Abstract
Nuclear factor-κB (NF-κB) is a pleiotropic mediator of inducible and specific gene regulation involving diverse biological activities including immune response, inflammation, cell proliferation, and death. The fine-tuning of the NF-κB DNA binding activity is essential for its fundamental function as a transcription factor. An increasing body of literature illustrates that this process can be elegantly and specifically controlled at multiple levels by different protein subsets. In particular, the recent identification of a non-Rel subunit of NF-κB itself provides a new way to understand the selective high-affinity DNA binding specificity of NF-κB conferred by a synergistic interaction within the whole complex. Here, we review the mechanism of the specification of DNA binding activity of NF-κB complexes, one of the most important aspects of NF-κB transcriptional control.
The NF-κB transcription factor regulates a variety of processes, including immune responses and cell death. “Specifier” proteins, such as RPS3, may fine-tune its DNA binding.
Nuclear factor-κB (NF-κB), a collective term for a family of transcription factors, was originally detected as a transcription-enhancing, DNA-binding complex governing the immunoglobulin (Ig) light chain gene intronic enhancer (Sen and Baltimore 1986; Lenardo et al. 1987). NF-κB is evolutionarily and structurally conserved and has representative members in a wide range of species. In essentially all unstimulated nucleated cells, NF-κB complexes are retained in latent cytoplasmic form through binding to a member of the inhibitor of NF-κB (IκB) proteins (Lenardo and Baltimore 1989; Lenardo et al. 1989; Hayden and Ghosh 2004; Hayden and Ghosh 2008). NF-κB induction typically occurs following the activation of the IκB kinase (IKK) signalosome, resulting in the phosphorylation and subsequent dispatch of the inhibitory IκBs to the proteasome for protein degradation (Hacker and Karin 2006). This cytoplasmic “switch” liberates NF-κB complexes for subsequent nuclear translocation and target gene transcription (Scheidereit 2006). It provides a pre-established genetic switch that is independent of new protein synthesis and triggered by a biochemical change in the cell. This adaptability and versatility no doubt underlies its broad use. A diverse spectrum of modulating stimuli can activate this pleiotropic transcription factor; furthermore, the fundamental use of NF-κB has been highlighted with an ever-increasing array of genetic targets, responsible for diverse biological activities including immune response, inflammation, cell proliferation, and death (Grilli et al. 1993) (also see http://www.nf-kb.org).
The best known subunits of mammalian NF-κB consist of five proteins in the Rel family: RelA (p65), RelB, c-Rel, p50, and p52, which are capable forming homo- and hetero-dimeric complexes in almost any combination (Hayden and Ghosh 2004). Each of these subunits harbors a prototypical amino-terminal sequence of roughly 300 amino acids, termed the Rel homology domain (RHD), that mediates dimerization, DNA-binding, nuclear localization, and cytoplasmic retention by IκBs (Rothwarf and Karin 1999; Chen and Greene 2004). In contrast, the transcription activation domain (TAD) necessary for the target gene expression is present only in the carboxyl terminus of p65, c-Rel, and RelB subunits. NF-κB complexes have long been thought to function dimerically; but functional and biochemical information belied this simple conceptualization. The native complex of NF-κB from nuclear extracts is more than 200 kDa, significantly higher than that reconstituted from purified p50 and p65 proteins (115 kDa) (Urban et al. 1991). Moreover, native NF-κB complexes have a >100-fold higher affinity for Ig κB motif DNA than reconstituted p65–p50 heterodimers (Phelps et al. 2000). A new study shows that another essential subunit of NF-κB complex, ribosomal protein S3 (RPS3), cooperates with Rel dimers to achieve full binding and transcriptional activity (Wan et al. 2007). As an integral component, RPS3 plays a critical role in determining the DNA binding affinity and specificity of NF-κB, which will be discussed in more detail in the following discussion (Wan et al. 2007). Therefore, the molecular machine known as NF-κB consists of both Rel and non-Rel subunits that actually comprise multiple protein complexes with different gene activation specificities, masquerading as a single NF-κB complex in the nucleus.
NF-κB exerts its fundamental role as transcription factor by binding to variations of the consensus DNA sequence of 5′-GGGRNYY YCC-3′ (in which R is a purine, Y is a pyrimidine, and N is any nucleotide) known as κB sites (Chen et al. 1998). How NF-κB selectively recognizes a small subset of relevant κB sites from the large excess of potential binding sites (about 1.4×104 estimated in human genome) is a critical step for stimulus-specific gene transcription. Increasing evidence suggests that specific chromatin modifications and configurations are required for NF-κB proteins to access the chromosomally embedded cognate κB motifs (Natoli et al. 2005; Natoli 2006). The presence of κB sites, however, appears to be a minimal requirement for NF-κB regulation but not sufficient for gene induction (Wan et al. 2007). We will attempt to decipher the elegant but recondite control of DNA binding activity of NF-κB proteins at multiple levels, which is one of the most important, yet complex, aspects of NF-κB function. This process, more abstruse than initially considered, involves IκBs, Rel subunits, Rel-associating proteins, and non-Rel subunits in both the cytoplasm and the nucleus, as well as complicated associations in the nuclear chromatin. This synopsis will highlight our current knowledge of the DNA binding activity of NF-κB complex, focusing on its liberation from cytoplasmic sequestration complexes to recruitment to cognate κB regulatory sites in the genome.
IκB FAMILY PROTEINS
The evidence that induction of NF-κB activity does not require new protein synthesis, and that the detergent deoxycholate liberated active κB-site DNA binding activity in cytosolic extracts from unstimulated cells, revealed a key property of this gene regulatory system (Baeuerle and Baltimore 1988). This established a new regulatory paradigm involving the specific and reversible DNA binding of NF-κB proteins governed by NF-κB inhibitor protein(s), known as IκB. The IκB protein family currently consists of eight members: IκBα, IκBβ, IκBγ, IκBε, IκBζ, Bcl-3, and the Rel protein precursors p105 and p100, all of which possess a characteristic structural feature of ankyrin repeats. The centerpiece of both classical and alternative NF-κB pathways is the IκBs, functioning at the primary level to regulate the DNA binding activity of NF-κB proteins.
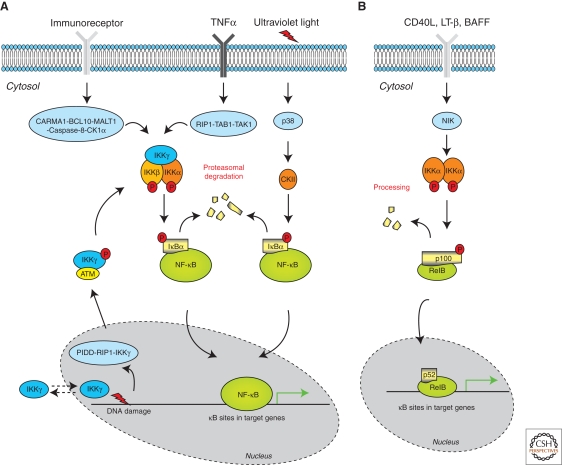
Specific modification and subsequent degradation of IκBs is critical in regulating NF-κB DNA binding activity, particularly in the classic or canonical pathway (Table 1). In unstimulated cells, NF-κB complexes are sequestered with various binding preferences by the IκBs in the cytoplasm because IκBs physically mask the nuclear localization signal (NLS) of NF-κB. On stimulation, whether intra- or extracellular, multiple intracellular signaling pathways converge on a tripartite IκB kinase (IKK) complex consisting of two functionally nonredundant kinases IKKα and IKKβ, as well as a regulatory subunit IKKγ (Fig. 1A). These mediate the phosphorylation of IκBs at specific amino acid residues (for instance, Ser-32 and Ser-36 in IκBα) predominantly through the action of the IKKβ subunit in the activated IKK complex. These phosphorylation events are a prerequisite for their successive ubiquitinylation by β-transducin repeat-containing protein (βTrCP) and degradation in the 26S proteasome. The removal of IκBs liberates NF-κB for nuclear translocation and binding to cognate sites in target genes.
