Abstract
We devised a strategy of 14-3-3 affinity capture and release, isotope differential (d0/d4) dimethyl labeling of tryptic digests, and phosphopeptide characterization to identify novel targets of insulin/IGF1/phosphatidylinositol 3-kinase signaling. Notably four known insulin-regulated proteins (PFK-2, PRAS40, AS160, and MYO1C) had high d0/d4 values meaning that they were more highly represented among 14-3-3-binding proteins from insulin-stimulated than unstimulated cells. Among novel candidates, insulin receptor substrate 2, the proapoptotic CCDC6, E3 ubiquitin ligase ZNRF2, and signaling adapter SASH1 were confirmed to bind to 14-3-3s in response to IGF1/phosphatidylinositol 3-kinase signaling. Insulin receptor substrate 2, ZNRF2, and SASH1 were also regulated by phorbol ester via p90RSK, whereas CCDC6 and PRAS40 were not. In contrast, the actin-associated protein vasodilator-stimulated phosphoprotein and lipolysis-stimulated lipoprotein receptor, which had low d0/d4 scores, bound 14-3-3s irrespective of IGF1 and phorbol ester. Phosphorylated Ser19 of ZNRF2 (RTRAYpS19GS), phospho-Ser90 of SASH1 (RKRRVpS90QD), and phospho- Ser493 of lipolysis-stimulated lipoprotein receptor (RPRARpS493LD) provide one of the 14-3-3-binding sites on each of these proteins. Differential 14-3-3 capture provides a powerful approach to defining downstream regulatory mechanisms for specific signaling pathways.
Activated tyrosine kinase receptors generally drive cells to assimilate nutrients; regulate partitioning of the assimilate to make storage polymers and biosynthetic precursors and for energy production; and promote cellular survival, growth, division, movement, and differentiation. From this spectrum, each cell displays a specific subset of responses depending on the hormone, specific receptors, cross-talk from other signaling pathways, metabolic conditions, and cellular complement of effector proteins. For example, insulin stimulates glucose uptake and glycogen synthesis in skeletal muscle, whereas IGF11 promotes survival, growth, and proliferation of many cell types (1, 2).
Many of these cellular responses are mediated via PI 3-kinase, which generates phosphatidylinositol 3,4,5-trisphosphate, promoting the activation of AGC protein kinases such as PKB/Akt and other signaling components (1, 3). PI 3-kinase is activated by binding to tyrosine-phosphorylated receptors such as the platelet-derived growth factor receptor or via adaptor molecules such as insulin receptor substrates, which are phosphorylated by the activated insulin receptor. Deregulated PI 3-kinase and downstream signaling has been linked to problems with wound healing, immune responses, neurodegeneration, and cardiovascular disease; decreased PI 3-kinase signaling may underlie insulin resistance and type II diabetes; and this pathway is often activated in human tumors (4, 5). To help pinpoint drug targets for these diseases we must define the mechanisms linking PI 3-kinase and other signaling pathways to downstream effectors and understand specificity with respect to different hormone/cell type combinations.
Many missing substrates of PI 3-kinase/AGC kinases must be found to explain all the cellular responses to insulin and growth factors (3). Several targets of PI 3-kinase/PKB signaling, including TSC2 (6), PRAS40 (7), AS160 (8), and FYVE domain-containing phosphatidylinositol 3-phosphate 5-kinase (9) were identified using the anti-PAS antibody, which loosely recognizes the minimal phosphorylated consensus for PKB, which is RXRXX(pS/pT) where pS is phosphoserine and pT is phosphothreonine. Another helpful feature for identifying new downstream targets is that phosphorylation by PKB sometimes creates binding sites for 14-3-3s, which are dimeric proteins that bind to specific phosphorylated sites on target proteins. Thus PKB promotes the binding of 14-3-3s to proteins including PFKFB2 cardiac PFK-2 (10, 11), BimEL (12), β-catenin (13), p27(Kip1) (14), PRAS40 (7), FOXO1 (15), Miz1 (16), TBC1D4 (AS160 (17, 18), and TBC1D1 (19). Functionally 14-3-3s can trigger changes in the conformations of their targets and alter how targets interact with other proteins. Consistent with 14-3-3/target interactions being important in cellular responses to growth factors and insulin, reagents that compete with targets for binding to 14-3-3s inhibit the IGF1-stimulated increase in the glycolytic stimulator fructose-2,6-bisphosphate (10) and PKB-dependent cell survival (20).
Some 14-3-3-binding sites on the above named proteins can also be phosphorylated by other basophilic protein kinases (21). For example, AS160 and TBC1D1 are two related RabGAPs (GTPase-activating protein for Rabs) regulated by multisite phosphorylation that regulate trafficking of GluT4 transporter to the plasma membrane for uptake of glucose. The two 14-3-3-binding sites on AS160 can be phosphorylated by PKB, p90RSK, serum- and glucocorticoid-inducible kinase, and other kinases, whereas one of the 14-3-3-binding sites on TBC1D1 is also a substrate of the energy-sensing kinase AMP-activated protein kinase (17–19). Thus, the relative sensitivity of glucose trafficking to insulin and AMP-activated protein kinase activators in different tissues may depend in part on the distribution of AS160 and TBC1D1. Other insulin-regulated 14-3-3 targets, such as myosin 1C (22), are also convergence points for phosphorylation by more than one AGC and/or Ca2+/calmodulin-dependent protein kinase.
Here many more proteins than those already identified were found to display 14-3-3 and/or PAS binding signals when the PI 3-kinase pathway was activated in cells against a “background” of other proteins whose 14-3-3 and PAS binding status was unaffected by PI 3-kinase signaling. We aimed to pick out the PI 3-kinase-regulated proteins, which was challenging given the hundreds of 14-3-3 binding partners in mammalian cells (10, 23–27). We used 14-3-3 affinity capture and release, identified phosphopeptides, and devised a quantitative proteomics approach in which 14-3-3-binding proteins from insulin-stimulated versus unstimulated cells were labeled with formaldehyde containing light or heavy isotopes, respectively. Biochemical checking of candidates from these screens, which included proteins with links to diabetes, cancers, and neurodegenerative disorders, confirmed the identification of novel downstream targets of PI 3-kinase, some of which are also convergence points for regulation by MAPK/p90RSK signaling.
EXPERIMENTAL PROCEDURES
Materials
Synthetic peptides were from Graham Bloomberg (University of Bristol). Oligonucleotides were from MWG-Biotech. IGF1 was from BIOSOURCE. Microcystin-LR was from Linda Lawton (Robert Gordon's University, Aberdeen, Scotland, UK). Vivaspin concentrators were from Vivascience. Tissue culture reagents were from Invitrogen. Protease inhibitor mixture tablets (catalog number 1697498) and sequencing grade trypsin were from Roche Applied Science. Precast SDS-polyacrylamide gels were from Invitrogen. Protein G-Sepharose and chromatographic matrices were from GE Healthcare. Formaldehyde-d0 and -d2 were from Sigma-Aldrich, and unless stated other chemicals were from BDH Chemicals or Sigma-Aldrich.
Stimulation and Extraction of HeLa and HEK293 Cells
To our knowledge, HeLa S3 and HEK293 cells do not express insulin receptors, although they have IGF1 receptors that bind insulin with lower affinity than the cognate receptor (28). Insulin, which is less expensive than IGF1, was therefore used for large scale experiments with HeLa S3 cells, which were cultured in suspension in Dulbecco's modified Eagle's medium containing 10% FCS, 1% glutamine, 1% penicillin-streptomycin, and 1% sodium pyruvate. Trial experiments indicated that serum starvation of HeLa S3 cells for 4 h followed by 20-min stimulation with 50 milliunits/ml (300 nm) insulin gave maximal phosphorylation of PKB (Thr(P)308 and Ser(P)473) and AS160 (Thr(P)642). Cells were harvested by centrifugation and snap frozen.
For Fig. 5, HEK293 cells (European Collection of Cell Cultures) were cultured in 15-cm dishes in Dulbecco's modified Eagle's medium containing 10% (v/v) FCS, 2 mm l-glutamine, non-essential amino acids, and 1% penicillin-streptomycin. At ∼60% confluency, cells were transfected using 30 μl of 1 mg/ml polyethylenimine for 5 μg of DNA. After 36 h, cells were rinsed with warm PBS and serum-starved in Dulbecco's modified Eagle's medium for 8 h. Where indicated, cells were preincubated with PI-103 (1 μm for 30 min) and BI-D1870 (10 μm for 30 min) and stimulated for 20 min with 50 ng/ml IGF1 and 100 ng/ml PMA. Cells were lysed in 0.5 ml of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.5, 1 mm EGTA, 1% Triton X-100, 1 mm sodium orthovanadate, 50 mm sodium fluoride, 5 mm sodium pyrophosphate, 0.27 m sucrose, 0.1% (by volume) 2-mercaptoethanol, “Complete” proteinase inhibitor mixture (one tablet/50 ml). Cell lysates were clarified by centrifugation at 4 °C for 20 min at 15,000 rpm.
