Abstract
Asymmetric cell division generates cell types with different specialized functions or fates. This type of division is critical to the overall cellular organization and development of many multicellular organisms. In plants, regulated asymmetric cell divisions are of particular importance because cell migration does not occur. The influence of extrinsic cues on asymmetric cell division in plants is well documented. Recently, candidate intrinsic factors have been identified and links between intrinsic and extrinsic components are beginning to be elucidated. A novel mechanism in breaking symmetry was revealed that involves the movement of typically intrinsic factors between plant cells. As we learn more about the regulation of asymmetric cell divisions in plants, we can begin to reflect on the similarities and differences between the strategies used by plants and animals. Focusing on the underlying molecular mechanisms, this article describes three selected cases of symmetry-breaking events in the model plant Arabidopsis thaliana. These examples occur in early embryogenesis, stomatal development, and ground tissue formation in the root.
Asymmetric division is important in plants because cells cannot migrate. Intrinsic and extrinsic cues that trigger asymmetry are beginning to be elucidated.
Plant cells are surrounded by rigid cell walls that prevent cell movement. Precisely oriented cell division is especially important in the absence of cell migration. When a plant cell divides, a new wall forms between the daughter cells; this permanently delineates their relative positions. As a consequence, the selection of the division plane is crucial for plant tissue organization and overall organ architecture.
The selection of the division plane is unique in plants compared with other organisms (for a detailed review of plant cytokinesis see Jürgens 2005). In budding yeast, landmarks at previous bud-sites position the division plane (see Slaughter et al. 2009), whereas in fission yeast it is positioned by the nucleus. The position of the centrosomes and the mitotic spindle determine division plane orientation in animal cells (Balasubramanian et al. 2004; see Munro and Bowerman 2009). In plants, the division plane is positioned by a ring of microtubules and F-actin, called the preprophase band that forms at the cell periphery (Mineyuki 1999). Despite these striking differences, similarities are beginning to emerge. For example, in budding yeast and animals, GTPase activation plays an important role in positioning division planes (Balasubramanian et al. 2004; see McCaffrey and Macara 2009; Slaughter et al. 2009; Munro and Bowerman 2009; Prehoda 2009). In plants, a GTPase was recently found to be localized at the preprophase band and may provide spatial information during division (Xu et al. 2008). Therefore, common molecular mechanisms may underlie division plane selection in plants and animals.
In all organisms, asymmetric cell division is a common method to obtain distinct cell types from a single progenitor cell. Before an asymmetric division a cell must first establish an internal asymmetry; this process is termed breaking symmetry. Breaking symmetry can be generally defined as the asymmetric organization of cellular components along one axis leading to cellular polarization as a prerequisite to division. An asymmetric cell division can generate two cells of different morphology (i.e., size and/or shape) or different specialized functions or fates. Specifically, the products of asymmetric cell divisions fall into two major categories. In one case, a mother cell can divide asymmetrically to produce a daughter cell to replace it and a daughter cell with a distinct fate, as in the case of stem cell divisions. Alternatively, a mother cell can divide asymmetrically to produce two distinct daughter cells at the expense of mother cell identity. Following division, there are two basic mechanisms by which a mother cell generates daughters that are distinct from itself. One mechanism is intrinsic to the cell and is defined by segregation of cellular determinants before division. The other mechanism is extrinsic to the cell and is defined by external cues that direct daughter cell fate (Horvitz and Herskowitz 1992). Intrinsic and extrinsic mechanisms are not necessarily independent and often work in combination to direct asymmetric division and specification of daughter cells.
The first zygotic division of fucoid brown algae, such as Fucus, is one of the earliest studied examples of symmetry breaking in plants regulated by both extrinsic and intrinsic mechanisms. Fucus zygotes are an excellent organism in which to examine the cellular biology underlying asymmetric cell division (for a recent review see Homble and Leonetti 2007). In the presence of extrinsic cues, such as light, the asymmetric zygotic division is oriented perpendicular to the light gradient so that one cell resides on the “shady” side of the gradient (Kropf 1992, 1997; Alessa and Kropf 1999). In the absence of environmental cues, the division is oriented relative to the sperm entry site and therefore appears random within a field of cells (Hable and Kropf 2000). This indicates that Fucus zygotes have an intrinsic mechanism to break symmetry in the absence of extrinsic cues. This intrinsic mechanism occurs rapidly after fertilization, however it can later be overridden by extrinsic cues. Although much is known about the role of the cytoskeleton, membrane depolarization, and ion flux in Fucus embryo polarization, the molecular components regulating these events remain unknown because of a lack of molecular tools. To address the molecular mechanisms regulating asymmetric cell divisions in plants, the use of a more genetically tractable plant is necessary.
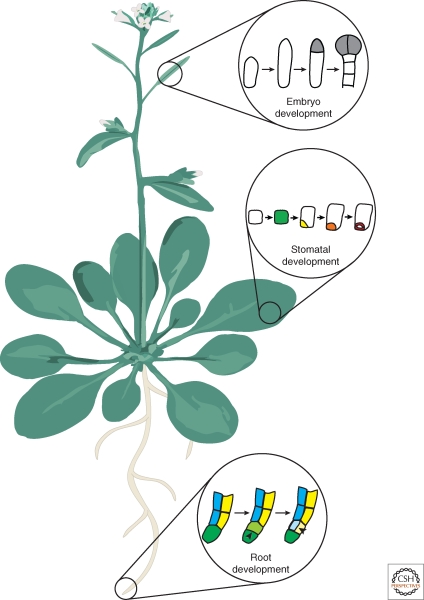
In the model plant Arabidopsis thaliana, many genes that are involved in asymmetric cell division have been identified. Interestingly, many of these genes are thought to function in both asymmetric division and subsequent cell fate specification suggesting that, in plants, these processes are tightly linked. Despite these advances, relatively little is known about the cellular processes of breaking symmetry in plants. Because of these two caveats, the term breaking symmetry must be defined more generally in plants. We use this term to describe a process in which a cell divides asymmetrically to form a daughter cell with a distinct fate. Here we discuss three examples in which the molecular mechanisms that participate in symmetry-breaking events in plants are beginning to be elucidated. These examples occur in early embryogenesis, formation of stomata, and root development (Fig. 1). We confine the topics of this article to Arabidopsis thaliana because the majority of the experimental work, particularly using molecular genetics, is focused on this species.