Table 1.
Post-translational modification of IκB family proteins
| IκB family proteins | Target residues | Enzymes | References |
| Phosphorylation | |||
| IκBα | S32 and S36 | IKKβ | (Hayden and Ghosh 2004) |
| IκBα | S283, S289, T291, S293, and T299 | CKII | (Lin et al. 1996; McElhinny et al. 1996; Schwarz et al. 1996) |
| IκBα | Y42 | p56-lck, Syk, and c-Src | (Koong et al. 1994) |
| Y42 | Unknown | (Schoonbroodt et al. 2000) | |
| Y42 and Y305 | Unknown | (Waris et al. 2003) | |
| IκBα | Unknown | PI3K/Akt | (Sizemore et al. 1999) |
| IκBβ | S19 and S23 | IKKβ | (Hayden and Ghosh 2004) |
| IκBβ | S313 and S315 | CKII | (Chu et al. 1996) |
| IκBε | S18 and S22 | IKKβ | (Hayden and Ghosh 2004) |
| Bcl-3 | S394 and S398 | GSK3β | (Viatour et al. 2004) |
| p100 | S99, S108, S115, S123, S866, S870, and S872 | IKKα | (Senftleben et al. 2001; Xiao et al. 2001; Xiao et al. 2004) |
| p105 | S927 and S932 | IKKβ | (Lang et al. 2003) |
| p105 | S903 and S907 | GSK3β | (Demarchi et al. 2003) |
| p105 | S337 | PKAc | (Hou et al. 2003) |
| Ubiquitination | |||
| IκBα | K21 and K22 | βTrCP | (Hayden and Ghosh 2004) |
| IκBβ | K6 | βTrCP | (Hayden and Ghosh 2004) |
| IκBε | K6 | βTrCP | (Hayden and Ghosh 2004) |
| p100 | K856 | βTrCP | (Amir et al. 2004) |
| p105 | Multiple Ks | βTrCP | (Cohen et al. 2004) |
| Sumoylation | |||
| IκBα | K21 | Unknown | (Desterro et al. 1998) |
| p100 | K90, K298, K689, and K863 | Ubc9 | (Vatsyayan et al. 2008) |
| Acetylation | |||
| p100 | Unknown | P300 | (Hu and Colburn 2005; Deng et al. 2006) |
| p105 | K431, K440, and K441 | P300 | (Furia et al. 2002; Deng and Wu 2003) |
| S-nitrosylation | |||
| p105 | C62 | (Matthews et al. 1996) |
IKKβ, IκB kinase β; CKII, Casein kinase II; p56-lck, lymphocyte-specific protein tyrosine kinase; Syk, spleen tyrosine kinase; c-Src, normal cellular Src kinase; PI3K, phosphoinositide 3-kinases; GSK3β, glycogen synthase kinase 3 β; IKKα, IκB kinase α; PKAc, catalytic subunit of protein kinase A; βTrCP, β-transducin repeat-containing protein; Ubc9, E2 small ubiquitin-like modifier (SUMO)-conjugating enzyme Ubc9; p300, E1A binding protein p300.
Figure 1.
A schematic representation of how NF-κB DNA binding is regulated by IκB degradation and processing following activating stimuli. (A) IκB degradation mechanism that depends on site-specific phosphorylation of IκBs mediated by activated IKKβ in the IKK complex as well as CKII. Unique intracellular signaling complexes (which contains more components as shown, e.g., CARMA1-BCL10-MALT1-Caspase-8-CK1α complex downstream of T cell receptor [TCR] ligation [Bidere et al. 2009]) phosphorylate and activate the IKK complex depending on the initial stimulus (e.g., TNF, immunoreceptors, and DNA damage). IKKβ phosphorylates serine residues located in the amino-terminus of IκBs following TNF and TCR stimulation. In contrast, NF-κB induction by ultraviolet light and select chemotherapeutic agents requires the MAP kinase p38 and CKII. CKII phosphorylates serine residues located in the carboxyl terminus of IκBs. Phosphorylation of IκBs leads to successive ubiquitinylation and proteasomal degradation. The removal of IκBs liberates NF-κB complexes for nuclear translocation and binding to cognate regions in target genes. (B) The alternative pathway, which is induced by a few stimuli including CD40L, LT-β, and BAFF. NIK and IKKα are critical kinases for the phosphorylation of p100 at regulatory serine residues within its carboxyl terminus. This causes proteasomal degradation of the carboxyl terminus of the p100 molecule, generating a DNA binding NF-κB complex containing RelB and processed p52.
Additionally, various kinases other than the IKK complex may also mediate site-specific modification of IκBs, contributing to the diversity of signals regulating the latent DNA binding activity of NF-κB. In ultraviolet (UV) light-induced NF-κB activation, the phosphorylation-dependent IκBα degradation process does not rely on the IKK complex but the alternative serine/threonine kinase, casein kinase II (CKII) (Fig. 1A). Several studies have shown that CKII phosphorylates IκBα at a cluster of serine and threonine residues in the carboxyl terminus through a p38-MAPK kinase-dependent pathway, which is critical for degradation of IκBα and induced NF-κB DNA binding activity in cells exposed to UV light (Lin et al. 1996; McElhinny et al. 1996; Schwarz et al. 1996). Moreover, tyrosine kinases p56-lck, Syk, and c-Src and the serine/threonine kinase GSK3β have also been reported to phosphorylate IκBs and control the DNA binding activity of NF-κB (Table 1) (Koong et al. 1994; Viatour et al. 2004). Along with phosphorylation, modifications such as sumoylation, acetylation, and s-nitrosylation of the IκBs at specific residues can also trigger the degradation process (Table 1). However, these modifications lead to the final common pathway of ubiquitination of IκBs at specific lysine residues as the essential requirement for IκB degradation and release of NF-κB for nuclear translocation (Table 1).
p100 and p105, the precursor proteins for NF-κB subunits p52 and p50 respectively, contain multiple ankyrin repeats (similar to those of the IκB family) in their carboxy-terminal domains. In some circumstances, these keep NF-κB complexes cytoplasmically anchored. The regulation of NF-κB DNA binding activity by p105 mirrors IκBs of the classical pathway by undergoing inducible, complete degradation, particularly when it is bound to NF-κB complexes (Heissmeyer et al. 2001). By contrast, partial processing, but not full degradation, of p100 represents another mechanism by which IκB regulates the DNA binding activity of NF-κB proteins. This was discovered recently in an alternative pathway of NF-κB regulation involving RelB and p52 (Fig. 1B) (Hayden and Ghosh 2004). A few select stimuli including CD40 ligand (CD40L), lymphotoxin β (LT-β), and B-cell activating factor (BAFF) phosphorylate p100 on carboxy-terminal serine residues, triggering successive polyubiquitination and proteasomal degradation of the carboxyl terminus of the p100 molecule. The resulting complex, containing RelB and processed p52, functions as the main NF-κB species with selected DNA binding activity in the alternative pathway. Of interest, the kinase signalosome mediating p100 phosphorylation contains the NF-κB-inducing kinase (NIK) and IKKα, and functions completely independently of IKKβ and IKKγ. Moreover, recent evidence suggests that IKKα phosphorylates p100 at several serine residues within the RHD of p52, and these modifications can augment p52 DNA binding, in addition to inducing the efficient processing of p105 (Xiao et al. 2004).