Fig. 5.
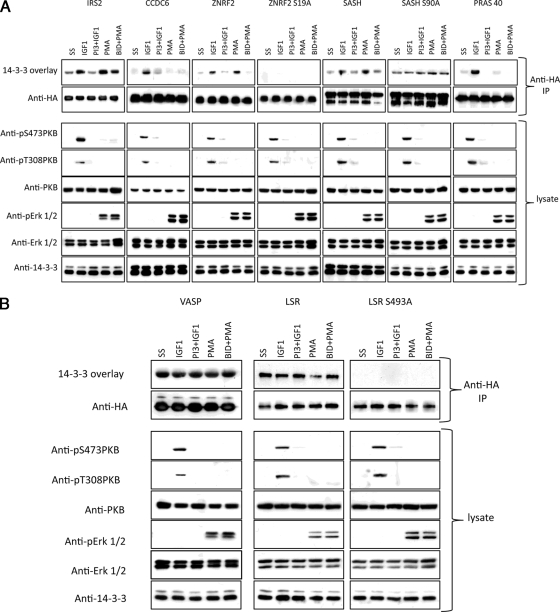
Cellular regulation of 14-3-3 binding of IRS2, CCDC6, ZNRF2, SASH1, PRAS40, VASP, and LSR. HA-IRS2, HA-CCDC6, HA-ZNRF2 (wild type and S19A mutant), HA-SASH1 (wild type and S90A mutant), HA-PRAS40, HA-VASP, and HA-LSR (wild type and S493A mutant) were isolated from transfected HEK293 cells that were stimulated as indicated, and anti-HA immunoprecipitates (IP) were analyzed by 14-3-3 overlay and Western blotting. Note that at higher exposures and in other experiments it was clear that 14-3-3 binding is not completely abolished with the ZNRF2 S19A mutation. As controls for the efficacy of stimuli and inhibitors, lysates were analyzed with antibodies against phospho-Thr308 and phospho-Ser473 of PKB/Akt, total PKB/Akt, phospho-Erk1/2 (pErk), and total Erk1/2. PI3, PI-103; BID, BI-D1870.
14-3-3 Affinity Chromatography
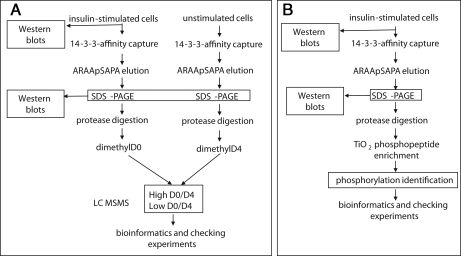
14-3-3 affinity chromatography involved binding of protein to 14-3-3-Sepharose (mixed BMH1 and BMH2, the 14-3-3 isoforms from Saccharomyces cerevisiae) and elution of specifically bound proteins by competition with the 14-3-3-binding synthetic ARAApSAPA phosphopeptide (Fig. 2) as in Pozuelo Rubio et al. (10) except that the high salt wash was only 500 ml and the mock peptide elution was omitted.
Fig. 2.
Experimental strategy for identifying proteins whose phosphorylation and binding to 14-3-3s is stimulated by insulin. A, as detailed under “Experimental Procedures,” HeLa suspension cell cultures were serum-starved and stimulated or not with insulin. Proteins were captured on a 14-3-3-Sepharose column, which was washed until protein was undetectable in the flow-through, and specifically bound proteins were eluted by competition with a 1 mm concentration of the 14-3-3-binding synthetic phosphopeptide ARAApSAPA. Eluates were concentrated, digested with trypsin, and incubated with formaldehyde containing no deuterium (d0 for the preparation from insulin-stimulated cells) or two deuteriums (to give d4 for each dimethyl group added onto the peptides from unstimulated cells). d0/d4 ratios were determined as outlined under “Experimental Procedures.” B, strategy for identifying phosphopeptides derived from proteins isolated by 14-3-3 capture and release from extracts of insulin-stimulated cells. Phosphopeptides were enriched by titanium dioxide affinity as detailed further under “Experimental Procedures.”
Western Blots, 14-3-3 Far-Western Overlays, and Immunoprecipitations
Sheep anti-HA was raised against the peptide YPYDVPDYA, and sheep anti-AS160 was raised against KAKIGNKP (17). Anti-phospho-Erk1/2 (Thr(P)202/Tyr(P)204), anti-phospho-Thr308 PKB, and anti-PKB/Akt were from Cell Signaling Technology. For Western blots the indicated antibodies were used at 1 μg/ml. Western blots and 14-3-3 overlays (using digoxigenin-labeled 14-3-3s in place of primary antibody) were visualized by ECL reagent or the Odyssey Infrared Imaging System (LI-COR, Inc.) as indicated. For immunoprecipitations with anti-HA, 4 μg of antibody/mg of lysate was mixed at 4 °C for 1 h, and then Protein G-Sepharose (30 μl of a 50% suspension in lysis buffer) was added and mixed for a further 1 h. The suspension was centrifuged at 12,000 × g for 1 min between washes.
Tryptic Digestion, Dimethylation, and Phosphopeptide Enrichment
14-3-3-binding proteins that had been purified from unstimulated or insulin-stimulated HeLa cells were denatured in lithium dodecyl sulfate sample buffer (Invitrogen) containing 10 mm DTT at 95 °C for 5 min, cooled, and alkylated with 50 mm iodoacetamide for 30 min in the dark at room temperature. The protein samples were loaded on adjacent lanes of a NuPAGE 4–12% gradient gel (Invitrogen) and electrophoresed at 160 V for 60 min, and the gel was stained with colloidal Coomassie Blue (Invitrogen). The gel lanes were each cut into seven equal sections (with band 1 at the top of the gel) that were washed successively with 50 mm triethylammonium bicarbonate; 50% acetonitrile, 50 mm triethylammonium bicarbonate (twice); and acetonitrile (15 min each wash) before drying in a SpeedVac (Eppendorf). Trypsin (5 μg/ml trypsin gold; Promega) in sufficient 25 mm triethylammonium bicarbonate to cover the gel pieces was added for 12 h at 30 °C. The supernatant was transferred to a fresh tube to which two 50% acetonitrile washes of the gel pieces were also added. The digested samples were split into two equal fractions and dried in a SpeedVac. One half was enriched for phosphopeptides using titanium dioxide, and the other half was dimethylated with formaldehyde using a modified version of the procedure described previously (29). Individual tryptic digests were redissolved in 2 μl of 25 mm sodium acetate buffer, pH 5.5, 30 mm sodium cyanoborohydride containing 0.2% (v/v) formaldehyde (d0 for the preparation from insulin-stimulated cells and d2 for the preparation from unstimulated cells) and incubated at room temperature for 15 min. The dimethylated digests were mixed pairwise for corresponding gel sections, diluted 25-fold with strong cation exchange (SCX) loading buffer (25% acetonitrile, 0.2% formic acid), and loaded onto 5 μl of Poros 50 HS beads equilibrated in the same buffer. The slurry was loaded on a Millipore SCX ZipTip and washed three times with 60 μl of SCX loading buffer. Peptides were eluted with 2 × 40 μl of 50% isopropanol, 0.2 m ammonium hydroxide and dried under vacuum.
For phosphopeptide enrichment, tryptic digests of the preparations from insulin-stimulated cells were dissolved in 200 mg/ml 2,3-dihydrobenzoic acid (Sigma-Aldrich) in 80% (v/v) acetonitrile, 5% (v/v) TFA (loading buffer). Titanspheres (5 mg of 5-μm spheres; Hichrom Ltd.) equilibrated in loading buffer were added to each digest and agitated for 10 min. The slurry was loaded in a C18 StageTip (Proxeon); washed three times with 80% (v/v) acetonitrile, 5% (v/v) TFA; eluted with 40 μl of 1 m ammonium hydroxide, 50% (v/v) acetonitrile; 40 μl of 50% (v/v) acetonitrile; and 40 μl of 0.5% (v/v) formic acid, 50% (v/v) acetonitrile; combined; and dried under vacuum.
LC-MS Analysis
Tryptic digests were analyzed using Ultimate 3200 nanoflow chromatography (LC Packings) coupled to an LTQ-Orbitrap (Thermo Finnigan) mass spectrometer equipped with a dynamic NanoSpray source (Optron). The dimethylated peptide mixtures were separated using an LC Packings Integrated System (Dionex, Camberley, UK) consisting of a WPS3000T microautosampler, FLM3200 microcolumn switching module, UltiMate LPG3600 micropump, a PepMap C18 column (75 μm, 15 cm; LC Packings), and mobile phases of 2% acetonitrile, 0.1% formic acid in water (A) and 90% acetonitrile, 0.085% formic acid in water (B). The column was equilibrated in 2% B at a flow rate of 300 nl/min.
