Abstract
Plants start their life as a single cell, which, during the process of embryogenesis, is transformed into a mature embryo with all organs necessary to support further growth and development. Therefore, each basic cell type is first specified in the early embryo, making this stage of development excellently suited to study mechanisms of coordinated cell specification—pattern formation. In recent years, it has emerged that the plant hormone auxin plays a prominent role in embryo development. Most pattern formation steps in the early Arabidopsis embryo depend on auxin biosynthesis, transport, and response. In this article, we describe those embryo patterning steps that involve auxin activity, and we review recent data that shed light on the molecular mechanisms of auxin action during this phase of plant development.
Auxin has emerged as a major regulator of pattern formation in the plant embryo. By activating transcriptional regulators, this hormone promotes the specification of various cell fates.
ESTABLISHMENT OF THE EMBRYO BODY PLAN
All plants start as a zygote and generate a mature embryo with one or two cotyledons, an embryonic stem, and embryonic root. However, the trajectories of cell division leading to the final shape can be dramatically different between species (Johri et al. 1992). As the cell division patterns, and hence cell ontogeny relationships are particularly regular in Arabidopsis, this species has been used as a model for most embryogenesis research. For this reason, we focus our discussion on Arabidopsis.
Two axes are formed during embryogenesis: The apical–basal (upper–lower) and radial (outer–inner) axes, and each serves as a reference for postembryonic development. The future apical–basal axis is already apparent before fertilization by the intrinsic polarity of the egg cell (Mansfield and Briarty 1991; Laux and Jürgens 1997). This cell has its nucleus and most of the cytoplasm at one side of the cell and a large vacuole at the opposite end. After fertilization, the zygote elongates and divides asymmetrically in a smaller apical cell and a larger, highly vacuolated basal cell. The apical cell goes through two rounds of longitudinal divisions followed by a transverse division to form the eight-cell proembryo. A number of transverse divisions of the basal cell produce a cell file called the suspensor (Fig. 1).
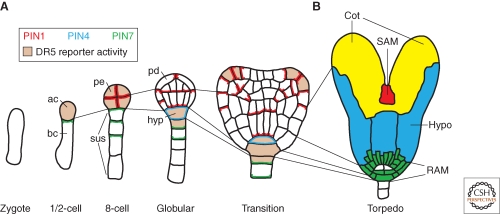
Figure 1.
Cell lineages, PIN protein localization, and auxin response maxima during Arabidopsis embryogenesis. Arabidopsis embryos follow a regular and stereotypical cell division pattern. Lineages are indicated by thin lines between individual stages. PIN protein localization at membranes is marked with red (PIN1), blue (PIN4), and green (PIN7) lines and DR5 reporter activity is indicated by pink color. (A) After division of the zygote, the one-cell embryo (same for two-cell embryos) expresses PIN7 in the basal daughter cell (bc), with the protein on the side of the apical cell (ac), which expresses the DR5 reporter. After two more cell-division rounds, all proembryo (pe) cells express PIN1 without apparent polarity and show DR5 reporter activity. Basal suspensor (sus) cells express PIN7, which is polarly localized on the proembryo side. At the globular stage, basal PIN1 polarity is established in the central lower cells of the proembryo, whereas PIN1 localizes apically in outer protoderm (pd) cells. At the same time, PIN7 polarity reverses in suspensor cells and PIN4 is activated in the uppermost suspensor cell. This cell now expresses the DR5 reporter and is specified as hypophysis (hyp). During the transition stage, PIN1 polarity at the flanks of apical embryo half converges in adjacent cells accompanied by the appearance of new DR5 maxima. These sites mark the initiation of the cotyledons. (B) At the torpedo stage, primordia for each of the seedling organs can be distinguished. Discrete regions of the embryo give rise to the root apical meristem (RAM, green; note, white cells correspond to the future quiescent center), the hypocotyl (hypo, blue), the cotyledons (cot, yellow), and the shoot apical meristem (SAM, red).
At the eight-cell stage, three regions can be distinguished along the apical–basal axis: The upper tier of the proembryo, the lower tier of the proembryo, and the extraembryonic suspensor cells. The apical tier of the proembryo will give rise to the shoot meristem and most of the cotyledons, whereas the lower tier of the proembryo will form the abaxial part of the cotyledons, the hypocotyl, root, and root meristem initials. The suspensor pushes the embryo into the lumen of the ovule and serves as a connection between the developing embryo and the maternal tissues.
The radial axis emerges when cells of the eight-cell proembryo divide periclinally to give rise to the protoderm and ground tissue lineages (Jürgens 1995). The proembryo now consists of 16 cells and is at the dermatogen stage. At the 32-cell globular stage, the uppermost suspensor cell, called the hypophysis, divides asymmetrically to form an upper lens-shaped cell and a lower cell. These cells will be incorporated into the embryo to form the quiescent center and the columella root cap cells, respectively. At the transition stage of embryogenesis, the radial symmetry of the apical region of the embryo changes into a bilateral symmetry when the cotyledons arise from the flanks of the apical domain. At approximately the same time, the shoot meristem is established between the emerging cotyledons.
