coordinated migration, once mesoderm cells reach the dorsal ectoderm, multiple signaling inputs control which differentiation programs are adopted. For instance, in addition to FGF signaling, TGF-β signaling is also required to induce mesoderm cells that reach the dorsal-most regions of the ectoderm to adopt cardial and dorsal somatic mesoderm lineage choices (Fig. 4B).
CONCLUDING REMARKS
Much work has been focused on the study of the Dorsal patterning network. Classical genetic studies have uncovered several Dorsal target genes, as well as many of the components of the upstream pathway. Conventional promoter analysis methods using reporter genes have revealed the inputs and minimal enhancers that regulate several Dorsal target genes. Genome-level studies have increased this knowledge fivefold, from the standpoint of numbers of target genes and numbers of enhancers known. ChIP-chip studies have uncovered multiple enhancer sites and cross talk between AP and DV systems. All of these data together reveal several layers of regulation in this system, including multiple inputs to each of the Dorsal target genes (see Figs. 2C and 3B).
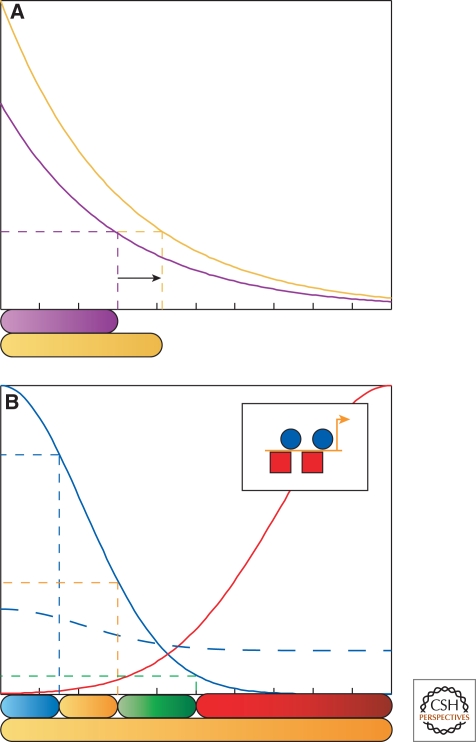
The existence of this regulation is not surprising, as physical arguments show that a lone morphogen is not sufficient to explain developmental patterning (Lander 2007; Jaeger et al. 2008). In particular, a single-morphogen model fails to account for the remarkable insensitivity of developmental patterns with respect to many natural and experimental perturbations to the system (see Fig. 5A). Therefore, any viable model of morphogen-mediated patterning must include multiple levels of regulation on target gene expression (see Figs. 2 and 3).
Figure 5.
Revisiting the morphogen gradient model. (A) Depiction of classical morphogen gradient (purple). Horizontal dashed line denotes the concentration threshold that defines one cell fate boundary (shown below the graph). Gold curve denotes the same morphogen simulated with a 50% increase in morphogen production. The classical morphogen model predicts that the cell fate boundary, in response, will also shift by roughly 50% (arrow). This is an unacceptably sensitive system, and does not comport with experimental evidence. (B) Illustration of the affect of multiple inputs to cis-regulatory modules. In this hypothetical tissue, a primary morphogen (blue) initiates expression of several target genes (blue, orange, green, red) within the tissue. A secondary morphogen (red) is expressed in the red cells, but is repressed in the rest of the tissue by the primary morphogen, and acts as a repressor to other target genes (Inset). Consider a case in which the primary morphogen is present in a shallow gradient (dotted blue), at a concentration above green threshold, yet below the orange threshold, so that no secondary morphogen is present. The classical morphogen gradient model would predict all cells to turn green. However, because the secondary morphogen also serves as input to the target genes (and is not present in this case), it is possible that instead the orange gene is ubiquitously expressed.
However, this is simply a necessary condition for developmental reliability. It remains to be seen whether the abundance of multiple inputs and feedback regulation seen in the Dorsal patterning system, and in morphogen systems in general, is sufficient to explain the exquisite precision of threshold outputs and the remarkable reliability seen in tissue patterning. Systems-level analyses of patterning mechanisms present an opportunity to address this question.