The dried digests were resolubilized in 50% (v/v) acetonitrile, 0.1% (v/v) TFA; diluted 10-fold with 0.1% (v/v) TFA; and loaded onto a C18 capillary trap (Michrom Bioresources, Auburn, CA) equilibrated in buffer A at a flow rate of 20 μl/min. After 8 min, the capillary cartridge was switched in line with the analytical column and eluted with the following gradient: 2–50% buffer B (8–80 min), 50–85% B (80–85 min), 85–2% B (85–90 min), and 2% B (90–100 min). The column eluate was electrosprayed with a voltage of 1200 V applied to a Picotip (FS360-50-15-N, New Objective, Woburn, MA).
Mass spectra were acquired using two different acquisition methods. For protein identification, the LTQ-Orbitrap was programmed to perform two FT scans (60,000 resolution) on 300–800- and 800–1800-amu mass ranges with the top five ions from each scan selected for LTQ-MS/MS. FT spectra were internally calibrated using a single lock mass (445.1200 atomic mass units). For phosphopeptide analysis, the same two FT scans were performed, but multistage activation was performed on the selected ions with a neutral loss of 97.98, 48.99, 32.66, and 24.50. For these two methods, target ion numbers were 500,000 for FT full scan on the orbitrap and 10,000 MSn on the LTQ.
Peptide and Protein Identification
Raw files were converted to peak lists in Mascot generic format (MGF) files using raw2msm v1.7 software (Matthias Mann) using default parameters and without any filtering, charge state deconvolution, or deisotoping. MGF files were searched using a Mascot 2.2 in-house server against the Internation Protein Index human 3.26 database (57,846 sequences; 26,015,783 residues). For the quantitative dimethyl labeling experiments, search parameters were as follows: digestion with trypsin; two missed cleavages permitted; fixed modification, carbamidomethyl cysteine; variable modifications, oxidized methionine, dimethyl N terminus, and dimethyllysine; a precursor mass tolerance of 10 ppm with a possible wrong picking set to two isotopes; and an MS/MS mass tolerance of 0.8 Da. The Mascot integrated decoy database search calculated a false discovery rate of 1.39% (38 reverse database peptide matches from a total of 2719 peptide matches) when searching was performed on the concatenated MGF files with an ion score cutoff of 20 and a significance threshold of p < 0.05. Only peptides with ion scores over 20 were considered, and only proteins with at least one unique peptide (red bold in Mascot) were considered. This ion score threshold is enough to keep the false discovery rate under 2%. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. When a protein was identified with only one peptide or with only one unique peptide (one red bold peptide), the MS2 spectrum was manually inspected and annotated (supplemental data).
For the phosphorylation site mapping experiments, Mascot search parameters were the same except for variable modifications, which included oxidized methionine and phosphorylation of serine/threonine/tyrosine. The Mascot integrated decoy database search calculated a false discovery rate of 0.45% (12 reverse database peptide matches for a total number of 2675 peptide matches) when searching was performed on the concatenated MGF files with an ion score cutoff of 20 and significance threshold of p < 0.05. Only phosphopeptides with ion scores over 20 were considered. This ion score threshold is enough to keep the false discovery rate under 2%. For more confidence, MS2 spectra were manually inspected and annotated (given in the supplemental figures).
Isotope-based Quantification
Quantification was done manually using Qual Browser v2.0.7 software (Thermo Finnigan). Only unique peptides (red bold peptides in Mascot) with signal intensity over 105 counts/s were used for the quantification. The maximum height of the extracted ion chromatogram with a mass tolerance of 10 ppm was used to evaluate the ratio of each calculated light/heavy dimethylated peptide pair. The reaction incorporates two molecules of formaldehyde per labeling, resulting in a mass difference of 4.02511 Da per labeling comparing light and heavy peptides. d0/d4 ratios for three (when possible) peptides were used to rank each protein listed in supplemental Table 1.
RESULTS
Visualization of Proteins Whose Phosphorylation and Binding to 14-3-3s Is Enhanced by IGF1 and Inhibited by PI 3-Kinase Inhibitors
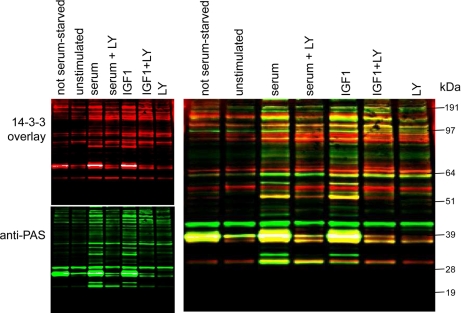
A 14-3-3-Sepharose-binding fraction from extracts of unstimulated cells contained many proteins that displayed 14-3-3 and PAS binding signals (Fig. 1). The number and intensities of such signals increased for preparations from cells stimulated with serum and IGF1 and decreased to the basal levels when the PI 3-kinase signaling pathway was blocked with the inhibitors LY294002 (Fig. 1), wortmannin, and PI-103 (not shown). Some 14-3-3 and PAS binding signals coincided with each other, such as the most prominent protein of ∼40 kDa, which is PRAS40 (7, 30), whereas others did not align with each other (Fig. 1), indicating PAS-binding proteins that do not bind directly to 14-3-3s or do not refold well and therefore perform poorly in the 14-3-3 binding assay. Conversely we observed some binding of 14-3-3s to non-PAS sites. Note that although raised to detect PKB/Akt substrates, the PAS antibody is also likely to detect residues phosphorylated by kinases with similar substrate specificities.
Fig. 1.
Proteins that bind to 14-3-3s in response to IGF1 in a PI 3-kinase-dependent manner. HEK293 cells cultured on 10-cm-diameter dishes in medium containing 10% (v/v) serum (labeled not serum-starved) were serum-starved for 4 h (unstimulated) and then stimulated as indicated with IGF1 at 50 ng/ml for 15 min and serum at 10% (v/v) for 15 min. Where indicated, cells were incubated with LY294002 (LY; 100 mm for 1 h) prior to stimulation. Cells were lysed in 0.3 ml/dish ice-cold lysis buffer, and 3 mg of each extract was added to 100 μl of a 50% (v/v) slurry of 14-3-3-Sepharose and mixed end over end for 4 h. After washing, the protein bound to 14-3-3-Sepharose was extracted into SDS sample buffer, separated by SDS-PAGE using a 4–12% gradient gel, and analyzed by Far-Western 14-3-3 overlay (red), which detects proteins on the blot that can bind directly to digoxigenin-labeled 14-3-3 proteins. Western blotting was also performed with the anti-PAS antibody (green).
Differential Screen to Identify Proteins Whose Phosphorylation and Binding to 14-3-3s Is Stimulated by Insulin
To identify the unknown targets of insulin/IGF1 signaling indicated by Fig. 1, we devised a strategy based on capturing 14-3-3-binding proteins in the cell extracts and releasing them specifically by competition with a 14-3-3-binding phosphopeptide followed by SDS-PAGE, in-gel protease digestion, isotope differential dimethyl labeling of N-terminal and lysine amine groups on peptides, and quantitative analysis of the d0/d4 ratios of corresponding peptides from the two preparations (Fig. 2A).
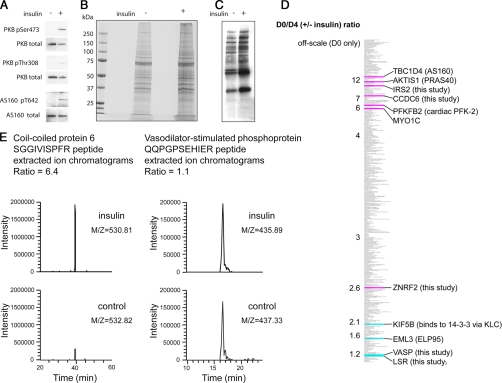
In several trial experiments, approximately twice as much protein was isolated by 14-3-3 capture from extracts of insulin- or serum-stimulated cells compared with serum-deprived, unstimulated cells. Although by eye we could not spot any proteins that looked selectively regulated on SDS-PAGE, Western blots revealed that AS160 was more abundant in the preparations from insulin-stimulated cells (Fig. 3, A, B, and C), whereas KLC2 was approximately equal in both preparations (not shown). These findings are consistent with 14-3-3 binding of AS160 being stimulated by insulin (17, 18) and findings that KLC2 binds to 14-3-3s irrespective of insulin stimulation.2 We therefore proceeded to digest proteins with trypsin to generate peptides for dimethyl labeling and analysis as outlined in Fig. 2A.