Figure 1.
An Arabidopsis plant. Schematic of an adult plant depicting roots, leaves, stems, and flowers. In this article, three symmetry-breaking events are discussed. These examples are taken from different organs of the plant. The first example is taken from embryo development, which occurs after fertilization of the flower in the silique (seed pod) in Arabidopsis. Next, symmetry breaking in the leaf epidermis during stomatal development is discussed. Finally, the asymmetric cell division that gives rise to the ground tissue in the root is considered.
ASYMMETRIC DIVISION OF THE ZYGOTE AND POLARIZATION OF THE EARLY EMBRYO
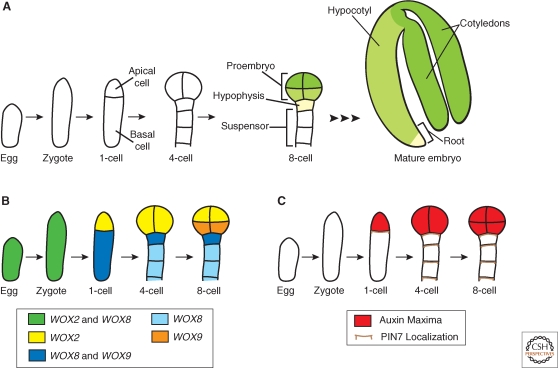
Embryogenesis is the process by which a zygote undergoes elaborate changes in cell number, fate, and morphology to ultimately form a mature embryo. An important step in embryogenesis is establishment of asymmetry (or polarity) along the longitudinal body axis. Initial embryo polarity is typically anterior–posterior (head–tail) in animals and apical–basal (shoot–root) in plants. In many multicellular organisms, breaking zygotic symmetry is critical for polarity establishment in the developing embryo because the resulting daughter cells have two distinct fates. Following fertilization, the Arabidopsis zygote expands longitudinally, then divides asymmetrically to generate a small apical cell and a larger basal cell (Fig. 2A). The invariant cell division pattern in Arabidopsis embryogenesis allows lineage analysis of each daughter cell (Mansfield and Briarty 1991). The apical cell undergoes a precise series of divisions generating a spherical eight-celled proembryo, which will give rise to the majority of the mature embryo. Apical–basal polarity is already recognizable in the eight-celled proembryo: The upper tier cells will give rise to the shoot and the lower tier cells will form the hypocotyl and a portion of the embryonic root (Fig. 2A) (Scheres et al. 1994). The basal cell undergoes repeated transverse divisions to produce an extraembryonic support structure, the suspensor, which connects the embryo to the maternal tissue (Jürgens 2001). The uppermost suspensor cell, the hypophysis, is later incorporated into the embryo to form a portion of the root (Fig. 2A) (Dolan et al. 1993; Scheres et al. 1994). The result of embryogenesis in higher plants is a simple juvenile form, in which most adult plant organs are absent (compare Fig. 1 to mature embryo in Fig. 2A). This is in contrast to embryogenesis in higher animals, which culminates in essentially a miniature form of the adult organism.
Figure 2.
Embryo development and the asymmetric localization of factors in the embryo. (A) Schematic of embryo development focusing on the events from the egg to the eight-cell stage. After fertilization, the zygote expands and then divides asymmetrically to produce the apical and basal cells. Embryo stages are based on the number of cells in the apical domain only, thus the first zygotic division results in a one-cell stage embryo. The apical cell undergoes a series of divisions to generate the eight-celled proembryo. The basal cell undergoes a series of strictly transverse divisions to generate the suspensor. Later in development, the uppermost cell of the suspensor, the hypophysis, is incorporated into the embryo. Therefore, by the eight-cell stage, four anatomically distinct cell types are present: The upper (green) and lower (yellow-green) tiers of the proembryo, the hypophysis (cream), and the suspensor (white). The suspensor remains extraembryonic, whereas the remaining cell types give rise to the cotyledons, hypocotyl, and root of the mature embryo as depicted by the corresponding color scheme in the eight-cell stage and mature embryo. (B) Schematic of the WOX gene expression patterns. By the eight-cell stage, the expression domains of the WOX genes coincide with the four distinct cell types present. (C) Schematic of the auxin maxima and PIN7 localization. PIN7 localization on the upper membrane of basal cells directs the flow of auxin from basal to apical cells. This polarized movement of auxin generates an auxin maximum in the apical domain as determined by expression of the DR5 reporter.
The invariant pattern of cell division in early Arabidopsis embryogenesis indicates a tight regulation of the asymmetric zygotic division and subsequent development of the apical and basal lineages up to the eight-cell stage (see Jenik et al. 2007 for detailed review of Arabidopsis embryogenesis). In this section, we highlight findings regarding the molecular mechanisms regulating asymmetric division of the zygote and cell fate specification of the daughter cells.
Asymmetric Zygotic Division and Suspensor Cell Fate Specification Require a MAPKKK Signaling Pathway
The mitogen-activated protein kinase kinase kinase (MAPKKK), YODA (YDA), is required for elongation and asymmetric division of the zygote and subsequent development of the basal cell lineage (Lukowitz et al. 2004). In yda loss-of-function mutants, zygotes fail to expand longitudinally and the first zygotic division produces cells of similar size. From the two-cell to eight-cell stage, yda defects are primarily observed in the basal cell lineage. Here, divisions appear in random orientations and suspensor identity markers are not expressed. Despite this, the apical lineage develops normally up to the eight-cell stage, after which it also shows cell division defects. Conversely, in yda gain-of-function mutants, the suspensor proliferates at the apparent expense of the apical lineage. These results indicate that polarity is important for embryo development. Additionally, these data suggest that a signaling network, including YDA, acts as a switch to activate suspensor cell fate. YDA also participates in asymmetric cell divisions later in plant development (see the following section). MAPKK kinases function in a variety of pathways but likely act within signaling modules downstream of extracellular receptors. Thus, it is tempting to speculate that external signals, possibly from maternal tissue, provide cues for zygotic asymmetry and specification of the first embryonic cell fate decision. However, other pathway components and how they may be differentially distributed before or after zygotic division remain unknown.