MECHANISMS OF PATTERN FORMATION
Embryogenesis starts with a single cell, the zygote, and ends with a mature embryo in which all cells have acquired a specific fate. The organized division and specification of cells during embryogenesis is called pattern formation. The processes that generate all of these different cell types from the zygote remain largely unknown. The highly organized cell divisions observed during embryogenesis in Arabidopsis do not seem to be critically important for axis formation and cell fate specification, as embryos mutant for the FASS gene have a completely altered pattern of cell division but still contain all cell types that build the basic body plan, and show a relatively normal axis (Torres-Ruiz and Jürgens 1994). What mechanisms are involved in pattern formation? Conceptually, when focusing on those patterning steps that involve asymmetric cell divisions, two mechanisms of pattern formation can be envisioned (Laux and Jürgens 1997). First, a cell with intrinsic polarity divides to generate two different daughter cells. Given the strong polarity of the egg cell and the hypophysis, this scenario could well apply to the division of these cells. Alternatively, initially identical daughter cells acquire different identities after division. This type of pattern formation requires cell–cell communication to ensure that the position, rather than lineage, of a cell determines its fate. The latter mechanism could also work over longer distances, to pattern fields of cells. In this context, an ever-increasing body of evidence shows that the plant hormone auxin is required for pattern formation (see Jenik et al. 2007 for a recent review). Importantly, all processes required for auxin activity—biosynthesis and transport, auxin perception by its receptor, and auxin response—are each required for pattern formation. In this article, we review the patterning steps that require auxin activity, focusing on recent findings that illuminate the mechanism of auxin action.
AUXIN CONTROLS MAJOR CELL SPECIFICATION EVENTS DURING EMBRYOGENESIS
Mutations in genes involved in auxin biosynthesis, transport, and response all result in embryo defects that can be grouped in a number of distinct phenotypes. These phenotypes reveal specific cell divisions and cell specification events that are controlled by auxin during embryogenesis and are summarized in Figure 2. The auxin-related phenotypes occur around three developmental stages. The first anomaly observed in a number of auxin-related mutants is the aberrant transverse division of the apical daughter cell just after zygote division. The second patterning step affected in auxin mutants is hypophysis division at the 32-cell stage. At the same time, the vascular precursor cells of the proembryo divide irregularly in a subset of auxin signaling mutants. Finally, from the transition stage onwards, the initiation, outgrowth, and correct separation of cotyledons is affected in many auxin mutants. The hypophysis defect often results in rootless seedlings, whereas the cotyledon initiation and separation defects result in seedlings with fused cotyledons, an aberrant number of cotyledons, asymmetric cotyledons, or the complete absence of cotyledons.
Figure 2.
Distinctive embryo phenotypes of auxin mutants. Characteristic defects of auxin-related mutants at three different stages of embryo development. The figures show examples of defects (red) that can occur at each developmental stage. (A) Transverse instead of longitudinal division of the apical cell at the one-cell stage. (B) At the globular stage, two phenotypes can be distinguished. The hypophysis divides aberrantly (left) resulting in a rootless seedling. In addition, vascular divisions may be incorrect. (C) At the heart stage, embryo cotyledon formation and/or separation are impaired, resulting in cotyledon fusion, an aberrant cotyledon number, or complete absence of cotyledons. Other auxin-related mutants have both cotyledon and root meristem defects. The root meristem defects in such mutants are the result of aberrant hypophysis division except for the plt1 plt2 plt3 plt4 mutant (*) in which the hypophysis descendants divide abnormally.
Auxin concentrations can not be visualized directly, which greatly hampers the understanding of auxin-dependent pattern formation. However, auxin induces the expression of the gene expression reporter DR5. Despite its shortcomings related to the limited sensitivity and the fact that auxin-dependent gene repression can not be visualized, DR5 reporter activity can be used to infer sites of auxin activity during embryogenesis (Friml et al. 2003) (Fig. 1). DR5 is first expressed in the apical cell after zygote division. DR5 expression rapidly increases in the descendants of the apical cell until the 32-cell stage, then the maximum of DR5 activity shifts basally into the uppermost suspensor cells, including the hypophysis. At the transition stage of embryogenesis, DR5 maxima appear at the flanks of the apical domain where the cotyledons initiate. The sites of DR5 activity align very well with the defective cell divisions in auxin mutants (Fig. 2), suggesting that the DR5 reporter does indeed reveal auxin responses that are relevant for pattern formation in the embryo.
In summary, regulated auxin maxima and activity are required for the organization of both the apical and basal embryo domains and are therefore critical for the establishment of the embryo pattern.