For example, tissue-level modeling has shown that the robustness of a pattern can generally be improved with negative feedback regulation (Eldar et al. 2003; Reeves et al. 2005). In the Dorsal system, negative feedback occurs through zygotic expression of WntD (Ganguly et al. 2005; Gordon et al. 2005), and perhaps Cactus as well (Araujo and Bier 2000). In another example involving the Bicoid patterning system, a recent study of gradient interpretation directly shows that absolute concentration thresholds of Bicoid are not the only determinate factors in this system, likely because there are multiple spatially dependent inputs to Bicoid target genes (see hypothetical scenario in Fig. 5B) (Ochoa-Espinosa et al. 2009). Is this also true for the Dorsal patterning system? This is likely to be the case in light of the current evidence, which shows that multiple inputs exist to control the expression of genes along the DV axis (Figs. 2 and 3) and that compensatory mechanisms may support expression in mutant backgrounds that show decreased Dorsal levels (Stathopoulos and Levine 2002b). This would imply that the absolute levels of Dorsal are less important than the sum total of several factors present within nuclei, integrated in a complex manner at each individual enhancer site (see Fig. 5B). Thus, the key to differential gene expression is combinatorial regulation through the interaction of multiple transcription factors. This underscores the need for a thorough understanding of the network of gene regulatory interactions that supports expression of genes.
Answering systems-level questions regarding the formation and effects of morphogen gradients will require integrating data from a wide variety of time, length, and concentration scales, ranging from the cis-regulatory code to tissue-wide signaling interactions. Subfields of traditional and post-genomic experimental biology, quantitative microscopy, and computational biology will each exert significant influence on future studies of morphogen gradients.
ACKNOWLEDGMENTS
This work was supported by grants to AS from the NIGMS (R01-GM077668) and the Searle Scholar Foundation. GTR is a fellow of The Jane Coffin Childs Memorial Fund for Medical Research, and was supported by a grant from The Jane Coffin Childs Memorial Fellowship for Medical Research.
Footnotes
Editors: James Briscoe, Peter Lawrence, and Jean-Paul Vincent
Additional Perspectives on Generation and Interpretation of Morphogen Gradients available at www.cshperspectives.org
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195 [DOI] [PubMed] [Google Scholar]
- Anderson KV 1998. Pinning down positional information: dorsal-ventral polarity in the Drosophila embryo. Cell 95:439–442 [DOI] [PubMed] [Google Scholar]
- Anderson KV, Jurgens G, Nusslein-Volhard C 1985. Establishment of dorsal-ventral polarity in the Drosophila embryo: Genetic studies on the role of the Toll gene product. Cell 42:779–789 [DOI] [PubMed] [Google Scholar]
- Araujo H, Bier E 2000. sog and dpp exert opposing maternal functions to modify toll signaling and pattern the dorsoventral axis of the Drosophila embryo. Development 127:3631–3644 [DOI] [PubMed] [Google Scholar]
- Beiman M, Shilo BZ, Volk T 1996. Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev 10:2993–3002 [DOI] [PubMed] [Google Scholar]
- Belvin MP, Anderson KV 1996. A conserved signaling pathway: The Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol 12:393–416 [DOI] [PubMed] [Google Scholar]
- Belvin MP, Jin Y, Anderson KV 1995. Cactus protein degradation mediates Drosophila dorsal-ventral signaling. Genes Dev 9:783–793 [DOI] [PubMed] [Google Scholar]
- Bergmann A, Stein D, Geisler R, Hagenmaier S, Schmid B, Fernandez N, Schnell B, Nusslein-Volhard C 1996. A gradient of cytoplasmic Cactus degradation establishes the nuclear localization gradient of the dorsal morphogen in Drosophila. Mech Dev 60:109–123 [DOI] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci 99:757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemar F, Nix DA, Piel J, Peterson B, Ronshaugen M, Sementchenko V, Bell I, Manak JR, Levine MS 2006. Comprehensive identification of Drosophila dorsal-ventral patterning genes using a whole-genome tiling array. Proc Natl Acad Sci 103:12763–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Jan LY, Jan YN 1990. Rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev 4:190–203 [DOI] [PubMed] [Google Scholar]
- Chen LY, Wang JC, Hyvert Y, Lin HP, Perrimon N, Imler JL, Hsu JC 2006. Weckle is a zinc finger adaptor of the toll pathway in dorsoventral patterning of the Drosophila embryo. Curr Biol 16:1183–1193 [DOI] [PubMed] [Google Scholar]