Besides their hallmark sequestration function in cytoplasm, IκBs can also regulate the DNA binding properties of NF-κB proteins in the nucleus. IκBζ localizes to the nucleus following its inducible expression (Yamamoto et al. 2004) and Bcl-3 is also found in the nucleus associated with p50- and p52-containing NF-κB complexes (Cogswell et al. 2000). IκBs such as IκBα, IκBβ, and IκBε are transcriptional targets of NF-κB itself, creating a negative feedback loop. This facilitates restoration of latent NF-κB complexes in the cytoplasm and maintains the cell's responsiveness to subsequent stimuli. With variant kinetics of degradation and resynthesis, IκBs compete NF-κB complexes off their chromosomal locations and export them back into cytoplasm. By affecting the cytoplasmic–nuclear shuttling of NF-κB proteins, these titrate the occupancy of gene regulatory elements in nuclear DNA. Strikingly, IκBβ may also function to directly regulate NF-κB DNA binding activity on relevant promoters or enhancers, as evident by its stable nuclear association with NF-κB complexes that are already bound to κB sites in chromatin (Thompson et al. 1995; Suyang et al. 1996). Moreover, both IκBζ and Bcl-3 primarily interact with specific NF-κB complexes and subvert their DNA binding activity. IκBζ was reported to associate preferentially with p50 homodimers and also negatively regulate p65-containing NF-κB complexes (Kang et al. 1992; Motoyama et al. 2005; Hayden and Ghosh 2008), which suggests it may possess the capability to selectively inhibit or activate specific NF-κB species. However, the role of Bcl-3 in regulating DNA binding seems controversial. It has been proposed to remove the repressive p50 homodimers from κB DNA, allowing transcriptionally active NF-κB species access to those cognate elements (Lenardo and Siebenlist 1994; Hayden and Ghosh 2008). However, recent evidence indicates that Bcl-3 might also stabilize p50 homodimers and inhibit NF-κB activation by preventing transcriptionally active NF-κB complexes from binding to κB DNA (Carmody et al. 2007). Regardless of the inhibited or enhanced NF-κB transactivation consequences, IκBs regulate the DNA binding activity of NF-κB proteins by controlling the high-affinity interactions of NF-κB species to cognate DNA regulatory sites.
POST-TRANSLATIONAL MODIFICATIONS OF REL PROTEINS
Like IκB proteins, Rel subunits are subject to numerous post-translational modifications, which represents another mechanism of regulating NF-κB activation that has been extensively studied recently. These modifications, within conserved RHD, TAD or linker sequences, convey various physiological functions of NF-κB (Viatour et al. 2005; Perkins 2006; Neumann and Naumann 2007; O'Shea and Perkins 2008). This includes control of NF-κB DNA binding activity through modifications, particularly phosphorylation and acetylation, of Rel subunits or adjusting the association of NF-κB complexes to either IκBs or κB DNA (Table 2). This could be caused by steric alternations of the Rel proteins themselves or to controlling their associations with other proteins in transcriptional regulatory complexes.
Table 2.
Post-translational modifications of Rel subunits that regulate NF-κB DNA binding activity
| Rel family proteins | Target residues | Enzymes | Functional effect on DNA binding activity | References |
| Phosphorylation | ||||
| p65 | T254 | Unknown | Enhancement | (Ryo et al. 2003) |
| S529 | CKII | Unknown | (Bird et al. 1997; Wang et al. 2000; O'Mahony et al. 2004) | |
| Unknown | PI3K/Akt | Enhancement | (Sizemore et al. 1999) | |
| S276 | MSK1 | Enhancement | (Reber et al. 2009) | |
| S536 | IKKα | Enhancement | (Jiang et al. 2003; O'Mahony et al. 2004) | |
| IKKβ | Enhancement | (Sakurai et al. 1999) | ||
| IKKε | Enhancement | (Buss et al. 2004; Adli and Baldwin 2006; Mattioli et al. 2006) | ||
| TBK1 | Enhancement | (Fujita et al. 2003; Buss et al. 2004) | ||
| RSK1 | Enhancement | (Bohuslav et al. 2004) | ||
| c-Rel | Unknown | TBK1 | Enhancement | (Harris et al. 2006) |
| Unknown | IKKε | Enhancement | (Sanchez-Valdepenas et al. 2006) | |
| p50 | S337 | PKAc | Enhancement | (Guan et al. 2005) |
| Acetylation | ||||
| p65 | K218, K221, and K310 | p300 | Enhancement | (Chen et al. 1998) |
| K122 and K123 | PCAF and p300 | Reduce | (Kiernan et al. 2003) | |
| p50 | K431, K440, and K441 | p300 | Enhancement | (Furia et al. 2002) |
CKII, Casein kinase II; MSK1, Mitogen- and stress-activated protein kinase-1; PI3K, Phosphoinositide 3-kinases; IKKα, IκB kinase α; IKKβ, IκB kinase β; IKKε, IκB kinase epsilon; TBK1, TANK-binding kinase 1; RSK1, Ribosomal protein S6 kinase; 90 kDa, polypeptide 1; PKAc, catalytic subunit of protein kinase A; p300, E1A binding protein p300; PCAF, p300/CREB binding protein-associated factor.
The phosphorylation of p65 under basal and activated conditions is by far the best-characterized modification among Rel proteins, and p65 is the transcriptionally active component of the NF-κB species that is most abundant and has the broadest function. So far, nine phosphorylation sites have been identified in p65 located in both the RHD and TAD, with several sites remarkably critical for DNA binding capability. Ser-536 phosphorylation by various kinases leads to a substantially weaker interaction of p65 with newly synthesized IκBα, leading to a dramatic decrease of nuclear p65 export, which enhances the duration of κB DNA access (Table 2). Furthermore, Ser-536-phosphorylated p65 does not interact with cytosolic IκBα at all, which could accelerate p65 nuclear localization and attachment to DNA (Sasaki et al. 2005). Phosphorylation of p65 at Ser-276 and Ser-536 together with successive acetylation at Lys-310 alters its affinity to IκB proteins, resulting in different kinetics of p65 cytoplasmic-nuclear shuttling and subsequent DNA binding activity (Chen et al. 2005). Phosphorylation of Thr-254 by the nuclear peptidylprolyl isomerase Pin1, followed by isomerization of specific amino acid residues, also strongly increases p65 nuclear translocation and DNA binding activity. Pin1-induced conformational changes increase the structural stability of p65 with dramatic decrease in IκBα affinity (Ryo et al. 2003). Recently, phosphorylation of Ser-276 by the mitogen- and stress-activated protein kinase-1 (MSK1) was shown to be essential for p65 binding to the κB intronic enhancer site of the mast cell growth factor SCF (stem cell factor) gene in inflammation (Reber et al. 2009). Phosphorylation of p65 can also repress the transcriptional activity of other Rel subunits. For instance, Ser-536-phosphorylated p65 was reported to specifically interact with RelB in the nucleus, forming a complex that cannot bind to κB DNA (Jacque et al. 2005).
Post-translational modification of c-Rel and its effect on DNA binding activity is similar to p65, but with fewer characterized kinase(s) or target residue(s). c-Rel can be specifically phosphorylated by the serine/threonine kinases NIK, TBK1, and IKKε on its carboxyl terminus, which dissociates the IκBα-c-Rel complex and enhances binding to cognate DNA (Harris et al. 2006; Sanchez-Valdepenas et al. 2006). Furthermore, tyrosine phosphorylation of c-Rel within its carboxyl terminus also increased its ability to bind κB sites in human neutrophils stimulated with granulocyte-colony-stimulating factor (Druker et al. 1994). More importantly, dysregulated Ser-525 phosphorylation located within c-Rel TAD, caused by a serine to proline point mutation discovered in two patients with follicular and mediastinal B-cell lymphoma, might slightly enhance c-Rel DNA binding activity, at least for κB motifs (Starczynowski et al. 2007). These data show that the carboxy-terminal modifications, especially in the TAD, are vital in regulating c-Rel DNA binding activity.