The Role of Homeodomain Transcription Factors in the Asymmetry of the Zygote
Members of the Arabidopsis homeodomain transcription factor family also have roles in asymmetric cell division and cell fate specification following the first zygotic division. WUS-RELATED HOMEOBOX2 (WOX2) and WOX8 are expressed in both the egg cell and zygote. Following the first zygotic division, WOX2 and WOX8 expression is restricted to the apical and basal cell lineages, respectively (Fig. 2B) (Haecker et al. 2004). Another WOX gene, WOX9, is also expressed in the basal cell, but its expression is later restricted to the hypophysis and then expands into the central domain of the proembryo (Fig. 2B) (Haecker et al. 2004; Wu et al. 2007). The expression pattern of these three WOX genes coincides with the four anatomically distinct cell types present at the eight-cell stage: the upper tier, lower tier, hypophysis, and suspensor (Fig. 2). However, the mechanisms regulating expression of WOX2, WOX8, and WOX9 in the early embryo are unknown.
Consistent with their asymmetric expression patterns, these WOX genes have critical roles in the apical and basal lineages. The embryos of wox2 mutants show cell division defects specifically in the apical domain. However, these defects are mild because of expression of related, partially redundant WOX genes in the apical domain (Haecker et al. 2004; Breuninger et al. 2008). In wox8 and wox9 single mutants, embryo development is largely normal (Wu et al. 2007; Breuninger et al. 2008). More severe defects are observed in wox8 wox9 double mutants, in which abnormal cell divisions occur in both the apical and basal cell lineages. Several genetic markers of apical cell fate, including WOX2, are undetectable in wox8 wox9 embryos. These results indicate WOX8/WOX9 activity in the basal lineage is required for formation of the apical lineage, and suggest communication between the lineages is important in the early embryo (Breuninger et al. 2008). Additionally, in wox8 wox9 embryos, aspects of apical and basal lineage development can be restored by ectopic WOX2 expression. However, in these embryos the zygote fails to elongate and the first division is more symmetrical, closely resembling yda embryos. Despite this similarity, yda wox8 wox9 triple mutant embryos completely arrest development after the first nearly symmetrical zygotic division. This phenotype is much more severe than yda or wox8 wox9 alone and indicates that these genes do not function in a linear pathway (Breuninger et al. 2008). This also suggests that signaling through the YDA pathway and WOX-regulated transcription act independently in breaking zygotic symmetry. WOX transcription networks are clearly involved in asymmetric cell division and fate specification in the apical and basal lineages, however details about how WOX genes become differentially expressed after the zygotic division are required to understand their role in this process.
The Role of Auxin in Maintaining Asymmetry in the Early Embryo
The plant hormone auxin is one of the most ubiquitous plant signaling molecules and it appears to be required for many diverse developmental processes (see Jenik and Barton 2005 and Leyser 2006 for recent reviews). Auxin movement occurs directionally from one cell to another and is mediated by a family of functionally redundant transmembrane efflux carriers, called PINs, named after their founding member PINFORMED1 (Galweiler et al. 1998; Petrásek et al. 2006). PIN localization on the cell membrane indicates the direction of auxin movement and mutations in PINs result in abnormal auxin transport (reviewed in Paponov et al. 2005). For instance, PIN7 is localized on the apical membrane of suspensor cells and expression of a synthetic auxin-dependent transcriptional reporter, DIRECT REPEAT5 (DR5), is expressed only in the apical lineage from the first zygotic division to the eight-cell stage (Fig. 2C) (Friml et al. 2003). These data suggest auxin flow is directed from basal to apical cells to set up the embryo axis. In support of this hypothesis, mutations in multiple PIN genes result in strong embryonic phenotypes, such as embryos without apparent polarity or suspensor-like structures that lack a proembryo (Friml et al. 2003). Additionally, mutations in either of two auxin response genes, MONOPTEROS (MP) and BODENLOS (BDL), result in abnormal orientation of apical cell division planes (Berleth and Jürgens 1993; Hamann et al. 1999). These results suggest that asymmetric auxin transport and response is required for establishment or maintenance of polarity after the zygotic division and specifying embryonic fate in the apical lineage. How the asymmetric distribution of auxin is initially established is unknown.
Putting it all Together in the Early Embryo
The phenotypic similarity between wox8 wox9 embryos and embryos with mutations in multiple PIN genes suggests a link between WOX transcriptional regulation and auxin movement or signaling. WOX8/WOX9 are predicted to regulate a signal, possibly auxin, which moves from the basal to the apical lineage and is required for apical development (Breuninger et al. 2008). Indeed, auxin flux (measured by PIN localization) and auxin response (assayed by DR5 expression) are defective in wox8 wox9 embryos. However, it is unclear whether WOX transcription factors directly regulate auxin movement or response (Breuninger et al. 2008). Interestingly, the role for auxin in specification of embryonic fate in the apical lineage appears to be complementary to YDA's role in specification of suspensor fate in the basal lineage. This suggests that the auxin and YDA signaling pathways function independently of each other (Lukowitz et al. 2004). Together these data indicate that asymmetric division is not sufficient for development of the apical and basal lineages, instead it appears that intercellular signaling is required to maintain the distinct cell fate of each lineage after the first zygotic division.