- Cowden J, Levine M 2002. The Snail repressor positions Notch signaling in the Drosophila embryo. Development 129:1785–1793 [DOI] [PubMed] [Google Scholar]
- Cowden J, Levine M 2003. Ventral dominance governs sequential patterns of gene expression across the dorsal-ventral axis of the neuroectoderm in the Drosophila embryo. Dev Biol 262:335–349 [DOI] [PubMed] [Google Scholar]
- Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Wieschaus EF 2005. Folded gastrulation, cell shape change and the control of myosin localization. Development 132:4165–4178 [DOI] [PubMed] [Google Scholar]
- DeLotto R, DeLotto Y, Steward R, Lippincott-Schwartz J 2007. Nucleocytoplasmic shuttling mediates the dynamic maintenance of nuclear Dorsal levels during Drosophila embryogenesis. Development 134:4233–4241 [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF 2007. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLos Biol 5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ 1997. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev 11:2952–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson S, Gurdon JB 1998. The interpretation of position in a morphogen gradient as revealed by occupancy of activin receptors. Cell 93:557–568 [DOI] [PubMed] [Google Scholar]
- Eldar A, Rosin D, Shilo BZ, Barkai N 2003. Self-enhanced ligand degradation underlies robustness of morphogen gradients. Dev Cell 5:635–646 [DOI] [PubMed] [Google Scholar]
- Erives A, Levine M 2004. Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci 101:3851–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Anderson KV 1992a. Decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell 71:451–461 [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Anderson KV 1992b. Localized enhancement and repression of the activity of the TGF-beta family member, decapentaplegic, is necessary for dorsal-ventral pattern formation in the Drosophila embryo. Development 114:583–597 [DOI] [PubMed] [Google Scholar]
- Francois V, Solloway M, O’Neill JW, Emery J, Bier E 1994. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev 8:2602–2616 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Jiang J, Ip YT 2005. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development 132:3419–3429 [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM 1996. heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev 10:3003–3017 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crespo S, Levine M 1993. Interactions between dorsal and helix-loop-helix proteins initiate the differentiation of the embryonic mesoderm and neuroectoderm in Drosophila. Genes Dev 7:1703–1713 [DOI] [PubMed] [Google Scholar]
- Gordon MD, Dionne MS, Schneider DS, Nusse R 2005. WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature 437:746–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans J, Schnorrer F, Nusslein-Volhard C 1999. Oligomerisation of Tube and Pelle leads to nuclear localisation of Dorsal. Mech Dev 81:127–138 [DOI] [PubMed] [Google Scholar]
- Gryzik T, Müller HJ 2004. FGF8-like1 and FGF8-like2 encode putative ligands of the FGF receptor Htl and are required for mesoderm migration in the Drosophila gastrula. Curr Biol 14:659–667 [DOI] [PubMed] [Google Scholar]
- Hashimoto C, Kim DR, Weiss LA, Miller JW, Morisato D 2003. Spatial regulation of developmental signaling by a serpin. Dev Cell 5:945–950 [DOI] [PubMed] [Google Scholar]
- Hecht PM, Anderson KV 1993. Genetic characterization of tube and pelle, genes required for signaling between Toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embryo. Genetics 135:405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AM, Rusch J, Levine M 1997. An anteroposterior Dorsal gradient in the Drosophila embryo. Genes Dev 11:1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JD, Schwyter DH, Shirokawa JM, Courey AJ 1993. The interplay between multiple enhancer and silencer elements defines the pattern of decapentaplegic expression. Genes Dev 7:694–704 [DOI] [PubMed] [Google Scholar]
- Ip YT, Kraut R, Levine M, Rushlow CA 1991. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell 64:439–446 [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M 1992a. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev 6:1728–1739 [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M 1992b. Dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 6:1518–1530 [DOI] [PubMed] [Google Scholar]
- Jaeger J, Irons D, Monk N 2008. Regulative feedback in pattern formation: Towards a general relativistic theory of positional information. Development 135:3175–3183 [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C 1999. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96:563–573 [DOI] [PubMed] [Google Scholar]