Fig. 3.
d0/d4 (± insulin) ratios of 14-3-3 affinity-purified proteins from unstimulated and insulin-stimulated cells. A, comparison of PKB/Akt and AS160 total protein levels and phosphorylation status in lysates of unstimulated and insulin-stimulated HeLa cells used for 14-3-3 phosphoproteomics screens. B, Coomassie-stained SDS gel of proteins isolated by 14-3-3 capture and release from extracts of unstimulated and insulin-stimulated cells (5 times these amounts were run on a parallel gel and used for the MS analyses). C, 14-3-3 Far-Western overlay assay of proteins isolated by 14-3-3 capture and release from extracts of unstimulated and insulin-stimulated cells. D, thumbnail of the d0/d4 (±insulin) rankings for 14-3-3 affinity-purified proteins where off-scale indicates the proteins that were only detected in the preparation from insulin-stimulated cells. Highlighted in pink are proteins already known to be phosphorylated and to bind to 14-3-3s in response to insulin/IGF1 as well as proteins that were discovered to be insulin/IGF1-responsive in this study. In blue are proteins that we already knew do not respond to insulin/IGF1 as well as proteins found not to be insulin/IGF1-responsive in this study (see text). E, examples of d0/d4 ratio quantification for peptides from CCDC6 (d0/d4 = 6.4 for this peptide, contributing to the overall ratio of 7.1 for three quantified peptides from this protein (Table I)) and VASP (d0/d4 = 1.1 for this peptide with an overall ratio of 1.3 for the protein (supplemental Table 1)) showing extracted ion chromatograms after analysis in the LTQ-Orbitrap mass spectrometer.
Supplemental Tables 1 and 4 list the 296 proteins that were identified and ranked in order of their d0/d4 ratios where high d0/d4 ratios indicate proteins that appear to be more enriched in the preparation of 14-3-3-binding proteins from insulin-stimulated cells. The d0/d4 (±insulin) ratios are also summarized in Fig. 3D with examples of d0/d4 data for two proteins given in Fig. 3E. Two proteins with low d0/d4 scores were kinesin heavy chain (KIF5B), which binds 14-3-3 indirectly via phosphorylated kinesin light chains (KLCs), and EML3 (ELP95) (Fig. 3D). In other projects, we have no indication of insulin regulation of KLCs or EML3.3 Strikingly, however, four proteins that are already known to bind to 14-3-3s in response to insulin/IGF1 via PI 3-kinase signaling, namely PFKFB2 (cardiac PFK-2), AKT1S1 (PRAS40), TBC1D4 (AS160), and MYO1C (myosin 1C) (7, 11, 17, 18, 22) scored relatively high d0/d4 ratios, meaning that these proteins were more highly represented among the 14-3-3-binding proteins from insulin-stimulated compared with unstimulated cells (Fig. 3D). This clustering of known “positives” was encouraging, and instead of refining the primary screen at this stage, we decided to test its predictive power by examining the cellular regulation of individual proteins with high and low d0/d4 ratios, respectively. The results are given later. We also used a second screening approach.
Phosphorylated Residues on Proteins Isolated by 14-3-3 Affinity Capture from Lysates of Insulin-stimulated Cells
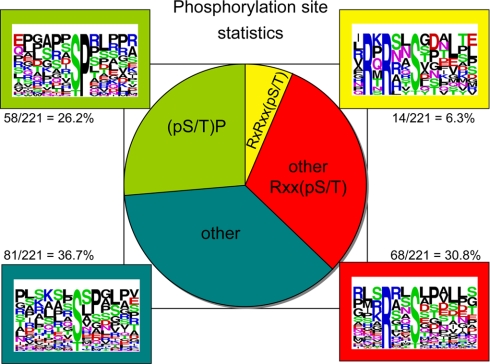
In the second screen, we identified phosphorylated residues in 14-3-3-binding proteins from lysates of insulin/IGF1-stimulated cells using the strategy in Fig. 2B. The full results are in supplemental Tables 2 and 3 with a summary in Fig. 4. Of 221 phosphorylated residues identified, 82 (37%) were derived from RXX(pS/pT) motifs, including 14 (6.3%) that conform to the RXRXX(pS/pT) PAS motif. Basic residues at −3 are characteristic of sites phosphorylated by basophilic AGC and Ca2+/calmodulin-dependent protein kinases and are often found in 14-3-3-binding sites. We also note some LXRXX(pS/pT) sites, which can be phosphorylated by kinases on several branches of the Ca2+/calmodulin-dependent protein kinase subfamily (31, 32), as well as phosphopeptides with Leu at +4, which is preferred by AMP-activated protein kinase and other Ca2+/calmodulin-dependent protein kinases (33). We identified 58 (26%) (Ser(P)/Thr(P))-Pro motifs, which are not generally associated with 14-3-3 binding. The remaining 81 phosphosites (37%) varied but included sequences with Pro at +2 as did the peptides in the RXRXX(pS/pT) and RXX(pS/pT) categories. Although not essential for 14-3-3 binding, a +2 Pro is found in the canonical 14-3-3-binding motifs where it adopts a cis conformation that twists the peptide out of the docking site (34, 35). Only 17 proteins, represented by 44 phosphopeptides, were found in both the differential dimethyl labeling (±insulin) and phosphopeptide screens, showing that these analyses are far from saturated (Table I).
Fig. 4.
Phosphopeptides derived from 14-3-3-binding proteins isolated from insulin-stimulated cells. The phosphorylated sites were classified according to whether they are found within motifs that conform to RXRXX(pS/pT), RXX(pS/pT) other than RXRXX(pS/pT), (pS/pT)P, or none of these (other). One phosphorylated site that matches both RXX(pS/pT) and (pS/pT)P is included in the RXX(pS/pT) group.
Table I. 14-3-3 affinity-captured proteins identified by both their d0/d4 ratios and phosphopeptides.
Seventeen proteins, represented by 44 phosphopeptides, were found in both the d0/d4 (±insulin) ratio screen (supplemental Table 1) and phosphorylation site identification screen (marked by * beside the gene symbols in supplemental Tables 1 and 3). The sequence within the square brackets indicates the phosphopeptide identified by mass spectrometry, whereas the extended sequence outside of the brackets is shown to aid identification of motifs that may be recognized by the protein kinases that phosphorylate these sites.
| Approved gene symbol | d0/d4 (±insulin) ratio | Phosphorylated residues as in supplemental Tables 2 and 3 with phosphorylated residues and oxidized methionines in lowercase letters |
|---|---|---|
| AKT1S1 (PRAS40) | 9.9 | LPRPR[LNtSDFQK]a |
| QQYAK[sLPVSVPVWGFK] | ||
| CAD | 6.7 | R[IHRAsDPGLPAEEPK] |
| PDGRFHLPPR[IHRAsDPGLPAEEPKEK] | ||
| CCDC6 | 7.1 | K[LDQPVSAPPsPR]D |
| R[ILQEKLDQPVsAPPsPR]D | ||
| DUT | 1.4 | M[PCSEETPAIsPSKR]A |
| EML3 | 1.6 | KLSRK[AIsSANLLVR]S |
| KLSR[KAIsSANLLVR]S | ||
| AGPAPATPSR[TPsLSPASSLDV] | ||
| GKDASIFQWR[VLGAGGAGPAPATPsR] | ||
| GKDASIFQWR[VLGAGGAGPAPAtPSRTPSLSPASSLDV] | ||
| FAM122B | 3.5 | DKPEKLYSPK[RIDFTPVsPAPSPTR] |
| DKPEKLYSPK[RIDFTPVSPAPsPTR] | ||
| DKPEKLYSPK[RIDFTPVsPAPsPTR] | ||
| APSPTRGFGK[MFVSSSGLPPsPVPSPR] | ||
| DKPEKLYSPK[RIDFTPVSPAPSPtRGFGK] | ||
| DKPEKLYSPK[RIDFTPVsPAPSPtRGFGK] | ||
| FOXK1 | 0.3 | PLSSR[sAPASPTHPHLmSPR] |
| HSPB1 | 2.7 | RALSR[QLsSGVSEIR]H |
| IRS2 | 9.0 | RSYR[RVsGDAAQDLDR]G |
| LMO7 | 2.1 | GIMR[RGEsLDNLDSPR]S |
| RQR[SAsVNKEPVSLPGImR]Ra | ||
| SASLPRSYRK[TDTVRLTsVVTPRPFGSQTR] | ||
| LSR | 1.2 | K[NLALsRESLVV]- |
| R[GPALTPIRDEEWGGHsPR]S | ||
| RRPR[ARsVDALDDLTPPSTAESGSR]a | ||
| SMRVLYYMEK[ELANFDPsRPGPPsGRVER] | ||
| EDDWRSRPSR[GPALTPIRDEEWGGHsPRsPR] | ||
| RARRPR[ARsVDALDDLTPPsTAESGSRSPTSNGGR]Sa (same RXRXXpS site in both phosphopeptides) | ||
| MICALL1 | L = only found in +insulin preparation | K[LQELASPPAGRPtPAPR]K |
| R[VEQmPQAsPGLAPR]T | ||
| PFKFB2 | 6.2 | PVRMR[RNsFTPLSSSNTIR]a |
| SSSNTIRRPR[NYSVGSRPLKPLsPLR] | ||
| TPI1 | 3.2 | KMNGR[KQsLGELIGTLNAAK]V |
| WDR20 | 1.5 | K[FATLsLHDR]K |
| YAP1 | 10.6 | KSHSR[QAsTDAGTAGALTPQHVR] |
| PPEPK[SHsRQAStDAGTAGALTPQHVR] | ||
| IVHVR[GDsETDLEALFNAVmNPK] | ||
| QVRPQELALR[SQLPtLEQDGGTQNPVSsPGmSQELR] | ||
| ALTPQHVR[AHSSPAsLQLGAVSPGTLTPTGVVSGPAATPTAQHLR] | ||
| ALTPQHVR[AHSSPAsLQLGAVsPGTLTPTGVVSGPAATPTAQHLR] | ||
| ZNRF2 | 2.6 | RTR[AYsGSDLPSSSSGGANGTAGGGGGAR]Aa |
| R[FPAQVPSAHQPsASGGAAAAAAAPAAPAAPR]S |
a RXRXX(pS/pT) motifs.