ASYMMETRIC CELL DIVISIONS IN STOMATAL DEVELOPMENT
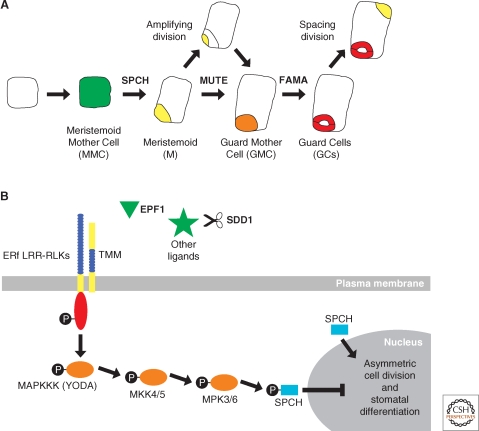
Another example of symmetry breaking in plants can be seen in the asymmetric cell divisions of the epidermis that generate stomata. Stomata are pores in the leaf epidermis that function in gas and water exchange. In Arabidopsis, formation of stomata is initiated by a dispersed population of multipotent stem cells, called meristemoid mother cells (MMCs). First, an MMC divides asymmetrically to form two cells of unequal size (Fig. 3A). The larger cell acts as a spacer between adjacent stomata, whereas the smaller cell, called a meristemoid (M), undergoes several asymmetric cell divisions to generate more meristemoids before becoming a guard mother cell (GMC) (Bünning 1953; Rasmussen 1981; Geisler et al. 2000; Bergmann and Sack 2007). Finally, the GMCs divide symmetrically to produce two guard cells (GCs), which act as valves surrounding a pore (Fig. 3A) (Pant and Kidwai 1967; Geisler et al. 2000; Bergmann and Sack 2007). Thus, stomata are formed progressively through a series of initially asymmetric and a final symmetric cell division.
Figure 3.
Asymmetric cell division in stomatal development. (A) Schematic of a cell progressing through stomatal development. (A, from left to right) An undifferentiated leaf epidermal cell in Arabidopsis acquires Meristemoid Mother Cell fate (MMC, green) before undergoing an asymmetric cell division to produce a Meristemoid (M, yellow) and a larger sister cell (white). The M cell may then go through a series of asymmetric cell divisions, called amplifying divisions, before differentiating into a Guard Mother Cell (GMC, orange). The GMC divides symmetrically to produce a pair of Guard Cells (GCs, red) that together comprise a stomate. This process may reiterate when the larger sister cell divides asymmetrically in a spacing division to produce another M cell that is separated from existing stomata by one cell. The intrinsic factors SPCH, MUTE, and FAMA act sequentially to regulate asymmetric cell division and stomatal development. (B) Extrinsic factors and the signaling pathway proposed to negatively regulate stomatal development. First, a ligand binds to the leucine-rich repeats (LRR, blue) of a putative heterodimer complex between TMM and one of the ERf LRR-RLKs, which possess an intracellular kinase domain (red). This interaction is thought to initiate a cascade of phosphorylation events (black circles) involving downstream MAPK signaling proteins (orange), including YODA. Candidate ligands (green) acting to trigger this signaling cascade include small peptides such as EPF1 and putative unknown proteins that may be processed by SDD1. Biochemical evidence for this model is lacking, except for the final phosphorylation of SPCH by MPK3 and MPK6. SPCH phosphorylation ultimately results in repression of asymmetric cell division and stomatal differentiation. In contrast, when SPCH is unphosphorylated, it promotes asymmetric cell division and meristemoid fate, as shown in (A).
In Arabidopsis, these coordinated divisions result in stomates that are separated from each other by an intervening epidermal cell (Sachs 1978, 1991; Geisler et al. 2000). This one-cell spacing pattern is derived from an asymmetric cell division, called a spacing division, that occurs in a cell that borders an existing M, GMC, or stomate (Fig. 3A) (Geisler et al. 2000). For example, when an MMC is flanked by a cell that is already progressing through stomatal development, a new division plane is oriented such that the resulting meristemoid is separated from existing stoma by the larger sister cell (Sachs 1978, 1991; Larkin et al. 1997; Croxdale 2000). Thus, new meristemoid cells are always physically separated from each other. The orientation and placement of the division plane in the MMC strongly suggests the involvement of extrinsic cues from neighboring cells rather than from intrinsic cues (Geisler et al. 2000).
Cell–cell Signaling and Breaking of Symmetry in the Epidermis
Cell–cell signaling was further implicated in coordinating asymmetric cell divisions of the stomatal lineage when the gene corresponding to the too many mouths (tmm) mutation was found to encode a leucine-rich repeat receptor-like protein (LRR-RLP). LRR-RLPs are similar to transmembrane receptor kinases but lack an intracellular kinase domain (Geisler et al. 2000; Nadeau and Sack 2002). In tmm mutants, the stomata are positioned randomly relative to one another because of a failure to properly orient asymmetric cell divisions. As a consequence, tmm stomata are frequently in direct contact because the spacing divisions are not properly executed (Geisler et al. 2000). Based on these data, it was hypothesized that TMM transduces extracellular signals in a manner similar to that proposed for other LRR-RLPs (Jeong et al. 1999; Trotochaud et al. 1999; Nadeau and Sack 2002; Dievart and Clark 2004; Bergmann and Sack 2007). Specifically, TMM signal transduction would occur through heterodimerization of TMM and another transmembrane protein with an intracellular signaling domain, such as a receptor-like kinase (RLK). As a complex, these proteins are predicted to perceive and transduce extracellular signals controlling the orientation and number of asymmetric cell divisions (Fig. 3B).
The discovery that stomatal spacing defects occur in plants with mutations in the ERECTA gene family (ERf), which encode LRR-RLKs, provided support for this hypothesis and candidate interaction partners for TMM (Masle et al. 2005; Shpak et al. 2005). Furthermore, expression and mutant analyses indicate that TMM and ERf are part of a genetic pathway in which TMM prevents signaling through ERECTA-like1 (ERL1) upon heterodimerization (Shpak et al. 2005). Additionally, mutations in YODA (YDA), a MAPKKK gene, are phenotypically similar to ERf mutants. Genetic analyses suggest that YDA acts downstream of TMM, as gain of function mutations in YDA do not produce stomata and can repress the clustered stomata observed in tmm mutants (Bergmann et al. 2004; Shpak et al. 2005). This indicates that MAP kinase (MAPK) signaling is involved in asymmetric cell division during stomatal development, in addition to its role in embryogenesis (as previously discussed). A signaling cascade downstream of YDA was subsequently identified that involves MAP kinase kinase4 (MKK4) and MKK5 followed by the MAP kinase3 (MPK3) and MPK6 (Wang et al. 2007). Double mutants and/or RNA interference knockdown of both MKK4 and MKK5 or MPK3 and MPK6 resulted in an epidermis composed entirely of stomata indicating a severe disruption in both regulated asymmetric divisions and cell specification. These data led to a hypothesis that a signaling cascade beginning at the membrane with TMM and ERf followed by YDA, MKK4/MKK5, and MPK3/MPK6, sequentially phosphorylate downstream targets to regulate both asymmetric cell divisions and cell fate specification in the stomatal lineage (Fig. 3B) (Wang et al. 2007; Lampard et al. 2008).