The RelB monomer is easily degraded and preferentially binds to p52 and p50, making it an unusual NF-κB subunit (Hayden and Ghosh 2004). Although the kinases have not been identified yet, phosphorylation of RelB at multiple residues including Thr-84, Ser-254, Ser-368, and Ser-552, have been reported as essential for its dimerization and degradation (Viatour et al. 2005; Neumann and Naumann 2007). It certainly deserves further investigation whether these or other unknown modifications of RelB regulate DNA binding, given the crucial role of RelB in the alternative pathway of NF-κB activation. Although inducible phosphorylation of p50, similar to its precursor p105, has been noticed in various cell types (Neumann and Naumann 2007), site-specific modifications of transactivation repressive p50 and p52 have been rarely reported. One recent study showed the catalytic subunit of PKA (PKAc) specifically phosphorylated mature p50 at Ser-337 located within RHD and augmented DNA binding activity, which may be important for maintaining stable negative regulation of NF-κB gene expression in unstimulated cells (Guan et al. 2005).
Acetylation is another key modification of Rel subunits that regulates their association with IκBs and direct binding to cognate DNA (Table 2). Five residues, Lys-122, Lys-123, Lys-218, Lys-221, and Lys-310, can be specifically acetylated in p65 in response to TNFα stimulation. Acetylation at Lys-218 and Lys-221 markedly diminishes the binding of p65 to IκBα thereby increasing its DNA binding (Chen et al. 2002). In particular, Lys-221 of p65 directly interacts with Met-279 of IκBα in the crystal structure of p65-p50-IκBα complex (Huxford et al. 1998); therefore, acetylation of Lys-221 may result in a conformational change in p65 and lessen its interaction with IκBα (Chen et al. 2002). In contrast, acetylation of p65 at Lys-122 and Lys-123 accelerates its dissociation from DNA and successive nuclear export by IκBα, attenuating NF-κB transactivation (Kiernan et al. 2003). In addition, acetylation of Rel subunits may modulate their binding with DNA directly. Acetylation of p65 at Lys-221 apparently causes a conformation change favoring κB DNA binding. This is supported by the crystal structure of p65-p50-κB DNA complex illustrating that Lys-221 directly contacts the DNA backbone (Chen et al. 1998). Various mutational analyses strengthen the concept that this acetylation influences DNA-binding properties. Mutation of Lys-221 produces a sharp decline in the DNA binding affinity of p65 homodimers and a substantial decrease in that of p65-p50 heterodimers (Chen et al. 2002). Strikingly, p50 can be acetylated in vitro by p300/CBP at Lys-431, Lys-440, and Lys-441, and this modification augments its DNA binding properties, as evidenced by pull-down assays using κB oligonucleotides (Furia et al. 2002). Collectively, site-specific acetylation of Rel proteins, at least p65 and p50, augments their intrinsic DNA binding activity, as shown for other transcription factors (Boyes et al. 1998).
REL SUBUNIT-ASSOCIATING PROTEINS
For full NF-κB transactivation to occur at a given target DNA locus, a successive enhanceosome complex needs to be assembled by multiple coactivators, corepressors, other transcription factors, and basal transcription machinery proteins through protein–protein interactions with Rel subunits (Hayden and Ghosh 2004; Hayden and Ghosh 2008). For instance, over 100 proteins can modulate NF-κB association with chromatin or the assembly of an NF-κB enhanceosome via their interaction with full length, RHD, TAD, or the central region of p65 (O'Shea and Perkins 2008). Until a short time ago, Rel subunits were widely regarded as the only executers to recognize and bind κB DNA. However, several Rel-associating proteins, albeit with no κB DNA binding capability themselves, have recently been illustrated to be essential in regulating NF-κB binding to cognate DNA (Table 3). These Rel-associating proteins feature restricted tissue distribution, specific expression profiles, or unique pathophysiological circumstances and inhibit, as currently reported, the DNA binding activity of NF-κB proteins. This establishes Rel-associating proteins as a group of tissue- or cell context-specific negative regulators of NF-κB beyond the IκB family, adding another level of complexity to the exquisite spatiotemporal control of DNA binding activity of NF-κB proteins.
Table 3.
Rel-associating proteins that regulate NF-κB DNA binding activity
| NF-κB subunits | Associating/interacting proteins | Domain/portion of Rel proteins sufficient for binding | Evidence for integration into DNA binding complexes | Functional effect on DNA binding activity | References |
| p65 | |||||
| RAI | RHD, aa 176–405 | No | Inhibition | (Yang et al. 1999) | |
| AhR | Not mapped | No | Inhibition | (Tian et al. 1999; Kim et al. 2000; Ruby et al. 2002) | |
| β-Catenin | Not mapped | No | Inhibition | (Deng et al. 2002) | |
| Cdk9 | Not mapped | No | Inhibition | (Amini et al. 2002) | |
| PIAS1 | TAD, aa 299–551 | No | Inhibition | (Liu et al. 2005) | |
| VP1686 | Not mapped | No | Inhibition | (Bhattacharjee et al. 2006) | |
| Myocardin | RHD, aa 1–276 | No | Inhibition | (Tang et al. 2008) | |
| RPS3a | RHD, aa 21–186 | Yes | Enhancement | (Wan et al. 2007) | |
| p50 | |||||
| β3-Endonexin | Not mapped | No | Inhibition | (Besta et al. 2002) | |
| Cdk9 | Not mapped | No | Inhibition | (Amini et al. 2002) | |
| β-Catenin | Not mapped | No | Inhibition | (Deng et al. 2002) |
RAI, RelA associated inhibitor; AhR, Aryl hydrocarbon receptor; Cdk9, Cyclin-dependent kinase 9; PIAS1, Protein inhibitor of activated STAT1; VP1686, Vibrio parahaemolyticus type III secretion protein; RPS3, Ribosomal protein S3; β3-Endonexin, Integrin β 3 binding protein.
aRPS3, an integral subunit of certain NF-κB DNA binding complex that also physically interacts with p65, is listed here to compare with Rel-associating proteins in their DNA-binding regulatory functions.
Some Rel-associating proteins attenuate NF-κB DNA binding in cotransfection experiments. RelA-associated inhibitor (RAI), isolated in a yeast two-hybrid screen utilizing the central region of p65 as bait, interacts with p65 in vitro and in vivo, and specifically inhibits p65 binding to the cognate DNA when cotransfected in human embryonic kidney 293 cells (Yang et al. 1999). A similar function was ascribed to the aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor. When cotransfected in 293 cells, AhR specifically inhibited the DNA binding activity of p65, but not the p50–p50 complex (Tian et al. 1999; Ruby et al. 2002), suggesting that synergic interaction between AhR and NF-κB, mainly the p65 subunit, is critical for suppression of immune responses and xenobiotic metabolism. Some pathogen-encoded proteins also possess the ability to regulate host NF-κB binding to cognate DNA, hampering immune responses directed against the microbes that express them. For instance, Vibrio parahaemolyticus is a causative agent of human gastrointestinal diseases and significantly suppresses the induction of the DNA binding activity of NF-κB via the physical interaction between its effector protein VP1686 and p65. Such attenuation of NF-κB DNA binding activity by VP1686 is sufficient to sensitize infected-macrophages for death, because of the diminished expression of many antiapoptosis-related NF-κB target genes (Bhattacharjee et al. 2006).