Although the features of many of the identified phosphopeptides were suggestive, the next question was whether any of the identified sites are actually phosphorylated by basophilic kinases downstream of PI 3-kinase and/or responsible for these proteins binding to 14-3-3. A literature survey showed that several of the RXRXX(pS/pT) motifs identified here are known 14-3-3-binding sites that are phosphorylated by PKB and other AGC kinases, including sites on PFKFB2 (cardiac PFK-2), BAD, AKT1S1 (PRAS40), and NEDD4L (Nedd4-2) (Refs. 7, 11, 36, and 37; supplemental Table 3). FOXO1 also has PKB-phosphorylated 14-3-3-binding sites that were not identified here, although we identified a (Ser(P)/Thr(P))-Pro site on this protein. The phosphorylated residue we identified on TSC2 is also known, although whether 14-3-3s bind to this site has been controversial (38, 39). We also identified phosphorylated residues from proteins reported previously to bind 14-3-3s in response to other or unknown signaling pathways, namely M110/MYPT1, GIT1, Raf1, A-RAF, KSR, PI4KB, PTPH1, KLC2, ELM3 (ELP95), HDAC4, HDAC7A, MAP3K2, DOCK7, CRTC2, YAP1, and WWTR1 (Taz) (Refs. 10, 23, 26, and 40–49; supplemental Table 3). For example, phosphorylated Ser294 (LNRTNpSQP) on phosphatidylinositol 4-kinase IIIβ (PI4KB) has been pinpointed as a 14-3-3-binding site that is phosphorylated by protein kinase D and critical for regulation of Golgi trafficking (42). The yeast homologue PIK1 also binds to 14-3-3s at an analogous phosphorylated site (LKRTApSNP) (50).
Cellular Regulation of insulin receptor substrate 2 (IRS2), CCDC6, ZNRF2, SASH1, PRAS40, Vasodilator-stimulated Phosphoprotein (VASP), and Lipolysis-stimulated Lipoprotein Receptor (LSR)
We next determined whether there were any novel targets of PI 3-kinase signaling among the candidates with high d0/d4 ratios (Figs. 2 and 3 and supplemental Table 1) and/or for which basophilic phosphorylated sites were identified (Fig. 4 and supplemental Table 2). Far-Western 14-3-3 overlay assays were used to determine whether tagged forms of candidate proteins extracted from transfected HEK293 cells bind directly to 14-3-3s (which is useful because supplemental Table 1 also lists proteins, such as kinesin heavy chain, that bind to 14-3-3s indirectly via intermediary phosphoproteins (10, 26)). In addition to IGF1 (in the presence or absence of the dual PI 3-kinase/mammalian target of rapamycin inhibitor PI-103) we stimulated cells with PMA (in the presence or absence of the p90RSK inhibitor BI-D1870) as a first step to testing the responsiveness of targets to other signaling pathways.
The IRS2 had a d0/d4 ratio of 9, and phosphopeptides were identified (Fig. 3D, Table I, and supplemental tables). IRS1 is known to bind 14-3-3 (51–53), and here we found that IRS2 also binds directly to 14-3-3 in a response to IGF1 that is blocked by PI-103. The IRS2/14-3-3 interaction was also promoted by phorbol ester and partially inhibited by BI-D1870 (Fig. 5), suggesting that p90RSK phosphorylates one 14-3-3-binding site on this protein possibly with a second site phosphorylated by a different phorbol ester-stimulated protein kinase.
CCDC6 also had a high d0/d4 score of 7.1 (Fig. 3D, Table I, and supplemental tables), and phosphorylated sites on this protein were identified here (Table I) and in the literature. We found that HA-CCDC6 bound to 14-3-3s in an insulin-stimulated and PI-103-inhibited manner (Fig. 5). In contrast, there was only a trace 14-3-3 binding signal for CCDC6 from phorbol ester-stimulated cells.
ZNRF2 with a d0/d4 score of 2.6 and an RXRXX(pS/pT) motif (RTRAYpS19GS) (Fig. 3D, Table I, and supplemental tables) showed insulin-stimulated and PI-103-inhibited binding to 14-3-3s. In contrast to IRS2 and CCDC6, ZNRF2 also bound to 14-3-3s in response to PMA, and this response was largely inhibited by the p90RSK inhibitor BI-D1870 (Fig. 5). The IGF1-stimulated and PMA-stimulated binding of 14-3-3s to HA-ZNRF2 was markedly decreased by Ser19 mutation to Ala (Fig. 5).
An RXRXXpS phosphorylated motif was identified in SASH1 (within the sequence RKRRVpS90QD; supplemental Tables 2 and 3). Fig. 5 shows that HA-SASH1 binds to 14-3-3, its binding capability is increased by stimulation of cells with IGF1 and phorbol ester, and these regulations are partially inhibited by PI-103 and BI-D1870, respectively. With a S90A mutation, the IGF1- and PMA-responsive increases in binding to SASH1 were lost, although the relatively high “basal” binding to 14-3-3 remained in this mutant. These data indicate that a 14-3-3 dimer may bind to one basal site and to the IGF1- and PMA-responsive phospho-Ser90 on SASH1.
PRAS40 is a known target of PKB (7). Like CCDC6 this protein was selectively responsive to PI 3-kinase/PKB and did not bind to 14-3-3s in response to phorbol ester (Ref. 31 and Fig. 5).
The VASP had a low d0/d4 ratio of 1.3 (supplemental Table 1), indicating that its 14-3-3 binding was not regulated by insulin. Consistent with this ranking, HA-VASP from transfected HEK293 cells was able to bind directly to 14-3-3 whether or not the cells were stimulated with insulin, IGF1, or phorbol ester (Fig. 5 and data not shown).
The LSR also had a low d0/d4 score of 1.2, and phosphorylated sites were identified for this protein, including an RXRXX(pS/pT) site (RPRARpS493LD; Fig. 3D, Table I, and supplemental tables). HA-LSR gave a 14-3-3 binding signal even when extracted from unstimulated cells, and the signal was no stronger from cells stimulated with IGF1 and PMA (Fig. 5 and data not shown). Nevertheless the binding of LSR to 14-3-3s was abolished by S493A mutation within the identified RXRXXpS motif (Fig. 5). We note that Ser493 has the characteristics of a potential site of phosphorylation by various kinases in the Ca2+/calmodulin-dependent protein kinase subfamily, and that 13 phosphorylated residues have been identified in mouse LSR (54). Thus, insulin-independent kinases promote the 14-3-3 binding of LSR.
DISCUSSION
14-3-3s bind to hundreds of phosphoproteins, so defining which signaling pathways stimulate the phosphorylation of which 14-3-3-binding sites opens exciting opportunities for understanding the global regulation of diverse cellular processes. Here we combined 14-3-3 affinity capture and proteomics strategies to visualize and identify the subset of proteins whose 14-3-3 binding responds to insulin/IGF1. These screens have proven predictive for those proteins that we tested with several novel validated targets of the insulin/IGF1/PI 3-kinase signaling pathway identified (Fig. 5). Further mining of the data sets should be fruitful, and the many candidates that remain to be checked include proteins with intriguing links to cancer, diabetes, and neurodegenerative disorders.