The candidate extrinsic factors that trigger this signaling cascade appear to be small proteins. A subtilisin-like serine protease encoded by the STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) gene is important for the proper orientation and number of asymmetric cell divisions in stomatal patterning (Berger and Altmann 2000; von Groll et al. 2002). SDD1 may function to process protein ligand(s) involved in signaling upstream of TMM (Berger and Altmann 2000; von Groll et al. 2002). Secondly, a small-secreted peptide called EPIDERMAL PATTERNING FACTOR 1 (EPF1) has been shown to be involved in stomatal patterning (Hara et al. 2007). EPF1 and SDD1 do not appear to interact genetically, suggesting SDD1 is not involved in the processing of EPF1 (Fig. 3B) (Hara et al. 2007). It is tempting to speculate that EPF1 provides positional cues to inform subsequent asymmetric divisions perhaps by a means similar to extracellular gradients as observed in animal systems. However, it is unknown whether a gradient exists, whether or not EPF1 binds to any of the aforementioned receptors, or is the only ligand involved in breaking symmetry in the epidermis.
Intrinsic Factors Involved in the Asymmetric Cell Divisions of the Stomatal Lineage
Many intrinsic factors acting in asymmetric cell divisions of the stomatal lineage are transcription factors of the basic helix-loop-helix (bHLH) class. Three bHLHs, SPEECHLESS (SPCH), MUTE, and FAMA, appear to act sequentially in stomatal formation starting with asymmetric cell divisions and progressive cell fate specification, and ending with stomatal differentiation (Fig. 3A) (MacAlister et al. 2007; Pillitteri et al. 2007; Kanaoka et al. 2008; Nadeau 2009). SPEECHLESS (SPCH) is required first as asymmetric cell divisions and expression of early stomatal identity markers are not observed in spch mutants. This suggests SPCH promotes the initial breaking of symmetry in the epidermis and may promote the transition to meristemoid fate. Loss-of-function mutants in a second transcription factor, MUTE, exhibit excessive asymmetric cell divisions in the epidermis and the resulting cells fail to differentiate into stomata. MUTE expression is not detected in spch mutants, suggesting that it acts downstream of SPCH to arrest asymmetric cell divisions and allows the transition to guard mother cell fate (MacAlister et al. 2007; Pillitteri et al. 2007). A third bHLH, FAMA, promotes the transition to guard cell fate. FAMA expression is not detected in spch and mute mutants, also suggesting that it acts downstream of SPCH and MUTE (MacAlister et al. 2007; Pillitteri et al. 2007). Thus, the sequential action of SPCH and MUTE appears to promote and regulate the breaking of symmetry in the epidermis and together these three bHLHs specify the resulting daughter cells.
This model is somewhat oversimplified as other intrinsic components have been identified, including several that interact with SPCH, MUTE, and FAMA. Four other bHLHs and two R2R3 MYB-type transcription factors (TFs) have recently been implicated as intrinsic factors involved in stomatal patterning and cell specification. The expression pattern and mutant phenotype of FAMA resemble that of the R2R3 MYB-type TF FOUR LIPS (FLP) (Lai et al. 2005; Ohashi-Ito and Bergmann 2006). Although plant R2R3 MYB-type TFs have been shown to heterodimerize with bHLHs, FAMA does not heterodimerize with FLP (Lee and Schiefelbein 1999; Zimmermann et al. 2004; Ohashi-Ito and Bergmann 2006). Instead, FAMA interacts genetically, in vitro, and in planta with two additional bHLHs, bHLH071 and bHLH093 (Ohashi-Ito and Bergmann 2006). The important role of bHLH dimerization in symmetry breaking is supported by the recent demonstration that the bHLH proteins SCREAM (SCRM)/ICE1 or SCREAM2 (SCRM2) heterodimerize in vitro and in vivo with SPCH, MUTE, and FAMA. In scrm/ice1 scrm2 double mutants, the epidermis consists only of fully differentiated epidermal cells, just like spch mutants (Kanaoka et al. 2008). Taken together, these data indicate that the combinatorial and sequential action of intrinsic factors, such as bHLHs, is integral to the regulation of asymmetric cell division in the stomatal lineage.
A Connection between Cell–cell Signaling and Intrinsic Factors in Stomatal Formation
A link between extrinsic and intrinsic factors involved in breaking symmetry in the stomatal lineage is predicted based on genetic experiments that show spch is epistatic to yda (MacAlister et al. 2007; Lampard et al. 2008). However, the targeted phosphorylation of intrinsic factors like SPCH by a MAPK signaling cascade, including factors like YDA, was regarded as dubious because MAPK signaling is common to many pathways in plant development (Lampard et al. 2008). Despite this, the receptor/MAPK signaling system was recently shown to modulate SPCH in vivo through phosphorylation that appears to repress SPCH activity (Fig. 3B) (Lampard et al. 2008). The region of SPCH targeted for phosphorylation is not found in any other Arabidopsis protein, but is common to SPCH-related proteins in other plant species. This suggests an evolutionarily conserved mechanism for regulation of SPCH activity in a specific cell type, the stomata (Lampard et al. 2008). This shows a concrete connection between the cell–cell signaling pathway and the sequential action of TFs in the symmetry-breaking events of the stomatal lineage.
ASYMMETRIC DIVISION IN THE CORTEX-ENDODERMAL CELL LINEAGE
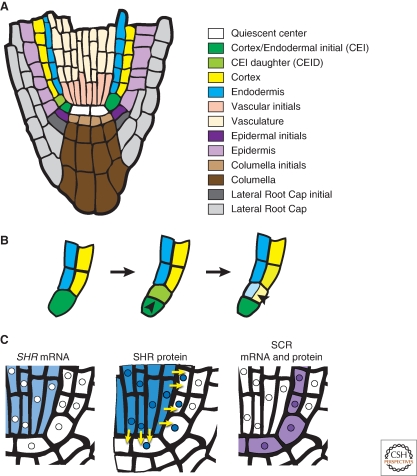
In the Arabidopsis root, an asymmetric cell division generates the two cell layers of the ground tissue. The ground tissue is located between the vasculature and epidermis and is comprised of two distinct cell types: the cortex and the endodermis (Fig. 4A). These two cell types are generated through asymmetric cell division of a single initial (stem) cell. The cortex/endodermal initial (CEI) undergoes a transverse asymmetric division to maintain the CEI and generate a CEI daughter (CEID). The CEID then divides asymmetrically, in a longitudinal orientation, to produce one cell for each ground tissue layer (Fig. 4B). Thus the asymmetric division of a single cell generates two morphologically and molecularly distinct cell types (Benfey et al. 1993; Dolan et al. 1993; Scheres et al. 1994; Di Laurenzio et al. 1996).