ROLE OF AUXIN BIOSYNTHESIS IN EMBRYO DEVELOPMENT
Auxin is synthesized from indole via tryptophan (Trp), or independent of tryptophan (Tao et al. 2008). Of the two, the Trp-dependent route is best understood, and bifurcates into at least three routes (Fig. 3A). Biosynthetic enzymes in two of these routes have nonredundant roles in embryogenesis. TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA1) and its closest homologs TRYPTOPHAN AMINOTRANSFERASE RELATED (TAR)1 and 2 function in one of these two Trp-dependent indole 3-acetic-acid (IAA) biosynthesis routes (Stepanova et al. 2008; Tao et al. 2008). The other branch of the Trp-dependent IAA biosynthesis pathway is represented by the YUCCA (YUC) family of flavin monooxygenases. Arabidopsis has 11 YUC genes of which YUC1, 4, 10, and 11 have been shown to be expressed in the embryo in overlapping patterns (Cheng et al. 2007). TAR1 and the YUC1, 4, 10, and 11 genes are all expressed from the globular stage on in the apical embryo region. Embryo defects in the yuc1 yuc4 yuc10 yuc11 quadruple mutant and in the taa1 tar1 tar2 triple mutant occur as early as the globular stage when the hypophysis divides abnormally. Seedlings do not have a root, a strongly reduced or no hypocotyl, and most of the seedlings have only one cotyledon.
Figure 3.
Auxin signaling involves auxin biosynthesis, transport, perception, and response. (A) In Arabidopsis, the auxin IAA (indole-3-acetic acid) is synthesized from indole through tryptophan or in a tryptophan-independent way. Biosynthetic enzymes in two tryptophan-dependent routes of IAA biosynthesis—the TAA and YUC proteins—have nonredundant roles in embryo development. (B) Auxin transport is mediated by the polar membrane localization of the PIN proteins (straight arrows crossing membrane bilayer), which in turn is regulated by several factors. The endocytosis of PIN proteins from the membrane to endosomes is clathrin-dependent (a), whereas the targeting of PIN proteins to the apical membrane depends on a (so far unknown) ADP ribosylation factor-guanine exchange factor (ARF-GEF) and the kinase PID (b). The recycling of PIN proteins to the basal membrane depends on the ARF-GEF GNOM and the PP2A phosphatases (c). (C) When auxin is perceived by its receptor TIR1, the affinity for the Aux/IAA proteins increases. The Aux/IAAs are subsequently ubiquitinated and degraded by the 26S proteasome. The ubiquitination of Aux/IAAs in Arabidopsis involves a ubiquitin activating enzyme (E1, not shown), a ubiquitin conjugating enzyme (E2), and the ubiquitin ligase (E3) SCFTIR1 that consists of a cullin protein, an RBX protein, one of the Arabidopsis SKP1(ASK) proteins, and the F-Box protein TIR1. Under low auxin concentrations, the Aux/IAA proteins bind and inhibit the ARF proteins by recruiting the TPL corepressor. When auxin levels rise, the Aux/IAA proteins are ubiquitinated and degraded, thereby releasing the ARF proteins to exert their function as transcriptional activators or repressors.
Interestingly, mutations in either of the two IAA biosynthesis pathways cause similar embryonic phenotypes (Fig. 2), suggesting that both IAA biosynthesis routes are required to provide sufficient auxin for correct regulation of division patterns in the apical and basal region of the embryo. Although defects in yuc1 yuc4 yuc10 yuc11 quadruple and taa1 tar1 tar2 triple mutants are observed in both apical and basal patterning, the genes are most prominently, if not exclusively, expressed in the embryo apex. The basal embryo phenotype could be explained by low but significant YUC and TAA/TAR gene expression in the basal half of the embryo. Alternatively, and perhaps more likely, auxin produced in the apical half of the embryo may be transported to elicit auxin responses in the lower half of the embryo.
ROLE OF AUXIN TRANSPORT IN EMBRYOGENESIS
Auxin is unique among plant hormones as it has a dedicated transport system. The direction of auxin transport is determined by the asymmetric membrane localization of the efflux carriers, the PIN proteins. Even before the identification of the PIN proteins, it was shown that pharmacological inhibition of auxin transport interferes with normal embryo patterning in several plant species (Liu et al. 1993; Hadfi et al. 1998), demonstrating a role for auxin transport in embryo patterning. At least four PIN proteins are dynamically expressed during embryogenesis (Fig. 1) (Friml et al. 2003). Immediately after the division of the zygote, PIN7 is localized to the apical side of the basal cell and its derivatives, presumably driving auxin transport into the apical cell. At the 32-cell stage, PIN7 polarity reverses to the basal membranes of the suspensor cells, probably resulting in auxin transport into the suspensor cells. PIN1 is expressed without any polarity in the embryo from the one-cell to the 16-cell stage. At the 32-cell stage, PIN1 becomes polarly localized to the basal membranes in the provascular cells next to the hypophysis, and transports auxin into the hypophysis. At the transition-stage of embryogenesis, PIN1 becomes polarly localized towards the flanks of the apical embryo domain, which likely results in auxin maxima at these sites. The PIN4 protein is expressed in the hypophysis cell and after division, in its uppermost daughter cell. The expression of PIN3 starts relatively late at the heart stage in the columella precursors.