In contrast to SILAC procedures where differential labeling is done at the cell stage, dimethyl labeling was performed later on in our procedure, although with adequate controls this should not be a problem, and our methods are applicable to cell and tissue types that cannot be conveniently SILAC labeled. Refining the quantitative aspects of the screens, defining changes in phosphorylation stoichiometries, filtering out proteins that show minor responses to stimuli, and so on will require many reiterative comparisons backed up by biochemical analyses such as those in Fig. 5. We noticed for example that most of the proteins identified had a d0/d4 (±insulin) ratio greater than 1 (Fig. 3B), suggesting that unknown underlying trends might have skewed the data. Possibilities are selective proteolysis of the 14-3-3-binding proteins from unstimulated cells and/or an insulin-stimulated modification of the endogenous 14-3-3s that lowers their avidity so that targets are more easily released and better able to bind to the 14-3-3-Sepharose.
We also note that the data fell into a range of d0/d4 rankings, not a discrete division into high and low ranked (insulin-stimulated binding or not) categories. This gradation could relate to the fact that 14-3-3s are dimers, which often bind to two phosphorylated sites on the same target. Thus, in some cases insulin could stimulate phosphorylation of both 14-3-3-binding sites, whereas other proteins might already have one site phosphorylated in the unstimulated state. For example, basal binding of 14-3-3s to TBC1d4 (AS160) due to phosphorylation of Ser341 is markedly enhanced when insulin stimulates phosphorylation of both Ser341 and Thr642 of AS160 (17). In 2002, Yaffe (55) introduced the concept of one phosphorylated site on a target providing a high affinity 14-3-3 binding “gatekeeper” that facilitates docking of a second lower affinity site on the same target into the other side of the 14-3-3 dimer. In such a configuration, a 14-3-3 dimer behaves as a logic gate, a device in which either (“or”) or both (“and”) of two inputs are needed to trigger an output. Comparing the data in Fig. 5 gives a sense of how targets differ in the “digital logic” of how their two phosphorylated sites engage with a 14-3-3 dimer presumably dependent on differences in affinities and avidities. For example, phospho-Ser19 appears to be a gatekeeper site that is dominant for binding of ZNRF2 to 14-3-3. In contrast, 14-3-3 displays one mode of binding to SASH1 that is not regulated by IGF1 and PMA but that is enhanced by phosphorylation of the IGF1/PI 3-kinase- and PMA/p90RSK-responsive Ser90. In cases where different kinases phosphorylate each site, 14-3-3s effectively become “coincidence detectors” for two different inputs. Perhaps the binding and functional effect of 14-3-3s can also depend on the temporal order of phosphorylation and dephosphorylation of the relevant sites. It will be interesting to explore the full range of digital behaviors of 14-3-3s more widely in the future.
Overall our aim to identify novel targets of the PI 3-kinase signaling pathway was achieved. Newly validated insulin-responsive targets include a protein involved in the insulin/PI 3-kinase signaling pathway itself (IRS2), a proapoptotic protein (CCDC6), an E3 ubiquitin ligase implicated in synaptic vesicle trafficking (ZNRF2), and a signaling adapter protein (SASH1). We also confirmed direct binding of 14-3-3s to the actin-associated protein VASP and LSR, a transmembrane protein receptor related to Lisch-like, which has recently been linked genetically to type II diabetes in mice. These findings give exciting prospects of insights into how insulin and growth factors control cell behavior.
CCDC6 is best characterized as a transforming fusion with the tyrosine kinase RET or phosphatase PTEN in papillary thyroid tumors (56–59) and tyrosine kinase domain of the platelet-derived growth factor β receptor in certain leukemias (60, 61), suggesting that the CCDC6 gene has a high propensity for recombination (62, 63). The normal CCDC6 protein was identified as a substrate of the kinase ataxia telangiectasia mutated in response to DNA damage (64). In wild-type cells, etoposide and ionizing radiation promoted phosphorylation on Thr434 by ataxia telangiectasia mutated, which stabilized the nuclear buildup of CCDC6 and promoted apoptosis (64, 65). We hypothesize that binding of 14-3-3 to CCDC6 in response to PI 3-kinase signaling may inhibit its proapoptotic activity.
Zinc and ring finger 2 is one of two E3 ubiquitin ligases (ZNRF1 and ZNRF2) with a zinc finger adjacent to a ring finger (66). E3 ligases generally act as specificity modules that bring substrates to the E2 for ubiquitylation. In yeast two-hybrid experiments, ZNRF2 was one of several E3 ubiquitin ligases that bind to UBC13, the active component of the E2 ligase for Lys63-linked ubiquitylation (67), and we found that UBC13 co-purifies with ZNRF2 from extracts of transfected HEK293 cells (data not shown) indicating a physiological partnership between ZNRF2 and UBC13. ZNRF1 and ZNRF2 are both highly expressed in nerve cell presynapses, and the catalytically inactive proteins inhibit Ca2+-dependent exocytosis in PC12 cells (66, 68). Serine 19 of ZNRF2, which is phosphorylated and binds to 14-3-3s in response to both IGF1/PI 3-kinase and phorbol ester, is N-terminal to a MAGE domain of unknown function (69), suggesting that regulated binding of 14-3-3s to ZNRF2 may affect Lys63 ubiquitylation via effects on the MAGE domain of ZNRF2.
SASH1 is a little characterized, ubiquitously expressed member of the sterile α motif- and SH3 domain-containing SLY1 family of signaling proteins whose down-regulation in colon cancers was found to have negative prognostic significance for metastases and survival (70).
For LSR, 14-3-3s bind to at least one phosphorylated site (phospho-Ser435 within RPRARpS435VDAL), and although the relevant kinase is unknown, AMP-activated protein kinase is a candidate. LSR had been isolated previously by its affinity for 14-3-3s (24, 27) and is a known target of growth factor signaling by virtue of its fibroblast growth factor-stimulated phosphorylation on a tyrosine residue (MRVLpYYMEK where pY is phosphotyrosine) (71). Levels of this protein are elevated in certain colon cancers (72), whereas knockdown of LSR increases motility of bladder cancer cells (73). LSR is a transmembrane protein with extracellular immunoglobulin-like domains and phosphorylated sites in the putative intracellular region (Pfam database). The name LSR refers to its proposed role as a cell surface lipoprotein receptor that is activated by free fatty acids and clears dietary triglyceride-rich lipoproteins from the blood into liver and other tissues (74–76). LSR is critical for liver and embryonic development (77). The other name for this protein, liver-specific basic helix-loop-helix leucine zipper transcription factor (LISCH7), appears to be a misnomer. LSR is encoded by one of three related human genes (LSR, C1orf32, and ILDR1), and genetic variation in the mouse form of C1orf32, named Lisch-like, was recently linked to susceptibility to type II diabetes (78). C1orf32 has a serine (ESRAHS466GFYQ) corresponding to Ser435 in LRS that although not in an RXRXXpS motif does look like a potential AMP-activated protein kinase site, but there is no matching serine residue in ILDR1. Given that elevated levels of fatty acids and triglyceride-rich lipoproteins are linked to inflammation and insulin resistance (79, 80), we need closer examination of the roles and regulation of this family of proteins.
In summary, 14-3-3 capture and quantitative proteomics led us to discover new downstream targets of insulin signaling with roles in apoptosis, vesicle trafficking, metabolism, and cancer. Future work on these proteins should lead to exciting mechanistic insights. Although insulin/PI 3-kinase/AGC signaling has a dramatic effect in regulating many 14-3-3-binding proteins (Fig. 1), of the targets that we followed up only CCDC6 and PRAS40 were selectively responsive to PI 3-kinase, whereas IRS2, ZNRF2, and SASH1 are convergence points for the PI 3-kinase and MAPK/p90RSK pathways (Fig. 5). Understanding how these two pathways exert widespread controls over cells is a fascinating problem made all the more pressing because drugs that target these pathways are promising anticancer therapies, although interplay between these two pathways is emerging and must be understood to devise optimal combination therapies. One ambitious goal is therefore to use 14-3-3 capture combined with sensitive mass spectrometry to define how the global 14-3-3-binding phosphoproteome responds to different extracellular stimuli and inhibitors. Such overviews of the changing 14-3-3-linked phosphoproteome of cells could be used to pinpoint biomarkers whose 14-3-3 binding status shows which signaling pathways are active in healthy and diseased tissues and how they respond to drugs.
Supplementary Material
Acknowledgments
We thank Claire Balfour for tissue culture support and the Division of Signal Transduction Therapy antibody production team coordinated by Dr. James Hastie for affinity purification of antibodies. We also thank Sandra Crowther, Rachel Naismith, and Rebekah Tillotson for assistance with data analysis.