Figure 4.
Symmetry breaking in ground tissue formation in the root. (A) Schematic of a longitudinal section of the Arabidopsis root tip with each cell type differentially colored. (B) Magnification of (A) focusing on the asymmetric cell divisions that generate the two cell layers of the ground tissue. (B, left) The CEI expands and then (B, center) divides transversely to regenerate itself and produce the CEID. (B, right) The CEID then divides longitudinally to generate the cells of the endodermis and cortex. (C) Schematic representation of the localization of SHR mRNA, SHR protein and SCR mRNA and protein. Yellow arrows depict the movement of SHR protein from the vasculature into the quiescent center, CEI, and endodermis. Note that SHR and SCR proteins are colocalized in the nuclei of these cell types. Nuclei are represented by small circles within the cells.
The asymmetric division of the CEID is regulated through activity of two transcription factors, SHORT‐ROOT (SHR) and SCARECROW (SCR). SHR and SCR are members of the GRAS family of transcriptional regulators and were found in a genetic screen to identify molecules with roles in radial patterning of the root (Benfey et al. 1993; Di Laurenzio et al. 1996; Pysh et al. 1999; Helariutta et al. 2000). Both shr and scr mutants each have only a single layer of ground tissue. In shr mutants, the CEID fails to undergo the longitudinal division resulting in a single ground tissue layer with only cortex features. In scr mutants, the CEID also fails to divide longitudinally, however, the resulting single layer of ground tissue has features of both the endodermis and cortex. These data suggest that SCR is required for the CEID division, whereas SHR is required both for CEID division and specification of endodermal cell fate.
SHR Reveals a Novel Mechanism to Regulate Symmetry Breaking
Differences in SHR mRNA and SHR protein localization suggested a novel mechanism for symmetry breaking in CEI cells. SHR mRNA is restricted to the vasculature (internal to the ground tissue), whereas SHR protein is found both in the vasculature and the adjacent cell layer (Helariutta et al. 2000; Nakajima et al. 2001). This adjacent cell layer includes the quiescent center, CEI, CEID, and endodermis (Fig. 3A,C) (Benfey et al. 1993; Dolan et al. 1993). Importantly, SHR protein is specifically required in the adjacent layer for asymmetric CEID division and endodermal specification (Nakajima et al. 2001; Sena et al. 2004; Cui et al. 2007). This is achieved though the regulated movement of SHR protein from the vasculature into the adjacent cells (Gallagher et al. 2004; Gallagher and Benfey 2009). Moreover, ectopic expression of SHR beyond the vasculature and endodermis results in supernumerary endodermal cell layers (Helariutta et al. 2000; Nakajima et al. 2001). The formation of these extra layers is dependent on SCR expression (Nakajima et al. 2001; Cui et al. 2007). These data indicate that SHR localization, together with SCR, are required for asymmetric cell division and endodermal specification.
In vivo molecular evidence supporting this conclusion was recently reported. First, SHR and SCR bind to the SCR promoter in chromatin immunoprecipitation experiments (Levesque et al. 2006; Cui et al. 2007). Secondly, SHR and SCR proteins interact as shown by coimmunoprecipitation experiments. Additionally, SHR and SCR have common transcriptional targets suggesting that they form a regulatory complex (Cui et al. 2007). Together these data have led to the following model for asymmetric CEID division and endodermal specification: SHR moves from the vasculature into the adjacent cell layer where it interacts with SCR and is sequestered into the nucleus. In the nucleus, SHR and SCR activate transcription of SCR and other shared downstream targets which function in the asymmetric division of the CEID. Additionally, nuclear sequestration confines SHR to the first cell layer adjacent to the vasculature, thereby restricting endodermal cell fate specification to a single layer (Cui et al. 2007).
One prediction of this model is that a sufficient amount of SCR is required to block SHR movement beyond the endodermis. A positive feedback mechanism by the SHR/SCR complex on SCR expression was proposed to achieve sufficient SCR levels to prevent further SHR movement. To test this, RNA interference was used to reduce SCR expression below a threshold level. Examination of plants with reduced SCR mRNA levels revealed SHR movement into additional layers. This resulted in activation of SCR and consequently additional asymmetric cell divisions and endodermal layers in these plants (Cui et al. 2007). These data show the importance of a SHR/SCR positive feedback loop in generating sufficient SCR quantities to restrict SHR movement to a single cell layer. Recent evidence has implicated additional players in the SHR/SCR model for regulation of asymmetric cell division, such as the zinc finger proteins JACKDAW, MAGPIE, and NUTCRACKER (Welch et al. 2007). Thus the proposed model for regulating symmetry breaking in the CEID is likely more complicated, involving the action of additional transcriptional regulators. In summary, a novel molecular mechanism of intracellular transcription factor movement regulates both the asymmetric division of the CEID and subsequent endodermal cell fate specification.
CONCLUDING REMARKS
These examples of asymmetric cell divisions in Arabidopsis indicate plant cells use both intrinsic and extrinsic mechanisms to regulate symmetry-breaking events. Positional information (extrinsic cues) has been previously shown to be important in plant development by clonal analysis and laser ablation studies (Irish and Sussex 1992; van den Berg et al. 1995, 1997; Kidner et al. 2000). Here, the prevalence of signaling pathways that include auxin, transmembrane and MAPK kinases, and protein movement indicate the significant role of extrinsic factors in symmetry breaking. Asymmetric cell divisions in plants also involve intrinsic factors such as the WOX, bHLH, and GRAS transcription factors. Recent work has begun to provide links between intrinsic and extrinsic factors suggesting that their cooperative function is necessary for asymmetric cell divisions. We have highlighted three examples of symmetry-breaking events that show this cooperation in embryogenesis, formation of stomata, and ground tissue formation. Below we compare and contrast these Arabidopsis examples with those found in metazoans.