The direction of auxin flow predicted by PIN protein localization corresponds well to the expression pattern of the auxin response reporter, suggesting that auxin response maxima reflect the concentration of auxin, and that this concentration pattern follows from active transport. Indeed, pin7 mutant embryos are affected in the DR5 activity in the early embryo and display associated cell division defects, suggesting that a proper auxin distribution and response is required for correct cell specification in the early embryo. Loss of PIN functionality disrupts embryo formation in a quantitative manner: The more PIN proteins are lost, the stronger the embryo phenotype (Fig. 2).
Several factors are important for correct PIN gene expression and protein localization (Fig. 3B). Mutations that cause altered PIN gene expression affect the same patterning processes described previously in more or less predictable ways (Izhaki and Bowman 2007; Ploense et al. 2009). It was found that PIN proteins are not statically localized in the membrane, but rather cycle between membrane and intracellular vesicles through endo- and exocytosis. The endocytosis of PIN proteins from the plasma membrane to endosomes is clathrin-dependent (Dhonukshe et al. 2007; Dhonukshe et al. 2008). The recycling of PIN proteins from endosomes to the basal plasma membrane requires the ARF-GEF protein GNOM (Mayer et al. 1993; Geldner et al. 2003). Other ARF-GEFs control the recycling of proteins to the apical plasma membrane (Kleine-Vehn et al. 2008). The serine-threonine kinase PINOID (PID) and the PROTEIN PHOSPHATASE 2A (PP2A) antagonistically control the phosphorylation status of PIN proteins (Bennett et al. 1995; Benjamins et al. 2001; Friml et al. 2004; Michniewicz et al. 2007). Phosphoryated PIN proteins are targeted to the apical plasma membrane, whereas dephosphorylated PIN proteins are targeted to the basal plasma membrane. There are at least three PID homologs that are also expressed during embryogenesis (Cheng et al. 2008).
Interference with any of these processes results in abnormal PIN polarity and altered expression patterns of the auxin response reporter DR5. The phenotypes associated with mutants that affect PIN localization are very similar to the phenotypes of the auxin efflux carrier mutants (Fig. 2). Although mutation in each of these components has a different effect on PIN protein distribution, generally, defects correspond well with the predicted auxin mis-distribution that would follow. In summary, changes in PIN expression or localization result in an altered auxin distribution, which in turn causes defects in embryo specification.
AUXIN PERCEPTION
Auxin elicits gene expression responses by binding to the F-box protein TIR1, hence increasing the affinity of TIR1 for the Aux/IAA family of transcriptional inhibitors. When the auxin concentration is high, auxin resides in the binding pocket of TIR1 and serves as a molecular glue to bring the Aux/IAAs and the auxin receptor together (Dharmasiri et al. 2005a; Kepinski and Leyser 2005; Tan et al. 2007). Once bound to the auxin receptor, the Aux/IAAs are ubiquitinated and subsequently degraded by the 26S proteasome (Fig. 3C). In Arabidopsis, TIR1 functions in the SCFTIR1 complex, which consists of the F-Box-protein TIR1, the ring finger protein RBX1, the cullin protein CUL1, and one of the ASK proteins (Gray et al. 2002; Shen et al. 2002; Liu et al. 2004). Related-to-ubiquitin (RUB) modification of the CUL1 subunit of the SCF complex is important for its function (Dharmasiri et al. 2003) and involves the activity of a heterodimeric RUB activating enzyme composed of ECR1 and AXR1 or the redundantly acting AXL, a RUB conjugating enzyme RCE1 and the RBX1 protein of the SCF complex, which serves as a RUB ligase (Fig. 3C) (Gray et al. 2002; Dharmasiri et al. 2007).
Mutations in the TIR1 subunit of the SCFTIR1 complex are not affected in embryo development because of redundancy with the closely related AUXIN SIGNALING F-BOX PROTEINS (AFB) 1, 2, and 3 (Dharmasiri et al. 2005b). Expression of TIR1 and AFB1, 2, and 3 is detected throughout embryogenesis starting in the preglobular embryo. The tir1 afb1 afb2 afb3 quadruple mutant often fails to make a root and hypocotyl and frequently has only one cotyledon (Fig. 2). As expected, levels of Aux/IAA proteins like IAA12/BDL are increased in the tir1 afb2 afb3 triple mutant (described later).