Footnotes
* This work was supported by the UK Medical Research Council via a Developmental Pathway Funding Scheme award and core funding, Diabetes UK, an Interdisciplinary Research Collaboration proteomic technology (Radical Solutions for Researching the Proteome (RASOR)) grant from the Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council, and the companies who support the Division of Signal Transduction Therapy at the University of Dundee, namely AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck-Serono, and Pfizer.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1 and 2 and Tables 1–4.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1 and 2 and Tables 1–4.
1 The abbreviations used are:
- IGF1
- insulin-like growth factor 1
- CCDC6
- coiled coil domain-containing 6
- IRS
- insulin receptor substrate
- LSR
- lipolysis-stimulated lipoprotein receptor (also known as LISCH7, liver-specific basic helix-loop-helix leucine zipper transcription factor)
- PAS
- phospho-Akt substrate
- PI
- phosphatidylinositol
- PKB
- protein kinase B, also known as Akt
- PMA
- the phorbol ester phorbol 12-myristate 13-acetate
- SASH1
- sterile α motif- and SH3 domain-containing protein 1
- VASP
- vasodilator-stimulated phosphoprotein
- YAP
- Yes-associated protein
- ZNRF
- zinc and ring finger
- MAPK
- mitogen-activated protein kinase
- HA
- hemagglutinin
- Erk
- extracellular signal-regulated kinase
- SCX
- strong cation exchange
- LTQ
- linear trap quadrupole
- MGF
- Mascot generic format
- KLC
- kinesin light chain
- SILAC
- stable isotope labeling with amino acids in cell culture
- E3
- ubiquitin-protein isopeptide ligase
- E2
- ubiquitin carrier protein
- AGC
- cAMP-dependent protein kinases A, cGMP-dependent protein kinases G, and phospholipid-dependent protein kinases G family of protein kinases
2 N. Wood, B. Wong, and C. MacKintosh, unpublished data.
3 N. Wood, B. Wong, K. Dissanayake, K. Geraghty, and C. MacKintosh, unpublished data.
REFERENCES
- 1.Cohen P. (2006) The twentieth century struggle to decipher insulin signalling. Nat. Rev. Mol. Cell Biol. 7, 867–873 [DOI] [PubMed] [Google Scholar]
- 2.Randhawa R., Cohen P. (2005) The role of the insulin-like growth factor system in prenatal growth. Mol. Genet. Metab. 86, 84– 90 [DOI] [PubMed] [Google Scholar]
- 3.Manning B. D., Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261– 1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkins P. T., Anderson K. E., Davidson K., Stephens L. R. (2006) Signalling through Class I PI3Ks in mammalian cells. Biochem. Soc. Trans. 34, 647– 662 [DOI] [PubMed] [Google Scholar]
- 5.Yuan T. L., Cantley L. C. (2008) PI3K pathway alterations in cancer: variations on a theme. Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 7.Kovacina K. S., Park G. Y., Bae S. S., Guzzetta A. W., Schaefer E., Birnbaum M. J., Roth R. A. (2003) Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 278, 10189–10194 [DOI] [PubMed] [Google Scholar]
- 8.Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. (2002) A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J. Biol. Chem. 277, 22115–22118 [DOI] [PubMed] [Google Scholar]
- 9.Berwick D. C., Dell G. C., Welsh G. I., Heesom K. J., Hers I., Fletcher L. M., Cooke F. T., Tavaré J. M. (2004) Protein kinase B phosphorylation of PIKfyve regulates the trafficking of GLUT4 vesicles. J. Cell Sci. 117, 5985–5993 [DOI] [PubMed] [Google Scholar]
- 10.Rubio M. Pozuelo, Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., Mackintosh C. (2004) 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 379, 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubio M. Pozuelo, Peggie M., Wong B. H., Morrice N., MacKintosh C. (2003) 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J. 22, 3514–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi X. J., Wildey G. M., Howe P. H. (2006) Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J. Biol. Chem. 281, 813–823 [DOI] [PubMed] [Google Scholar]
- 13.Tian Q., Feetham M. C., Tao W. A., He X. C., Li L., Aebersold R., Hood L. (2004) Proteomic analysis identifies that 14-3-3zeta interacts with beta-catenin and facilitates its activation by Akt. Proc. Natl. Acad. Sci. U.S.A. 101, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekimoto T., Fukumoto M., Yoneda Y. (2004) 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). EMBO J. 23, 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rena G., Prescott A. R., Guo S., Cohen P., Unterman T. G. (2001) Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem. J. 354, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanzel M., Kleine-Kohlbrecher D., Herold S., Hock A., Berns K., Park J., Hemmings B., Eilers M. (2005) Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nat. Cell Biol. 7, 30–41 [DOI] [PubMed] [Google Scholar]
- 17.Geraghty K. M., Chen S., Harthill J. E., Ibrahim A. F., Toth R., Morrice N. A., Vandermoere F., Moorhead G. B., Hardie D. G., MacKintosh C. (2007) Regulation of multisite phosphorylation and 14-3-3 binding of AS160 in response to IGF-1, EGF, PMA and AICAR. Biochem. J. 407, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramm G., Larance M., Guilhaus M., James D. E. (2006) A role for 14-3-3 in insulin-stimulated GLUT4 translocation through its interaction with the RabGAP AS160. J. Biol. Chem. 281, 29174–29180 [DOI] [PubMed] [Google Scholar]
- 19.Chen S., Murphy J., Toth R., Campbell D. G., Morrice N. A., Mackintosh C. (2008) Complementary regulation of TBC1D1 and AS160 by growth factors, insulin and AMPK activators. Biochem. J. 409, 449–459 [DOI] [PubMed] [Google Scholar]
- 20.Masters S. C., Fu H. (2001) 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 276, 45193–45200 [DOI] [PubMed] [Google Scholar]
- 21.Jacinto E., Lorberg A. (2008) TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 410, 19–37 [DOI] [PubMed] [Google Scholar]
- 22.Yip M. F., Ramm G., Larance M., Hoehn K. L., Wagner M. C., Guilhaus M., James D. E. (2008) CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metab. 8, 384–398 [DOI] [PubMed] [Google Scholar]
- 23.Angrand P. O., Segura I., Völkel P., Ghidelli S., Terry R., Brajenovic M., Vintersten K., Klein R., Superti-Furga G., Drewes G., Kuster B., Bouwmeester T., Acker-Palmer A. (2006) Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol. Cell. Proteomics 5, 2211–2227 [DOI] [PubMed] [Google Scholar]
- 24.Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., Zougman A., Woodgett J. R., Langeberg L. K., Scott J. D., Pawson T. (2004) Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 14, 1436–1450 [DOI] [PubMed] [Google Scholar]
- 25.Meek S. E., Lane W. S., Piwnica-Worms H. (2004) Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 279, 32046–32054 [DOI] [PubMed] [Google Scholar]
- 26.Ichimura T., Wakamiya-Tsuruta A., Itagaki C., Taoka M., Hayano T., Natsume T., Isobe T. (2002) Phosphorylation-dependent interaction of kinesin light chain 2 and the 14-3-3 protein. Biochemistry 41, 5566–5572 [DOI] [PubMed] [Google Scholar]
- 27.Benzinger A., Muster N., Koch H. B., Yates J. R., 3rd, Hermeking H. (2005) Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol. Cell. Proteomics 4, 785–795 [DOI] [PubMed] [Google Scholar]
- 28.Entingh-Pearsall A., Kahn C. R. (2004) Differential roles of the insulin and insulin-like growth factor-I (IGF-I) receptors in response to insulin and IGF-I. J. Biol. Chem. 279, 38016–38024 [DOI] [PubMed] [Google Scholar]
- 29.Hsu J. L., Huang S. Y., Chen S. H. (2006) Dimethyl multiplexed labeling combined with microcolumn separation and MS analysis for time course study in proteomics. Electrophoresis 27, 3652–3660 [DOI] [PubMed] [Google Scholar]
- 30.Harthill J. E., Rubio M. Pozuelo, Milne F. C., MacKintosh C. (2002) Regulation of the 14-3-3-binding protein p39 by growth factors and nutrients in rat PC12 pheochromocytoma cells. Biochem. J. 368, 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manke I. A., Nguyen A., Lim D., Stewart M. Q., Elia A. E., Yaffe M. B. (2005) MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell 17, 37–48 [DOI] [PubMed] [Google Scholar]
- 32.Stokoe D., Caudwell B., Cohen P. T., Cohen P. (1993) The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem. J. 296, 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. (2001) Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell 105, 257–267 [DOI] [PubMed] [Google Scholar]