Intrinsic and Extrinsic Factors in Embryonic Asymmetry
In the zygote and early embryo, both intrinsic and extrinsic factors appear to be required for the asymmetric zygotic division that establishes the plant body plan. Intrinsic factors, such as localized expression of transcription factors, are important for asymmetry in early animal embryo development and may also be important in the plant embryo. Well-known examples in animals include the homeobox (Hox) proteins. Hox genes are expressed in specific domains and are required for normal anterior–posterior patterning and cell type specification in both vertebrate and invertebrate embryos (Krumlauf 1994; Hughes and Kaufman 2002). The finding that homeobox family members in plants, the WOX proteins, also function in embryo patterning may suggest an evolutionarily important role for these proteins in establishment of asymmetry early in development (Mayer et al. 1991; Haecker et al. 2004). Despite the significant differences in plant and animal embryogenesis, a common mechanism may exist that involves differential localization of transcription factors after the first zygotic division.
Two lines of evidence support the role of extrinsic factors in asymmetry in the plant embryo. First, the defective asymmetric zygotic division in yda mutant embryos suggests that embryonic asymmetry may rely on external cues perceived and relayed by a MAPKKK-containing signaling pathway. Second, auxin signaling and transport are also implicated in maintaining asymmetry in the embryo; however, the site of auxin synthesis and how it is regulated remains unknown. Together these data implicate both intrinsic and extrinsic factors in breaking symmetry in the embryo. Future work will be necessary to identify additional players and elucidate the links between these extrinsic and intrinsic factors.
Cooperation of Intrinsic and Extrinsic Factors in Symmetry Breaking in the Epidermis
In stomatal formation, intrinsic factors interact in a consecutive and likely combinatorial fashion to regulate asymmetric cell division and cell fate specification. The action of these intrinsic factors appears to be controlled by extrinsic factors through a signal transduction cascade. This is similar to the formation of animal neurons and muscle fibers, where extrinsic signals regulate closely related bHLH proteins that act in a hierarchical fashion (Olson 1990, 1992; Weintraub et al. 1991; Campuzano and Modolell 1992; Jan and Jan 1993). Although there is similarity between bHLH action in plant stomatal and animal neural and muscle development, the evolutionary history of bHLH proteins suggests this relationship is distant. Plant bHLH proteins are believed to be part of an ancestral bHLH group, whereas many bHLHs involved in neurogeneis and myogenesis likely arose later in evolutionary history to perform animal specific functions (Atchley and Fitch 1997; MacAlister et al. 2007). These results suggest that plant and animal evolution may have arrived at a similar mechanism to regulate symmetry-breaking events.
The Distinction between Intrinsic and Extrinsic Factors is Blurred in Ground Tissue Formation
Asymmetric cell division and subsequent endodermal cell specification due to the movement of the transcription factor SHR constitutes a novel mechanism for symmetry breaking. The model for restricting SHR movement to a single cell layer requires SCR-mediated sequestration into the nucleus. SCR is downstream of SHR and a positive feed-forward loop is required to generate sufficient SCR to restrict SHR movement. In plants, the movement of several other developmentally important transcription factors beyond their transcriptional domains has been documented (Lucas et al. 1995; Perbal et al. 1996; Sessions et al. 2000; Wada et al. 2002; Kim et al. 2003). The requirement for movement of transcription factors in development may be a plant-specific mechanism that evolved as a byproduct of restricted cell movement. The intercellular movement of proteins customarily defined as intrinsic factors stimulates the provocative notion of one cell's intrinsic factor acting as an extrinsic factor in a neighboring cell.
Perspectives and Future Directions
Plants and animals represent the two main outgroups of multicellular eukaryotes; it is intriguing to contemplate the similarities and differences in their strategies to accomplish a fundamental process such as asymmetric cell division. As we learn more about the molecular mechanisms regulating symmetry breaking in plants, we can begin to draw interesting parallels to animal models. For example, partitioning of transcripts before cell division in animals is well documented, yet how WOX2/WOX8 transcripts in plants become differentially localized (or expressed) is unknown. On the other hand, because plants and animals independently evolved multicellularity, comparable molecular mechanisms regulating symmetry breaking must have also evolved independently. Additionally, there may be more to learn from the differences than the similarities. In the case of a transcription factor moving to regulate an asymmetric cell division, there is not a comparable example in animal systems. Future work may reveal whether this is a universal or plant-specific mechanism to regulate asymmetric cell division. Positioning of the division plane is another area of future interest. In plants, a clear functional link between the cell division machinery and the molecular mechanisms regulating asymmetric cell division has not been established. For example, in the mutants discussed in this article it is unclear if the defects are in asymmetric cell division, the orientation of the axis of symmetry relative to spatial cues, cell fate specification, or a combination of these. Continued work is required to distinguish between these possibilities as well as to determine just how far we can extend the apparent similarities and differences in regulated asymmetric cell divisions in plants and animals.
ACKNOWLEDGMENTS
We thank members of the Benfey Laboratory for critical reading of the article. Work in PNB's laboratory on root radial patterning is funded by grants from the National Institute of Health (NIH) RO1GM043778 and 1P50-GM081883. J.J.P. and J.M.V.N. contributed equally to this work.