The Arabidopsis genome encodes approximately 700 different F-box proteins (Gagne et al. 2002), of which four have been shown to bind to the Aux/IAA proteins. The other subunits of the SCF complex are not specific for auxin signaling. Nonetheless, all subunits of the SCF complex as well as the RUB modification of the cullin subunit are important for proper embryo development. The phenotypes of ubiquitin-ligase SCF complex mutants and mutants affecting RUB modification of CUL1 include defects in auxin regulated processes (Fig. 2). Collectively, these results show that auxin perception by its receptor and the subsequent degradation of Aux/IAAs are required for embryogenesis, and the defects observed when these components are inactive are the same as when biosynthesis or transport are impaired.
AUXIN RESPONSE IN THE EMBRYO IS MEDIATED BY AUXIN RESPONSE FACTORS
Auxin promotes the degradation of Aux/IAA proteins, short-lived nuclear proteins. When sufficiently abundant, Aux/IAA proteins bind to and inhibit the activity of another class of auxin response transcriptional regulators, the AUXIN RESPONSE FACTORS (ARFs). When auxin is perceived by its receptor, Aux/IAAs are degraded, thereby releasing the ARFs to exert their function as activators or repressors of transcription. Among the 23 ARFs in Arabidopsis, five have been shown to be transcriptional activators in carrot protoplasts, whereas the remaining 18 ARFs may act as repressors (Ulmasov et al. 1999; Tiwari et al. 2003). The transcriptional activator MONOPTEROS (MP)/ARF5 is critical for embryogenesis because the mp loss-of-function mutant shows defects in most auxin-dependent embryo patterning processes (Fig. 2). In addition, ARF7 and ARF17 contribute somewhat to cotyledon development, and it is possible that there is further redundancy between other members of the family. However, most of the defects seen in auxin biosynthesis, transport, or receptor mutants can be explained by the altered activity of MP/ARF5, which defines a complete pathway for auxin-dependent embryo development, from synthesis of the hormone to the activation of transcription.
In the next sections, we discuss the two auxin-dependent processes that have been studied in most detail: Root meristem formation and the specification of the shoot apical meristem and cotyledons.
ROLE OF AUXIN RESPONSE IN ROOT INITIATION
MP activity is required to specify the hypophysis, and a loss-of-function mp mutant shows aberrant hypophysis division resulting in a rootless phenotype (Fig. 2) (Berleth and Jürgens 1993; Weijers et al. 2006). The gain-of-function bdl/iaa12 mutant encoding a stabilized version of BDL/IAA12 has the same phenotype (Fig. 2) (Hamann et al. 1999; Hamann et al. 2002). It was shown that BDL binds to MP and inhibits its transcriptional activity. In the early globular stage, embryo MP and BDL proteins accumulate in all subepidermal cells, but not in the hypophysis (Weijers et al. 2006). Hence, MP acts non-autonomously in hypophysis specification as MP activity is required in the cells adjacent to the hypophysis to specify this cell (Fig. 4). Interestingly, among the nonautonomous signals that could mediate MP-dependent cell communication in hypophysis specification is auxin itself. DR5 activity in the hypophysis is lost in the mp mutant and PIN1 levels are reduced in the mp mutant. However, because exogenous auxin treatment does not restore hypophysis specification in mp or bdl mutant embryos, auxin alone does not seem sufficient for specifying the uppermost suspensor cell as hypophysis. Therefore, auxin is unlikely to be the only signal downstream of MP. The nature of the other signal(s) is currently unknown. The fact that auxin elicits a response in the future hypophysis suggests that there must be another auxin machinery consisting of an ARF and an Aux/IAA in this cell (Fig. 4).
Figure 4.
Hypophysis specification in the globular-stage embryo. MP activity is required non-cell-autonomously in the provascular cells (light blue) adjacent to the uppermost suspensor cell (pink) to specify this cell as hypophysis. In the provascular cells, high auxin levels release MP from its inhibitor, the Aux/IAA protein BDL, and the corepressor TPL. Subsequently, MP induces the expression of PIN1 in the provascular cells, resulting in auxin transport to the uppermost suspensor cell. MP also promotes the transport of a hypothetical signal (S) to the future hypophysis. Here, auxin releases another yet unidentified ARF from a so far unknown Aux/IAA protein to elicit an auxin response that converges with S to specify hypophysis fate.