The regulatory effects of Rel-associating proteins have been studied not only in the cellular context of NF-κB signaling, but also in in vitro biochemical investigations with purified proteins. One of the first studies described a surprising cross-regulation of NF-κB by β-catenin (Deng et al. 2002). Both p65 and p50 could complex with β-catenin independently of IκBα, but seemed to require additional cellular factors. β-catenin markedly attenuated the DNA binding of both p50–p65 and p50–p50 complexes as shown by electrophoretic mobility shift assays (EMSA), causing reduced NF-κB target gene expression. Of note, β-catenin is not directly integrated into the NF-κB-DNA complex in spite of its strong association with NF-κB subunits, suggesting it may interact with NF-κB proteins to disrupt their DNA binding ability (Deng et al. 2002). Inhibition of NF-κB DNA binding activity by β-catenin could be important in oncogenesis, as hinted at by an inverse correlation between the β-catenin expression levels and levels of the NF-κB target gene Fas in colon and breast tumor tissues (Deng et al. 2002). Furthermore, the inhibitory effects on NF-κB DNA binding by Cyclin-dependent kinase 9 (Cdk9), the PIAS1 (protein inhibitor of activated STAT [signal transducers and activator of transcription] 1) protein, and myocardin were also shown using EMSA, in which these proteins significantly reduced the DNA binding activity of NF-κB (Amini et al. 2002; Liu et al. 2005; Tang et al. 2008). Cdk9 markedly suppressed the association of the p50–p50, p65–p65, and p50–p65 complexes to κB DNA, and the ability of NF-κB to modulate HIV-1 gene transcription was controlled by this inhibitory function (Amini et al. 2002). Both PIAS1 and myocardin specifically inhibited the DNA binding activity of p65-containing complexes, serving as negative regulators of NF-κB in certain conditions. PIAS1 was originally identified in the Jak/STAT signaling pathway, whereas myocardin is expressed specifically in cardiac and smooth muscle cells (Liu et al. 2005; Tang et al. 2008). Of interest, their effects on NF-κB DNA binding appear essential for tuning cytokine-induced NF-κB target gene expression and cardiomyocyte proliferation and differentiation, respectively (Liu et al. 2005; Tang et al. 2008).
As described above, β-catenin and Cdk9 both complex with p50 and dramatically inhibit its DNA binding activity (Amini et al. 2002; Deng et al. 2002). β3-endonexin is another p50-interacting molecule that inhibits p50–p65 complex binding to κB DNA. Moreover, binding of β3-endonexin to p50 was inhibited in the presence of wild-type but not mutated κB oligonucleotides, suggesting a steric competition between β3-endonexin and κB DNA for the p50–p65 complex. Despite the association with the transcription repressive p50 subunit, β3-endonexin negatively regulates expression of the urokinase-type plasminogen activator receptor that is essential for endothelial migration (Besta et al. 2002).
INTEGRAL NON-REL SUBUNITS IN NF-κB COMPLEXES
As for the unresolved and important question of how regulatory specificity of NF-κB is achieved, it has long been regarded that the variability of κB sequences may govern the usage of certain Rel dimers at specific promoters. Principally, each κB site variant could preferentially recruit one type of Rel dimer over other species (Natoli et al. 2005; Natoli 2006). Selective NF-κB gene expression, however, cannot be completely explained by a simple correlation between the sequence of κB sites in target genes and the requirement for a specific Rel dimer (Hoffmann et al. 2003). However, κB sites may still impart a specific configuration for NF-κB binding, because a single nucleotide change in an NF-κB binding site affected the formation of productive interactions between Rel dimer and coactivators (Leung et al. 2004). Therefore, other protein components beyond Rel subunits could form integral parts of the NF-κB binding complex, thus controlling its recognition and action on target genes. Support for this hypothesis comes from observations made in seminal previous studies. First, the contradiction in the size of native NF-κB from nuclear extracts (> 200 kDa) and that reconstituted from purified p50 and p65 proteins (115 kDa) implies the presence of other proteins in the native complex (Urban et al. 1991). Second, reconstituted p65–p50 heterodimers from purified proteins have a >100-fold lower affinity for DNA than native NF-κB complexes, at least for binding to the Ig κB motif (Phelps et al. 2000). Third, distinct variants of the κB motifs display different responses to various NF-κB inducers in different cell types implying selective regulation. Finally, a large number of NF-κB binding motifs beyond either canonical or variant κB site sequences were revealed throughout the human genome by mapping with chromatin immunoprecipitation-coupled microarray or sequencing (Martone et al. 2003; Schreiber et al. 2006; Lim et al. 2007). Collectively, these findings strongly suggested that other non-Rel proteins could not only regulate NF-κB DNA binding activity, but also participate in DNA binding as an essential component.
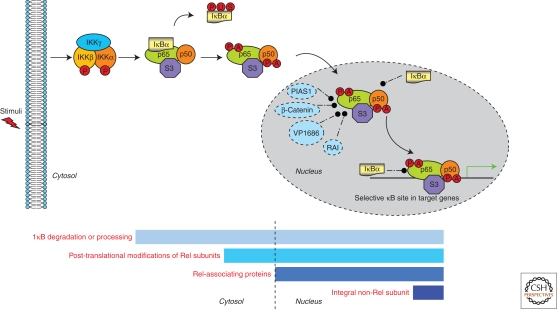
This hypothesis was confirmed by a recent study showing that RPS3, an integral non-Rel subunit in certain NF-κB DNA binding complexes, is essential for the recruitment of NF-κB p65 to selected κB sites (Wan et al. 2007). RPS3 prominently features a heterogeneous nuclear protein K (hnRNP K) homology (KH) domain, a structural motif that binds single-stranded RNA and DNA with some sequence specificity (Siomi et al. 1993). Indeed, the KH domain within RPS3 is essential for association with p65 (Wan et al. 2007). This study underscored the inherent complexity of NF-κB binding to κB sites by demonstrating that DNA binding capability is not conferred strictly by Rel subunits, as has been long assumed. Rather, the integral non-Rel subunit RPS3 represents a newly recognized subunit that potently contributes to DNA binding activity. These observations suggest a new regulatory paradigm in which DNA binding activity could be regulated within NF-κB complexes through synergistic interactions between Rel and non-Rel subunits (Fig. 2).
Figure 2.
Complex regulation of NF-κB DNA binding activity. As illustrated in one of the most abundant NF-κB complexes, the DNA binding activity of NF-κB is controlled in both the cytoplasm and nucleus at multiple levels: involving degradation or processing of inhibitory IκBs; the post-translational modification of Rel subunits; Rel-associating proteins that modulate NF-κB DNA binding potential; and an integral non-Rel subunit RPS3 required for selective NF-κB target gene transcription based on enhanced DNA binding affinity. The bars represent the intracellular locations where the DNA binding activity of NF-κB proteins is regulated by indicated proteins.
RPS3 was found to physically interact with p65 in a proteomic screen, and shown to be critical in NF-κB transactivation. Under conditions of reduced RPS3, the DNA binding activity of NF-κB complexes was significantly attenuated. Knockdown of RPS3 resulted in failed recruitment of p65 to selected endogenous gene regulatory sites and abortive induction, despite normal p65 nuclear translocation. Therefore, RPS3 facilitates p65 binding to cognate DNA, which is essential for normal expression of specific NF-κB target genes involved in key physiological processes. This was dramatically shown for the induction of immunoglobulin κ light chain gene expression in B cells and cell proliferation and cytokine secretion in T cells that were markedly impaired by RPS3 knockdown (Wan et al. 2007). Strikingly, purified RPS3 protein exerted a dramatic synergistic effect on the DNA binding activity of both p65–p65 and p50–p65, but not p50–p50 complexes in EMSA (Wan et al. 2007). As a DNA-binding facilitator, RPS3 dramatically stabilizes NF-κB association with certain cognate sites. This could explain the extremely high affinity of semipurified NF-κB complexes for DNA, which is not manifested by complexes formed solely of purified p50 and p65 subunits. By contrast, none of the aforementioned Rel-associating proteins has been shown to integrate into NF-κB DNA binding complexes, and all of them inhibit the p65 DNA binding activity, although several were assessed in vitro using recombinant proteins in EMSA (Amini et al. 2002; Deng et al. 2002; Liu et al. 2005; Tang et al. 2008).