- Jiang J, Levine M 1993. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72:741–752 [DOI] [PubMed] [Google Scholar]
- Jiang J, Kosman D, Ip YT, Levine M 1991. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev 5:1881–1891 [DOI] [PubMed] [Google Scholar]
- Jiang J, Rushlow CA, Zhou Q, Small S, Levine M 1992. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J 11:3147–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Cai H, Zhou Q, Levine M 1993. Conversion of a dorsal-dependent silencer into an enhancer: Evidence for dorsal corepressors. EMBO J 12:3201–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez F, Martin-Morris LE, Velasco L, Chu H, Sierra J, Rosen DR, White K 1995. vnd, a gene required for early neurogenesis of Drosophila, encodes a homeodomain protein. EMBO J 14:3487–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Guichet A, Ephrussi A, Casanova J 2000. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 14:224–231 [PMC free article] [PubMed] [Google Scholar]
- Kadam S, McMahon A, Tzou P, Payne S, Stathopoulos A 2009. FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development 136:739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y, Stahl S, Crews S 1998. Specification of the Drosophila CNS midline cell lineage: Direct control of single-minded transcription by dorsal/ventral patterning genes. Gene Expr 7:171–189 [PMC free article] [PubMed] [Google Scholar]
- Kirov N, Zhelnin L, Shah J, Rushlow C 1993. Conversion of a silencer into an enhancer: Evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J 12:3193–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov N, Childs S, O’Connor M, Rushlow C 1994. The Drosophila dorsal morphogen represses the tolloid gene by interacting with a silencer element. Mol Cell Biol 14:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt C, Knust E, Tietze K, Campos-Ortega JA 1989. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J 8:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Ip YT, Levine M, Arora K 1991. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 254:118–122 [DOI] [PubMed] [Google Scholar]
- Lander AD 2007. Morpheus unbound: reimagining the morphogen gradient. Cell 128:245–256 [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Tan Y-Q, Hashimoto C 2001. Activation of a protease cascade involved in patterning the Drosophila embryo. Proc Natl Acad Sci 98:5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M, Grunewald B 1990. Cell shape changes during gastrulation in Drosophila. Development 110:73–84 [DOI] [PubMed] [Google Scholar]
- Levine M, Davidson EH 2005. Gene regulatory networks for development. Proc Natl Acad Sci 102:4936–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456:400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Stathopoulos A 2009. Design Flexibility in cis-Regulatory Control of Gene Expression: Synthetic and Comparative Evidence. Dev Biol 327: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P, Roth S, Reichhart JM 2003. A serpin regulates dorsal-ventral axis formation in the Drosophila embryo. Curr Biol 13:2097–2102 [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci 99:763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M 2004. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131:2387–2394 [DOI] [PubMed] [Google Scholar]
- McMahon A, Supatto W, Fraser SE, Stathopoulos A 2008. Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science 322:1546–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel V, Schweisguth F 2000. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev 14:377–388 [PMC free article] [PubMed] [Google Scholar]
- Morisato D 2001. Spatzle regulates the shape of the Dorsal gradient in the Drosophila embryo. Development 128:2309–2319 [DOI] [PubMed] [Google Scholar]
- Moussian B, Roth S 2005. Dorsoventral axis formation in the Drosophila embryo–shaping and transducing a morphogen gradient. Curr Biol 15:887–899 [DOI] [PubMed] [Google Scholar]
- Nambu JR, Franks RG, Hu S, Crews ST 1990. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63:63–75 [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsigiros A, Struffi P, Small S 2009. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci 106: 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Huang JD, Coorey AJ 1991. Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev 5:1892–1901 [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Levine M 2005. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc Natl Acad Sci 102:4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks S, Wieschaus E 1991. The Drosophila gastrulation gene concertina encodes a G alpha-like protein. Cell 64:447–458 [DOI] [PubMed] [Google Scholar]