Exactly how MP activity is restricted to the cells adjacent to the hypophysis and what genes are activated by MP is currently unknown. In part, MP activity is controlled by the activity of BDL. It was recently shown that BDL interacts with TOPLESS (TPL), a transcriptional corepressor (Long et al. 2002; Long et al. 2006; Szemenyei et al. 2008). Importantly, the rootless phenotype of the bdl mutant is suppressed by mutations in the ubiquitously expressed TPL gene. Expression of a fusion protein containing the carboxyl terminus of TPL and the carboxy-terminal dimerization domain of BDL under the control of the BDL promoter results in mp-like phenotypes. This shows that BDL inhibits MP by recruiting the TPL co-repressor (Fig. 3C). tpl mutant embryos either lack cotyledons (Fig. 2) or show a transformation of the apical pole into a second root. The formation of an apical root does not appear to depend on MP, and it will be interesting to see the extent of overlap between TPL-dependent and MP-dependent transcriptional control.
There is only a limited number of genes whose function has been suggested to act downstream of MP in root initiation. Among the few are the PLETHORA (PLT) genes. Mutations in two or more of these AP2-type transcription factors PLT1, PLT2, PLT3, and PLT4/BABY BOOM (BBM) interfere with divisions of the hypophyseal derivatives, resulting in the absence of a quiescent center (QC) at early heart stage (Fig. 2) (Aida et al. 2004; Galinha et al. 2007). DR5 expression is not affected in plt1 plt2 double mutants, indicating that auxin response is not impaired. Being activated hours after auxin treatment, the PLT genes appear to be late auxin response genes and are therefore probably not direct targets of an ARF. Nonetheless, the expression of the PLT genes is partially dependent on MP activity, as PLT1 and PLT2 expression is lost in globular and heart-stage mp embryos, but not in the octant-stage mp embryo. Postembryonically, ectopic PLT expression induces the formation of ectopic roots in the shoot. Therefore, PLT genes are master genes in root meristem development and are part of the developmental program that is activated by auxin during root initiation.
Correct control of MP-dependent root initiation may also involve chromatin regulation, as mutations in the plant homeodomain finger (PHD) proteins OBERON (OBE) 1 and 2 affect root meristem initiation (Saiga et al. 2008; Thomas et al. 2009). obe1 obe2 double mutants show aberrant hypophysis division and are rootless (Fig. 2), have a disorganized shoot meristem, and leaf formation arrests before or after the formation of the first pair of leaves. PHD finger proteins specifically recognize a modification of histone H3 that marks active genes. OBE1 and 2 are expressed throughout the embryo from the four-cell stage on. The expression of the PLT genes but not MP is lost in the obe1 obe2 double mutant. Furthermore, MP is epistatic to OBE, but is not required for OBE expression. The establishment of auxin response maxima in obe1 obe2 double mutant embryos is largely similar to the wild-type pattern. This suggests that OBE functions downstream of MP to control root meristem development. It is possible that OBE modifies the chromatin at MP target loci to make the promoters more accessible.
ROLE OF AUXIN IN SHOOT MERISTEM FORMATION AND COTYLEDON FORMATION
At the transition stage of embryogenesis, the cotyledons initiate at the flanks of the apical embryo domain at the sites where the auxin response reporter DR5 is highly expressed (Benkova et al. 2003). In between, the shoot meristem is specified (Mayer et al. 1998). The CUP-SHAPED COTYLEDON (CUC) genes 1 and 2 redundantly regulate the initiation of the shoot meristem and the separation of cotyledons together with SHOOT MERISTEMLESS (STM) (Barton and Poethig 1993; Aida et al. 1997; Aida et al. 1999). CUC1, CUC2, and STM are expressed between the presumptive cotyledon primordia in the globular stage embryo. At the bending-cotyledon stage, CUC1 and 2 expression is restricted to the cotyledon margins, whereas STM is only expressed in the shoot apical meristem (SAM).
Mutations in MP, PID, and PIN1 all affect cotyledon separation and the bilateral symmetry of the cotyledons (Fig. 2). Examination of the CUC gene expression in mp, pin1, and pin1 pid embryos revealed that all these genes are required for the activation of CUC2 in cotyledon boundaries and the repression of CUC1 in cotyledons (Aida et al. 2002; Furutani et al. 2004). Importantly, mutation of CUC1 in the pin1 pid double mutant partially restores cotyledon development, suggesting that the cotyledon formation defect is at least in part the consequence of ectopic CUC1 activity. Presumably, PIN1 and PID ensure the generation of an auxin maximum that is required to control CUC gene expression during cotyledon initiation and separation. It is unclear if MP directly regulates the CUC genes in response to the auxin peak supplied by PID/PIN, or whether MP mainly acts to promote proper PIN1 gene expression.