Footnotes
Editors: Rong Li and Bruce Bowerman
Additional Perspectives on Symmetry Breaking in Biology available at www.cshperspectives.org
REFERENCES
- Alessa L, Kropf DL 1999. F-actin marks the rhizoid pole in living Pelvetia compressa zygotes. Development 126:201–209 [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM 1997. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci 94:5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol 14:R806–818 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA 1993. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 119:57–70 [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T 2000. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14:1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD 2007. Stomatal development. Annu Rev Plant Biol 58:163–181 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR 2004. Stomatal development and pattern controlled by a MAPKK kinase. Science 304:1494–1497 [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G 1993. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118:575–587 [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T 2008. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14:867–876 [DOI] [PubMed] [Google Scholar]
- Bünning E 1953. Entwicklungs- und Bewegungphysiologie der Pflanzen Springer Verlag, Berlin, Germany [Google Scholar]
- Campuzano S, Modolell J 1992. Patterning of the Drosophila nervous system: The achaete-scute gene complex. Trends Genet 8:202–208 [DOI] [PubMed] [Google Scholar]
- Croxdale JL 2000. Stomatal patterning in angiosperms. Amer J Bot 87:1069–1080 [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316:421–425 [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE 2004. LRR-containing receptors regulating plant development and defense. Development 131:251–261 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86:423–433 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119:71–84 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426:147–153 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN 2009. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J 57:785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN 2004. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14:1847–1851 [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282:2226–2230 [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD 2000. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12:2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable WE, Kropf DL 2000. Sperm entry induces polarity in fucoid zygotes. Development 127:493–501 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131:657–668 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G 1999. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126:1387–1395 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergman DC, Kakimoto T 2007. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21:1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101:555–567 [DOI] [PubMed] [Google Scholar]
- Homble F, Leonetti M 2007. Emergence of symmetry breaking in fucoid zygotes. Trends Plant Sci 12:253–259 [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I 1992. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell 68:237–255 [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC 2002. Hox genes and the evolution of the arthropod body plan. Evol Dev 4:459–499 [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM 1992. A fate map of the Arabidopsis embryonic shoot apical meristem. Development 115:745–753 [Google Scholar]
- Jan YN, Jan LY 1993. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell 75:827–830 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Barton MK 2005. Surge and destroy: The role of auxin in plant embryogenesis. Development 132:3577–3585 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W 2007. Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23:207–236 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11:1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G 2001. Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J 20:3609–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G 2005. Cytokinesis in higher plants. Annu Rev of Plant Biol 56:281–299 [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU 2008. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20:1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L 2000. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211:191–199 [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D 2003. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130:4351–4362 [DOI] [PubMed] [Google Scholar]
- Kropf DL 1992. Establishment and expression of cellular polarity in fucoid zygotes. Microbiol Rev 56:316–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropf DL 1997. Induction of polarity in fucoid zygotes. Plant Cell 9:1011–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R 1994. Hox genes in vertebrate development. Cell 78:191–201 [DOI] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, Lee E-K, Nakagawa T, Zhao L, Geisler M, Sack FD 2005. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17:2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, MacAlister CA, Bergmann DC 2008. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322:1113–1116 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Marks MD, Nadeau J, Sack F 1997. Epidermal cell fate and patterning in leaves. Plant Cell 9:1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J 1999. WEREWOLF, a MYB-related protein in Arabidopsis is a position-dependent regulator of epidermal cell patterning. Cell 99:473–483 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. 2006. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O 2006. Dynamic integration of auxin transport and signalling. Curr Biol 16:R424–433 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270:1980–1983 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C 2004. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116:109–119 [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC 2007. Transcription factor control of asymmetric divisions that establish the stomatal lineage. Nature 445:537–540 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG 1991. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69:461–476 [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD 2005. The ERECTA gene regulates plant transcription efficiency in Arabidopsis. Nature 436:866–870 [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Miséra S, Jürgens G 1991. Mutations affecting body organization in the Arabidopsis embryo. Nature 353:402–407 [Google Scholar]
- McCaffrey LM, Macara IG 2009. Widely conserved signaling pathways in the establishment of cell polarity. Cold Spring Harb Perspect Biol 1:a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineyuki Y 1999. The preprophase band of microtubules: Its function as a cytokinetic apparatus in higher plants. Int Rev of Cytol 187:1–49 [Google Scholar]
- Munro E, Bowerman B 2009. Cellular symmetry breaking during C. elegans development. Cold Spring Harb Perspect Biol 1:a003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA 2009. Stomatal development: New signals and fate determinants. Curr Opin Plant Biol 12:29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD 2002. Control of stomatal distribution of the Arabidopsis leaf surface. Science 296:1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413:307–11 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC 2006. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18:2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN 1990. MyoD family: A paradigm for development? Genes Dev 4:1454–1461 [DOI] [PubMed] [Google Scholar]
- Olson EN 1992. Interplay between proliferation and differentiation within the myogenic lineage. Dev Biol 154:261–272 [DOI] [PubMed] [Google Scholar]
- Pant DD, Kidwai PF 1967. Development of stomata in some Cruciferae. Ann Bot 31:513–521 [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K 2005. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci 10:170–177 [DOI] [PubMed] [Google Scholar]
- Perbal MC, Haughn G, Saedler H, Schwarz-Sommer Z 1996. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122:3433–3441 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. 2006. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312:914–918 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU 2007. Termination of asymmetric cell division and differentiation of stomata. Nature 445:501–505 [DOI] [PubMed] [Google Scholar]
- Prehoda KE 2009. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb Perspect Biol 1:a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN 1999. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18:111–119 [DOI] [PubMed] [Google Scholar]
- Rasmussen H 1981. Terminology and classification of stomata and stomatal development-A critical survey. Bot J Linn Soc 83:199–212 [Google Scholar]
- Sachs T 1978. The development of spacing patterns in the leaf epidermis. In The clonal basis of development, Subtelny S, Sussex IM eds New York: Academic Press, 161–183 [Google Scholar]
- Sachs T 1991. Pattern formation in plant tissues New York: Cambridge University Press [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P 1994. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120:2475–2487 [Google Scholar]
- Sena G, Jung JW, Benfey PN 2004. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131:2817–2826 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D 2000. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289:779–782 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Keiko TU 2005. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309:290–293 [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol 1:a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE 1999. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11:393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B 1995. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378:62–65 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390:287–289 [DOI] [PubMed] [Google Scholar]
- von Groll U, Berger D, Altmann T 2002. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14:1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K 2002. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129:5409–5419 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama Liu, Walker JC, Zhang S 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. 1991. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science 251:761–766 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B 2007. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21:2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chory J, Weigel D 2007. Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev Biol 309:306–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CS, Muller S, Meier I 2008. RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci 105:18637–18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34 [DOI] [PubMed] [Google Scholar]