- Peri F, Technau M, Roth S 2002. Mechanisms of Gurken-dependent pipe regulation and the robustness of dorsoventral patterning in Drosophila. Development 129:2965–2975 [DOI] [PubMed] [Google Scholar]
- Ratnaparkhi GS, Jia S, Courey AJ 2006. Uncoupling dorsal-mediated activation from dorsal-mediated repression in the Drosophila embryo. Development 133:4409–4414 [DOI] [PubMed] [Google Scholar]
- Ray RP, Arora K, Nusslein-Volhard C, Gelbart WM 1991. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development 113:35–54 [DOI] [PubMed] [Google Scholar]
- Reeves GT, Kalifa R, Klein DE, Lemmon MA, Shvartsman SY 2005. Computational analysis of EGFR inhibition by Argos. Dev Biol 284:523–535 [DOI] [PubMed] [Google Scholar]
- Roth S 1993. Mechanisms of dorsal-ventral axis determination in Drosophila embryos revealed by cytoplasmic transplantations. Development 117:1385–1396 [DOI] [PubMed] [Google Scholar]
- Rusch J, Levine M 1994. Regulation of the dorsal morphogen by the Toll and torso signaling pathways: A receptor tyrosine kinase selectively masks transcriptional repression. Genes Dev 8:1247–1257 [DOI] [PubMed] [Google Scholar]
- Rushlow C, Levine M 1990. Role of the zerknullt gene in dorsal-ventral pattern formation in Drosophila. Adv Genet 27:277–307 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE 2007. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev 21:436–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DS, Hudson KL, Lin TY, Anderson KV 1991. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev 5:797–807 [DOI] [PubMed] [Google Scholar]
- Schupbach T 1987. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell 49:699–707 [DOI] [PubMed] [Google Scholar]
- Sen J, Goltz JS, Stevens L, Stein D 1998. Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal-ventral polarity. Cell 95:471–481 [DOI] [PubMed] [Google Scholar]
- Simpson P 1983. Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics 105:615–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, DeLotto R 1994. Ventralizing signal determined by protease activation in Drosophila embryogenesis. Nature 368:548–551 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2002a. Dorsal gradient networks in the Drosophila embryo. Dev Biol 246:57–67 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2002b. Linear signaling in the Toll-Dorsal pathway of Drosophila: activated Pelle kinase specifies all threshold outputs of gene expression while the bHLH protein Twist specifies a subset. Development 129:3411–3419 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2004. Whole-genome analysis of Drosophila gastrulation. Curr Opin Genet Dev 14:477–484 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2005a. Genomic regulatory networks and animal development. Dev Cell 9:449–462 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2005b. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev Biol 280:482–493 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M 2002. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111:687–701 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M 2004. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev 18:687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Towb P, Chiem DN, Foster BA, Wasserman SA 2004. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. EMBO J 23:100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Fallon JF 1999. Fibroblast growth factors as multifunctional signaling factors. Int Review Cytol 185:45–106 [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW 2006. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133:1967–1977 [DOI] [PubMed] [Google Scholar]
- Thisse B, Stoetzel C, El Messal M, Perrin-Schmitt F 1987. Genes of the Drosophila maternal dorsal group control the specific expression of the zygotic gene twist in presumptive mesodermal cells. Genes Dev 5:1285–1298 [Google Scholar]
- Valentine SA, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey AJ 1998. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol 18:6584–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M 2007. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21:385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Sen J, Stevens L, Goltz JS, Stein D 2005. Drosophila pipe protein activity in the ovary and the embryonic salivary gland does not require heparan sulfate glycosaminoglycans. Development 132:3813–3822 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Cande J, Ronshaugen M, Papatsenko D, Levine M 2006a. Evolution of the ventral midline in insect embryos. Dev Cell 11:895–902 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Senger K, Levine M, Papatsenko D 2006b. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol 16:1358–1365 [DOI] [PubMed] [Google Scholar]