As indicated earlier, few (potential) target genes of auxin response pathway in the embryo are known. Recently, it was shown that DORNRÖSCHEN (DRN) is such a direct target (Chandler et al. 2007; Cole et al. 2009). DRN encodes an AP2-type transcription factor and acts redundantly with its paralog DRN-like (DRNL) in cotyledon development. Double homozygous drn drnl embryos have pinlike structures without cotyledons (Fig. 2). DRN has a very dynamic expression pattern in embryo development. From the two-cell till, the 16-cell stage DRN is expressed throughout the embryo, then becomes restricted to the apical domain at the sites where the cotyledon primordia will emerge, and finally is expressed at the tips of the cotyledons in the heart-stage embryo. The expression of the auxin response reporter DR5 and the localization of the PIN1 protein are changed in drn drnl double mutant embryos. Because DRN is an MP target, but also itself controls auxin transport, this gene acts both downstream of auxin and upstream of auxin transport. Even though MP and DRN expression overlap to a large extent, DRN expression only depends on MP in the tips of the cotyledons. Drn drnl embryos also show hypophyseal cell defects (Fig. 2), although the genes are not expressed in the basal region of the embryo. The nonautonomous action of these genes in root formation may depend on their influence on auxin transport.
Gain-of-function mutations in two Aux/IAA proteins also result in cotyledon defects. The gain-of-function iaa18 mutation that stabilizes IAA18 causes aberrant cotyledon outgrowth in 3–8% of the embryos (Fig. 2) (Ploense et al. 2009). IAA18 is expressed in the apical domain of the embryo from the 16-cell stage on. In the gain-of-function iaa18 mutant, PIN1 is asymmetrically expressed with stronger expression in one side of the embryo. This probably contributes to the cotyledon defects in iaa18 embryos. IAA18 can inhibit MP activity when ectopically expressed in the embryo, and overexpression of MP rescues several postembryonic leaf defects in the iaa18 mutant. However, MP can not be the only target of IAA18 as mp iaa18 mutants are more severe than the single mutants. The gain-of-function bdl mutant also has cotyledon defects. Likewise, mp bdl embryos have cotyledon defects, indicating that BDL must have other targets in addition to MP (Hamann et al. 1999).
CONVERGENCE OF ARF AND WOX TRANSCRIPTION FACTOR ACTIVITIES IN EARLY EMBRYOGENESIS
The prominent role of auxin in patterning the early embryo is becoming increasingly clear. However, several other pathways have also been shown to operate in patterning. It will be interesting to see how these are interwoven at the molecular level. One example of such integration is the convergence of MP and WOX functions.
The members of the WUSCHEL related homeobox (WOX) transcription factor gene family are differently expressed in the early embryo (Haecker et al. 2004). WOX2 and WOX8 are both expressed in the egg cell and the zygote. After the division of the zygote, expression of WOX2 marks the apical cell, whereas WOX8 is expressed in the basal cell. In the eight-cell embryo, WOX2, WOX8, and WOX9 are expressed in four different regions. The apical tier of the proembryo expresses WOX2, the lower tier of the proembryo expresses WOX9, the uppermost suspensor cell expresses both WOX8 and WOX9, and the other suspensor cells express only WOX8.
From the eight-cell stage to the 16-cell stage, WOX9 expression expands in the descendents of the lower tier of the eight-cell proembryo and is lost in the uppermost suspensor cell. This shift of WOX9 expression from the uppermost suspensor cell to the embryo does not occur in mp embryos, indicating that MP is required for both activation of WOX9 in embryo cells and down-regulation of WOX9 in the hypophysis (Breuninger et al. 2008). There are more points of convergence between MP and WOX genes. The wox2 mutant shows defects in cell divisions in the apical embryo domain, and this is strongly enhanced in the wox2 mp mutant. Similarly, the mp wox8 wox9 triple mutant shows synergistic phenotypes. The wox8 wox9 double mutant shows strong defects in both the apical and the basal cell lineage (Fig. 2), in part because the expression of WOX2 is lost. This is accompanied by loss of PIN1 expression and ubiquitous DR5 activity. These results show that WOX2, 8, and 9, and MP control the same embryo patterning processes. Therefore, embryo patterning is controlled by auxin in concert with other, potentially auxin-independent pathways.
ROLE OF AUXIN IN PATTERN FORMATION
As detailed previously, auxin controls several cell specification and pattern formation processes in the early embryo. The obvious question is how a molecule that is structurally so simple can elicit such different responses. In postembryonic auxin-dependent growth and patterning, auxin has been proposed to accumulate in concentration gradients, with cells converting different threshold concentrations to distinct responses (Galinha et al. 2007). However, evidence for concentration-dependent readout is still lacking. The other conceptual extreme of auxin activity would be that of a trigger that would elicit a predefined response above a certain threshold. Although in the former scenario, multiple distinct responses are specified by the perceived auxin concentration, in the latter, there would be an “all-or-nothing” response. Although no definite answer can be given at this moment, it appears that alterations in auxin concentrations can not bypass the predefined cell fate in the embryo. For example, when auxin levels are ubiquitously increased through expression of a biosynthesis gene and inhibition of transport, DR5 is activated throughout the embryo (Weijers et al. 2005). However, this is associated with fusion of cotyledons, but no ectopic specification of pattern elements. Likewise, removal of auxin causes a failure to specify the root or cotyledons, but no cell-fate transformations. A plausible scenario is that a prepattern determines the developmental potential that can be triggered by auxin, in which the concentration of auxin could determine response amplitude. However, reality may not be so simple, because a recent report suggests that inappropriate accumulation of auxin in cotyledon tips—either through expression of dominant–negative Rab5 or apolar PIN1—is sufficient to convert these cells to root identity (Dhonukshe et al. 2008). Nonetheless, a critical open question in auxin-dependent embryo patterning remains the mode of auxin action. The size of the Aux/IAA and ARF families, as well as the presence of six TIR1/AFB receptors would allow enormous combinatorial complexity to equip each cell with a unique response machinery to enable unique cellular auxin responses. Systematic analysis of Aux/IAA and ARF functions in the embryo, as well as identification of target genes and processes of auxin in the embryo will provide critical insight into the prominent role of auxin as an embryo patterning molecule.