It is important to recognize that RPS3 is not an NF-κB-associated transcriptional coactivator, which by definition reorganizes chromatin templates and recruits the basal transcriptional machinery to the promoter region. RPS3 possesses little, if any, intrinsic transcriptional activating ability in a standard coactivation assay (Wan et al. 2007). Furthermore, the ability of RPS3-specific antibodies to dramatically supershift or diminish p65-containing DNA complexes in EMSAs strongly suggests that RPS3 is an integral part of NF-κB DNA binding complexes (Wan et al. 2007). By contrast, administration of a specific antibody against p300, one of well-characterized transcriptional coactivators that complex with NF-κB, did not alter NF-κB-DNA complexes, suggesting that p300 is not incorporated into the DNA binding complex (Deng et al. 2002). Further lines of evidence support the notion that RPS3 is an integral subunit of NF-κB: RPS3 physically associates with p65, p50, and IκBα in resting cells (guided through its interaction with p65); RPS3 can specifically translocate to the nucleus in response to T cell receptor (TCR) and TNFα stimulation; RPS3 is recruited to κB sites in a large number of NF-κB-driven genes in vivo upon stimulation; and RPS3 and p65 are significantly correlated in transcribing a subset of TCR ligation-induced NF-κB genes (Wan et al. 2007). Whether the RPS3 subunit is essential in other stimuli-induced NF-κB signal pathways and whether it targets certain NF-κB complexes to specific κB sites in different cell types certainly deserves further investigation. Because RPS3 is only required for selected particular genomic κB sites to be activated under certain conditions and preferentially directs binding to κB sites with some sequence specificity, we call it a “specifier” subunit of NF-κB. This also lends credence to the idea that there are actually multiple molecular complexes containing RPS3-like “specifier” subunits with different gene activation specificities that all masquerade as single NF-κB complex in the nucleus. Indeed, another KH domain protein, Src-associated in mitosis, 68 kDa (Sam68), was found to be essential for some NF-κB gene transcription where RPS3 is not required (F. Wan and M. Lenardo, unpubl.). Sequence specificity preferred by various KH domains could confer different gene activation patterns via diverse NF-κB complexes. These findings may unveil a novel regulatory paradigm in which KH domain proteins serve as essential functional components in regulating the DNA binding activity of not only NF-κB, but also other transcription factors, because several other KH domain proteins have been shown to bind to DNA recognition motifs and to promote transcription (Tomonaga and Levens 1996; Ostrowski et al. 2003; Moumen et al. 2005).
CONCLUDING REMARKS
Since it was originally identified as a regulator of κ light chain expression in B cells over 20 years ago, NF-κB has served as a paradigm for signaling associated with inflammation, autoimmunity, and cancer. Recruitment of NF-κB proteins to regulatory DNA sites within the chromatin is fundamental and crucial for their target gene transcription. The complexity inherent in the DNA binding activity of NF-κB proteins is essential for achieving a fine-tuned regulatory specificity. An increasing body of literature illustrates that the DNA binding activity of NF-κB proteins can be elegantly and specifically controlled at multiple levels by different protein subsets, including IκBs, Rel subunits, Rel-associating proteins, and integral non-Rel subunits (Fig. 2). In particular, the expanding list of Rel-associating proteins constitutes a unique category of tissue- or cell type-specific negative regulators of NF-κB beyond IκBs that control DNA binding activity under certain circumstances. Furthermore, the recent identification of a non-Rel subunit of NF-κB itself provides a new way to understand the selective high-affinity DNA binding specificity of NF-κB conferred by a synergistic interaction within the whole complex.
Despite extensive studies on the control of NF-κB DNA binding activity, numerous issues are still unresolved and warrant further investigation. These include the identity of regulatory kinases and site-specific modulating target residues in IκBs that mediate their degradation/processing; additional post-translational modifications of Rel subunits beyond p65 and their affect on DNA binding; and specific associating and regulatory proteins for RelB and c-Rel. Because the inclusion of RPS3 does not fully explain the size of native NF-κB or its high affinity to cognate κB sites, the identification of other unknown non-Rel subunits of NF-κB is well worth further investigation. Undoubtedly, a more complete understanding of the complex control of the DNA binding activity of NF-κB proteins will not only revise or add to our fundamental knowledge of gene regulation, but also elucidate novel target molecules for pharmacological interventions.
ACKNOWLEDGMENTS
We thank Andrew Snow and Amanda Weaver for critical reading of the manuscript. The work in the authors' laboratory is supported by the Intramural Research Program of the National Institutes of Health (NIH), NIAID. F.W. is a recipient of NIH grant K99CA137171. We apologize to those who made also important contributions to the issues discussed and who could not be cited because of space limits.
Footnotes
Editors: Louis M. Staudt and Michael Karin
Additional Perspectives on NF-κB available at www.cshperspectives.org
REFERENCES
- Adli M, Baldwin AS 2006. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-κB activity via regulation of Ser-536 p65/RelA phosphorylation. J Biol Chem 281:26976–26984 [DOI] [PubMed] [Google Scholar]
- Amini S, Clavo A, Nadraga Y, Giordano A, Khalili K, Sawaya BE 2002. Interplay between cdk9 and NF-κB factors determines the level of HIV-1 gene transcription in astrocytic cells. Oncogene 21:5797–5803 [DOI] [PubMed] [Google Scholar]
- Amir RE, Haecker H, Karin M, Ciechanover A 2004. Mechanism of processing of the NF-κB2 p100 precursor: Identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(β-TrCP) ubiquitin ligase. Oncogene 23:2540–2547 [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D 1988. I κB: A specific inhibitor of the NF-κB transcription factor. Science 242:540–546 [DOI] [PubMed] [Google Scholar]
- Besta F, Massberg S, Brand K, Muller E, Page S, Gruner S, Lorenz M, Sadoul K, Kolanus W, Lengyel E, et al. 2002. Role of β(3)-endonexin in the regulation of NF-κB-dependent expression of urokinase-type plasminogen activator receptor. J Cell Sci 115:3879–3888 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee RN, Park KS, Kumagai Y, Okada K, Yamamoto M, Uematsu S, Matsui K, Kumar H, Kawai T, Iida T, et al. 2006. VP1686, a Vibrio type III secretion protein, induces toll-like receptor-independent apoptosis in macrophage through NF-κB inhibition. J Biol Chem 281:36897–36904 [DOI] [PubMed] [Google Scholar]
- Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, Davis RE, Lenz G, Anderson DE, Arnoult D, et al. 2009. Casein kinase 1α governs antigen-receptor-induced NF-κB activation and human lymphoma cell survival. Nature 458:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TA, Schooley K, Dower SK, Hagen H, Virca GD 1997. Activation of nuclear transcription factor NF-κB by interleukin-1 is accompanied by casein kinase II-mediated phosphorylation of the p65 subunit. J Biol Chem 272:32606–32612 [DOI] [PubMed] [Google Scholar]
- Bohuslav J, Chen LF, Kwon H, Mu Y, Greene WC 2004. p53 induces NF-κB activation by an IκB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. J Biol Chem 279:26115–26125 [DOI] [PubMed] [Google Scholar]
- Boyes J, Byfield P, Nakatani Y, Ogryzko V 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594–598 [DOI] [PubMed] [Google Scholar]
- Buss H, Dorrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M 2004. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases including IκB kinase (IKK)-α, IKKβ, IKKepsilon, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J Biol Chem 279:55633–55643 [DOI] [PubMed] [Google Scholar]
- Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH 2007. Negative regulation of toll-like receptor signaling by NF-κB p50 ubiquitination blockade. Science 317:675–678 [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC 2004. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol 5:392–401 [DOI] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391:410–413 [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. Embo J 21:6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC 2005. NF-κB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol 25:7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZL, McKinsey TA, Liu L, Qi X, Ballard DW 1996. Basal phosphorylation of the PEST domain in the IκBβ regulates its functional interaction with the c-rel proto-oncogene product. Mol Cell Biol 16:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr 2000. Selective activation of NF-κB subunits in human breast cancer: Potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 19:1123–1131 [DOI] [PubMed] [Google Scholar]
- Cohen S, Achbert-Weiner H, Ciechanover A 2004. Dual effects of IκB kinase β-mediated phosphorylation on p105 Fate: SCF(β-TrCP)-dependent degradation and SCF(β-TrCP)-independent processing. Mol Cell Biol 24:475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi F, Bertoli C, Sandy P, Schneider C 2003. Glycogen synthase kinase-3 β regulates NF-κB1/p105 stability. J Biol Chem 278:39583–39590 [DOI] [PubMed] [Google Scholar]
- Deng WG, Wu KK 2003. Regulation of inducible nitric oxide synthase expression by p300 and p50 acetylation. J Immunol 171:6581–6588 [DOI] [PubMed] [Google Scholar]
- Deng J, Miller SA, Wang HY, Xia W, Wen Y, Zhou BP, Li Y, Lin SY, Hung MC 2002. β-catenin interacts with and inhibits NF-κB in human colon and breast cancer. Cancer Cell 2:323–334 [DOI] [PubMed] [Google Scholar]
- Deng WG, Tang ST, Tseng HP, Wu KK 2006. Melatonin suppresses macrophage cyclooxygenase-2 and inducible nitric oxide synthase expression by inhibiting p52 acetylation and binding. Blood 108:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell 2:233–239 [DOI] [PubMed] [Google Scholar]
- Druker BJ, Neumann M, Okuda K, Franza BR Jr, Griffin JD 1994. rel Is rapidly tyrosine-phosphorylated following granulocyte-colony stimulating factor treatment of human neutrophils. J Biol Chem 269:5387–5390 [PubMed] [Google Scholar]
- Fujita F, Taniguchi Y, Kato T, Narita Y, Furuya A, Ogawa T, Sakurai H, Joh T, Itoh M, Delhase M, et al. 2003. Identification of NAP1, a regulatory subunit of IκB kinase-related kinases that potentiates NF-κB signaling. Mol Cell Biol 23:7780–7793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furia B, Deng L, Wu K, Baylor S, Kehn K, Li H, Donnelly R, Coleman T, Kashanchi F 2002. Enhancement of nuclear factor-κ B acetylation by coactivator p300 and HIV-1 Tat proteins. J Biol Chem 277:4973–4980 [DOI] [PubMed] [Google Scholar]
- Grilli M, Chiu JJ, Lenardo MJ 1993. NF-κB and Rel: Participants in a multiform transcriptional regulatory system. Int Rev Cytol 143:1–62 [DOI] [PubMed] [Google Scholar]
- Guan H, Hou S, Ricciardi RP 2005. DNA binding of repressor nuclear factor-κB p50/p50 depends on phosphorylation of Ser337 by the protein kinase A catalytic subunit. J Biol Chem 280:9957–9962 [DOI] [PubMed] [Google Scholar]
- Hacker H, Karin M 2006. Regulation and function of IKK and IKK-related kinases. Sci STKE 2006:re13. [DOI] [PubMed] [Google Scholar]
- Harris J, Oliere S, Sharma S, Sun Q, Lin R, Hiscott J, Grandvaux N 2006. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J Immunol 177:2527–2535 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S 2004. Signaling to NF-κB. Genes Dev 18:2195–2224 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S 2008. Shared principles in NF-κB signaling. Cell 132:344–362 [DOI] [PubMed] [Google Scholar]
- Heissmeyer V, Krappmann D, Hatada EN, Scheidereit C 2001. Shared pathways of IκB kinase-induced SCF(βTrCP)-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBα. Mol Cell Biol 21:1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Leung TH, Baltimore D 2003. Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. Embo J 22:5530–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Guan H, Ricciardi RP 2003. Phosphorylation of serine 337 of NF-κB p50 is critical for DNA binding. J Biol Chem 278:45994–45998 [DOI] [PubMed] [Google Scholar]
- Hu J, Colburn NH 2005. Histone deacetylase inhibition down-regulates cyclin D1 transcription by inhibiting nuclear factor-κB/p65 DNA binding. Mol Cancer Res 3:100–109 [DOI] [PubMed] [Google Scholar]
- Huxford T, Huang DB, Malek S, Ghosh G 1998. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell 95:759–770 [DOI] [PubMed] [Google Scholar]
- Jacque E, Tchenio T, Piton G, Romeo PH, Baud V 2005. RelA repression of RelB activity induces selective gene activation downstream of TNF receptors. Proc Natl Acad Sci U S A 102:14635–14640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Takahashi N, Matsui N, Tetsuka T, Okamoto T 2003. The NF-κB activation in lymphotoxin β receptor signaling depends on the phosphorylation of p65 at serine 536. J Biol Chem 278:919–926 [DOI] [PubMed] [Google Scholar]
- Kang SM, Tran AC, Grilli M, Lenardo MJ 1992. NF-κB subunit regulation in nontransformed CD4+ T lymphocytes. Science 256:1452–1456 [DOI] [PubMed] [Google Scholar]
- Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M 2003. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. J Biol Chem 278:2758–2766 [DOI] [PubMed] [Google Scholar]
- Kim DW, Gazourian L, Quadri SA, Romieu-Mourez R, Sherr DH, Sonenshein GE 2000. The RelA NF-κB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene 19:5498–5506 [DOI] [PubMed] [Google Scholar]
- Koong AC, Chen EY, Giaccia AJ 1994. Hypoxia causes the activation of nuclear factor κB through the phosphorylation of I κ B α on tyrosine residues. Cancer Res 54:1425–1430 [PubMed] [Google Scholar]
- Lang V, Janzen J, Fischer GZ, Soneji Y, Beinke S, Salmeron A, Allen H, Hay RT, Ben-Neriah Y, Ley SC 2003. βTrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol Cell Biol 23:402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo MJ, Baltimore D 1989. NF-κB: A pleiotropic mediator of inducible and tissue-specific gene control. Cell 58:227–229 [DOI] [PubMed] [Google Scholar]
- Lenardo M, Siebenlist U 1994. Bcl-3-mediated nuclear regulation of the NF-κB trans-activating factor. Immunol Today 15:145–147 [DOI] [PubMed] [Google Scholar]
- Lenardo M, Pierce JW, Baltimore D 1987. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science 236:1573–1577 [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Fan CM, Maniatis T, Baltimore D 1989. The involvement of NF-κB in β-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57:287–294 [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D 2004. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 118:453–464 [DOI] [PubMed] [Google Scholar]
- Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, et al. 2007. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol Cell 27:622–635 [DOI] [PubMed] [Google Scholar]
- Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J 1996. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol 16:1401–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, Cheng G, Wu H, Shuai K 2005. Negative regulation of NF-κB signaling by PIAS1. Mol Cell Biol 25:1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. 2003. Distribution of NF-κB-binding sites across human chromosome 22. Proc Natl Acad Sci U S A 100:12247–12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JR, Botting CH, Panico M, Morris HR, Hay RT 1996. Inhibition of NF-κB DNA binding by nitric oxide. Nucleic Acids Res 24:2236–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, Schmitz ML 2006. Inducible phosphorylation of NF-κB p65 at serine 468 by T cell costimulation is mediated by IKKepsilon. J Biol Chem 281:6175–6183 [DOI] [PubMed] [Google Scholar]