ACKNOWLEDGMENTS
We apologize to those whose valuable contributions we could not include because of space constraints. We wish to thank Cristina Llavata Peris, Annemarie Lokerse, and Eike Rademacher for helpful comments on this manuscript. This work was supported by a grant from the Dutch Organization for Scientific Research (NWO; ALW-VIDI 864.06.012).
Footnotes
Editors: Mark Estelle, Dolf Weijers, Karin Ljung, and Ottoline Leyser
Additional Perspectives on Auxin Signaling at www.cshperspectives.org
REFERENCES
- Aida M, Ishida T, Tasaka M 1999. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126:1563–1570 [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120 [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M 1997. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M 2002. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129:3965–3974 [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS 1993. Formation of the shoot apical mersitem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119:823–831 [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R 2001. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128:4057–4067 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jürgens G, Friml J 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602 [DOI] [PubMed] [Google Scholar]
- Bennett RM, Alvarez J, Bossinger G, Smyth DR 1995. Morphogenesis in pinoid mutants in Arabidopsis thaliana. Plant J 8:505–520 [Google Scholar]
- Berleth T, Jürgens G 1993. The role of the MONOPTEROS gene in organising the basal body region of the Arabidopsis embryo. Development 118:575–587 [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T 2008. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14:867–876 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Grewe B, Werr W 2007. The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134:1653–1662 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y 2007. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y 2008. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci 105:21017–21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W 2009. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136:1643–1651 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M 2005a. The F-box protein TIR1 is an auxin receptor. Nature 435:441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M 2003. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. Embo J 22:1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jürgens G, Estelle M 2007. AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52:114–123 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M 2005b. Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell 9:109–119 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J 2007. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17:520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Tanaka H, Goh T, Ebine K, Mahonen AP, Prasad K, Blilou I, Geldner N, Xu J, Uemura T, et al. 2008. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456:962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426:147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306:862–865 [DOI] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M 2004. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131:5021–5030 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD 2002. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci 99:11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G 2003. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112:219–230 [DOI] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M 2002. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14:2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfi K, Speth V, Neuhaus G 1998. Auxin-induced developmental patterns in Brassica juncea embryos. Development 125:879–887 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131:657–668 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G 1999. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126:1387–1395 [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki A, Bowman JL 2007. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W 2007. Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23:207–236 [DOI] [PubMed] [Google Scholar]
- Johri BM, Ambegaokar KB, Srivastava PS 1992. Comparative embryology of Angiosperms Springer-Verlag, Berlin, Germany [Google Scholar]
- Jürgens G 1995. Axis formation in plant embryogenesis: Cues and clues. Cell 81:467–470 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wisniewska J, Paciorek T, Benkova E, Friml J 2008. ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol 18:526–531 [DOI] [PubMed] [Google Scholar]
- Laux T, Jürgens G 1997. Embryogenesis: A new start in life. Plant Cell 9:989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu Z, Chua NH 1993. Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D 2004. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16:5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312:1520–1523 [DOI] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK 2002. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129:2797–2806 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG 1991. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69:461–476 [Google Scholar]
- Mayer U, Büttner G, Jürgens G 1993. Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the GNOM gene. Development 117:149–162 [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. 2007. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130:1044–1056 [DOI] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW 2009. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga S, Furumizu C, Yokoyama R, Kurata T, Sato S, Kato T, Tabata S, Suzuki M, Komeda Y 2008. The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development 135:1751–1759 [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P 2002. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell 13:1916–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–1386 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Schmidt D, Bayer EM, Dreos R, Maule AJ 2009. Arabidopsis plant homeodomain finger proteins operate downstream of auxin accumulation in specifying the vasculature and primary root meristem. Plant J 59:426–436 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Jürgens G 1994. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120:2967–2978 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1999. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci 96:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R 2005. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell: tpc.105.034637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G 2006. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10:265–270 [DOI] [PubMed] [Google Scholar]