- McElhinny JA, Trushin SA, Bren GD, Chester N, Paya CV 1996. Casein kinase II phosphorylates IκBα at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol Cell Biol 16:899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama M, Yamazaki S, Eto-Kimura A, Takeshige K, Muta T 2005. Positive and negative regulation of nuclear factor-κB-mediated transcription by IκB-zeta, an inducible nuclear protein. J Biol Chem 280:7444–7451 [DOI] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O'Connor MJ, Jackson SP 2005. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123:1065–1078 [DOI] [PubMed] [Google Scholar]
- Natoli G 2006. Tuning up inflammation: How DNA sequence and chromatin organization control the induction of inflammatory genes by NF-κB. FEBS Lett 580:2843–2849 [DOI] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I 2005. Interactions of NF-κB with chromatin: The art of being at the right place at the right time. Nat Immunol 6:439–445 [DOI] [PubMed] [Google Scholar]
- Neumann M, Naumann M 2007. Beyond IκBs: Alternative regulation of NF-κB activity. Faseb J 21:2642–2654 [DOI] [PubMed] [Google Scholar]
- O'Mahony AM, Montano M, Van Beneden K, Chen LF, Greene WC 2004. Human T-cell lymphotropic virus type 1 tax induction of biologically Active NF-κB requires IκB kinase-1-mediated phosphorylation of RelA/p65. J Biol Chem 279:18137–18145 [DOI] [PubMed] [Google Scholar]
- O'Shea JM, Perkins ND 2008. Regulation of the RelA (p65) transactivation domain. Biochem Soc Trans 36:603–608 [DOI] [PubMed] [Google Scholar]
- Ostrowski J, Kawata Y, Schullery DS, Denisenko ON, Bomsztyk K 2003. Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res 31:3954–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND 2006. Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene 25:6717–6730 [DOI] [PubMed] [Google Scholar]
- Phelps CB, Sengchanthalangsy LL, Malek S, Ghosh G 2000. Mechanism of κ B DNA binding by Rel/NF-κ B dimers. J Biol Chem 275:24392–24399 [DOI] [PubMed] [Google Scholar]
- Reber L, Vermeulen L, Haegeman G, Frossard N 2009. Ser276 phosphorylation of NF-κB p65 by MSK1 controls SCF expression in inflammation. PLoS ONE 4:e4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwarf DM, Karin M 1999. The NF-κB activation pathway: A paradigm in information transfer from membrane to nucleus. Sci STKE 1999:RE1. [DOI] [PubMed] [Google Scholar]
- Ruby CE, Leid M, Kerkvliet NI 2002. 2,3,7,8-Tetrachlorodibenzo-p-dioxin suppresses tumor necrosis factor-α and anti-CD40-induced activation of NF-κB/Rel in dendritic cells: p50 homodimer activation is not affected. Mol Pharmacol 62:722–728 [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP 2003. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 12:1413–1426 [DOI] [PubMed] [Google Scholar]
- Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W 1999. IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J Biol Chem 274:30353–30356 [DOI] [PubMed] [Google Scholar]
- Sanchez-Valdepenas C, Martin AG, Ramakrishnan P, Wallach D, Fresno M 2006. NF-κB-inducing kinase is involved in the activation of the CD28 responsive element through phosphorylation of c-Rel and regulation of its transactivating activity. J Immunol 176:4666–4674 [DOI] [PubMed] [Google Scholar]
- Sasaki CY, Barberi TJ, Ghosh P, Longo DL 2005. Phosphorylation of RelA/p65 on serine 536 defines an IκBα-independent NF-κB pathway. J Biol Chem 280:34538–34547 [DOI] [PubMed] [Google Scholar]
- Scheidereit C 2006. IκB kinase complexes: Gateways to NF-κB activation and transcription. Oncogene 25:6685–6705 [DOI] [PubMed] [Google Scholar]
- Schoonbroodt S, Ferreira V, Best-Belpomme M, Boelaert JR, Legrand-Poels S, Korner M, Piette J 2000. Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of IκBα in NF-κB activation by an oxidative stress. J Immunol 164:4292–4300 [DOI] [PubMed] [Google Scholar]
- Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA 2006. Coordinated binding of NF-κB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci U S A 103:5899–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EM, Van Antwerp D, Verma IM 1996. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: Requirement for degradation of free IκBα. Mol Cell Biol 16:3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D 1986. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell 47:921–928 [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M 2001. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 293:1495–1499 [DOI] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21:1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore N, Leung S, Stark GR 1999. Activation of phosphatidylinositol 3-kinase in response to interleukin-1 leads to phosphorylation and activation of the NF-κB p65/RelA subunit. Mol Cell Biol 19:4798–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starczynowski DT, Trautmann H, Pott C, Harder L, Arnold N, Africa JA, Leeman JR, Siebert R, Gilmore TD 2007. Mutation of an IKK phosphorylation site within the transactivation domain of REL in two patients with B-cell lymphoma enhances REL's in vitro transforming activity. Oncogene 26:2685–2694 [DOI] [PubMed] [Google Scholar]
- Suyang H, Phillips R, Douglas I, Ghosh S 1996. Role of unphosphorylated, newly synthesized IκBβ in persistent activation of NF-κB. Mol Cell Biol 16:5444–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RH, Zheng XL, Callis TE, Stansfield WE, He J, Baldwin AS, Wang DZ, Selzman CH 2008. Myocardin inhibits cellular proliferation by inhibiting NF-κB(p65)-dependent cell cycle progression. Proc Natl Acad Sci U S A 105:3362–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Phillips RJ, Erdjument-Bromage H, Tempst P, Ghosh S 1995. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell 80:573–582 [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA 1999. Ah receptor and NF-κB interactions, a potential mechanism for dioxin toxicity. J Biol Chem 274:510–515 [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Levens D 1996. Activating transcription from single stranded DNA. Proc Natl Acad Sci U S A 93:5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban MB, Schreck R, Baeuerle PA 1991. NF-κB contacts DNA by a heterodimer of the p50 and p65 subunit. Embo J 10:1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsyayan J, Qing G, Xiao G, Hu J 2008. SUMO1 modification of NF-κB2/p100 is essential for stimuli-induced p100 phosphorylation and processing. EMBO Rep 9:885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Dejardin E, Warnier M, Lair F, Claudio E, Bureau F, Marine JC, Merville MP, Maurer U, Green D, et al. 2004. GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol Cell 16:35–45 [DOI] [PubMed] [Google Scholar]
- Viatour P, Merville MP, Bours V, Chariot A 2005. Phosphorylation of NF-κB and IκB proteins: Implications in cancer and inflammation. Trends Biochem Sci 30:43–52 [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al. 2007. Ribosomal protein S3: A KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell 131:927–939 [DOI] [PubMed] [Google Scholar]
- Wang D, Westerheide SD, Hanson JL, Baldwin AS Jr 2000. Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J Biol Chem 275:32592–32597 [DOI] [PubMed] [Google Scholar]
- Waris G, Livolsi A, Imbert V, Peyron JF, Siddiqui A 2003. Hepatitis C virus NS5A and subgenomic replicon activate NF-κB via tyrosine phosphorylation of IκBα and its degradation by calpain protease. J Biol Chem 278:40778–40787 [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC 2001. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell 7:401–409 [DOI] [PubMed] [Google Scholar]
- Xiao G, Fong A, Sun SC 2004. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J Biol Chem 279:30099–30105 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, et al. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBzeta. Nature 430:218–222 [DOI] [PubMed] [Google Scholar]
- Yang JP, Hori M, Sanda T, Okamoto T 1999. Identification of a novel inhibitor of nuclear factor-κB, RelA-associated inhibitor. J Biol Chem 274:15662–15670 [DOI] [PubMed] [Google Scholar]