- 35.Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 36.Liang X., Butterworth M. B., Peters K. W., Walker W. H., Frizzell R. A. (2008) An obligatory heterodimer of 14-3-3beta and 14-3-3epsilon is required for aldosterone regulation of the epithelial sodium channel. J. Biol. Chem. 283, 27418–27425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.She Q. B., Solit D. B., Ye Q., O'Reilly K. E., Lobo J., Rosen N. (2005) The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell 8, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai S. L., Tee A. R., Short J. D., Bergeron J. M., Kim J., Shen J., Guo R., Johnson C. L., Kiguchi K., Walker C. L. (2006) Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 173, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Inoki K., Yeung R., Guan K. L. (2002) Regulation of TSC2 by 14-3-3 binding. J. Biol. Chem. 277, 44593–44596 [DOI] [PubMed] [Google Scholar]
- 40.Baljuls A., Schmitz W., Mueller T., Zahedi R. P., Sickmann A., Hekman M., Rapp U. R. (2008) Positive regulation of A-RAF by phosphorylation of isoform-specific hinge segment and identification of novel phosphorylation sites. J. Biol. Chem. 283, 27239–27254 [DOI] [PubMed] [Google Scholar]
- 41.Fanger G. R., Widmann C., Porter A. C., Sather S., Johnson G. L., Vaillancourt R. R. (1998) 14-3-3 proteins interact with specific MEK kinases. J. Biol. Chem. 273, 3476–3483 [DOI] [PubMed] [Google Scholar]
- 42.Hausser A., Link G., Hoene M., Russo C., Selchow O., Pfizenmaier K. (2006) Phospho-specific binding of 14-3-3 proteins to phosphatidylinositol 4-kinase III beta protects from dephosphorylation and stabilizes lipid kinase activity. J. Cell Sci. 119, 3613–3621 [DOI] [PubMed] [Google Scholar]
- 43.Kanai F., Marignani P. A., Sarbassova D., Yagi R., Hall R. A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L. C., Yaffe M. B. (2000) TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koga Y., Ikebe M. (2008) A novel regulatory mechanism of myosin light chain phosphorylation via binding of 14-3-3 to myosin phosphatase. Mol. Biol. Cell 19, 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller J., Ory S., Copeland T., Piwnica-Worms H., Morrison D. K. (2001) C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8, 983–993 [DOI] [PubMed] [Google Scholar]
- 46.Screaton R. A., Conkright M. D., Katoh Y., Best J. L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J. R., 3rd, Takemori H., Okamoto M., Montminy M. (2004) The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119, 61–74 [DOI] [PubMed] [Google Scholar]
- 47.Wang K., Degerny C., Xu M., Yang X. J. (2009) YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 87, 77–91 [DOI] [PubMed] [Google Scholar]
- 48.Zhang S. H., Kobayashi R., Graves P. R., Piwnica-Worms H., Tonks N. K. (1997) Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3beta protein. J. Biol. Chem. 272, 27281–27287 [DOI] [PubMed] [Google Scholar]
- 49.Nishino T. G., Miyazaki M., Hoshino H., Miwa Y., Horinouchi S., Yoshida M. (2008) 14-3-3 regulates the nuclear import of class IIa histone deacetylases. Biochem. Biophys. Res. Commun. 377, 852–856 [DOI] [PubMed] [Google Scholar]
- 50.Demmel L., Beck M., Klose C., Schlaitz A. L., Gloor Y., Hsu P. P., Havlis J., Shevchenko A., Krause E., Kalaidzidis Y., Walch-Solimena C. (2008) Nucleocytoplasmic shuttling of the Golgi phosphatidylinositol 4-kinase pik1 is regulated by 14-3-3 proteins and coordinates Golgi function with cell growth. Mol. Biol. Cell 19, 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Craparo A., Freund R., Gustafson T. A. (1997) 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem. 272, 11663–11669 [DOI] [PubMed] [Google Scholar]
- 52.Ogihara T., Isobe T., Ichimura T., Taoka M., Funaki M., Sakoda H., Onishi Y., Inukai K., Anai M., Fukushima Y., Kikuchi M., Yazaki Y., Oka Y., Asano T. (1997) 14-3-3 protein binds to insulin receptor substrate-1, one of the binding sites of which is in the phosphotyrosine binding domain. J. Biol. Chem. 272, 25267–25274 [DOI] [PubMed] [Google Scholar]
- 53.Xiang X., Yuan M., Song Y., Ruderman N., Wen R., Luo Z. (2002) 14-3-3 facilitates insulin-stimulated intracellular trafficking of insulin receptor substrate 1. Mol. Endocrinol. 16, 552–562 [DOI] [PubMed] [Google Scholar]
- 54.Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., Comb M. J. (2008) Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. U.S.A. 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaffe M. B. (2002) How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57 [DOI] [PubMed] [Google Scholar]
- 56.Grieco M., Cerrato A., Santoro M., Fusco A., Melillo R. M., Vecchio G. (1994) Cloning and characterization of H4 (D10S170), a gene involved in RET rearrangements in vivo. Oncogene 9, 2531–2535 [PubMed] [Google Scholar]
- 57.Grieco M., Santoro M., Berlingieri M. T., Melillo R. M., Donghi R., Bongarzone I., Pierotti M. A., Della Porta G., Fusco A., Vecchio G. (1990) PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell 60, 557–563 [DOI] [PubMed] [Google Scholar]
- 58.Puxeddu E., Zhao G., Stringer J. R., Medvedovic M., Moretti S., Fagin J. A. (2005) Characterization of novel non-clonal intrachromosomal rearrangements between the H4 and PTEN genes (H4/PTEN) in human thyroid cell lines and papillary thyroid cancer specimens. Mutat. Res. 570, 17–32 [DOI] [PubMed] [Google Scholar]
- 59.Tong Q., Li Y., Smanik P. A., Fithian L. J., Xing S., Mazzaferri E. L., Jhiang S. M. (1995) Characterization of the promoter region and oligomerization domain of H4 (D10S170), a gene frequently rearranged with the ret proto-oncogene. Oncogene 10, 1781–1787 [PubMed] [Google Scholar]
- 60.Drechsler M., Hildebrandt B., Kündgen A., Germing U., Royer-Pokora B. (2007) Fusion of H4/D10S170 to PDGFRbeta in a patient with chronic myelomonocytic leukemia and long-term responsiveness to imatinib. Ann. Hematol. 86, 353–354 [DOI] [PubMed] [Google Scholar]
- 61.Schwaller J., Anastasiadou E., Cain D., Kutok J., Wojiski S., Williams I. R., LaStarza R., Crescenzi B., Sternberg D. W., Andreasson P., Schiavo R., Siena S., Mecucci C., Gilliland D. G. (2001) H4(D10S170), a gene frequently rearranged in papillary thyroid carcinoma, is fused to the platelet-derived growth factor receptor beta gene in atypical chronic myeloid leukemia with t(5;10)(q33;q22). Blood 97, 3910–3918 [DOI] [PubMed] [Google Scholar]
- 62.Nikiforova M. N., Stringer J. R., Blough R., Medvedovic M., Fagin J. A., Nikiforov Y. E. (2000) Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science 290, 138–141 [DOI] [PubMed] [Google Scholar]
- 63.Savage J. R. (2000) Cancer. Proximity matters. Science 290, 62–63 [DOI] [PubMed] [Google Scholar]
- 64.Merolla F., Pentimalli F., Pacelli R., Vecchio G., Fusco A., Grieco M., Celetti A. (2007) Involvement of H4(D10S170) protein in ATM-dependent response to DNA damage. Oncogene 26, 6167–6175 [DOI] [PubMed] [Google Scholar]
- 65.Celetti A., Cerrato A., Merolla F., Vitagliano D., Vecchio G., Grieco M. (2004) H4(D10S170), a gene frequently rearranged with RET in papillary thyroid carcinomas: functional characterization. Oncogene 23, 109–121 [DOI] [PubMed] [Google Scholar]
- 66.Araki T., Milbrandt J. (2003) ZNRF proteins constitute a family of presynaptic E3 ubiquitin ligases. J. Neurosci. 23, 9385–9394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plans V., Scheper J., Soler M., Loukili N., Okano Y., Thomson T. M. (2006) The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell. Biochem. 97, 572–582 [DOI] [PubMed] [Google Scholar]
- 68.Araki T., Nagarajan R., Milbrandt J. (2001) Identification of genes induced in peripheral nerve after injury. Expression profiling and novel gene discovery. J. Biol. Chem. 276, 34131–34141 [DOI] [PubMed] [Google Scholar]
- 69.Xiao J., Chen H. S. (2004) Biological functions of melanoma-associated antigens. World J. Gastroenterol. 10, 1849–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rimkus C., Martini M., Friederichs J., Rosenberg R., Doll D., Siewert J. R., Holzmann B., Janssen K. P. (2006) Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br. J. Cancer 95, 1419–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hinsby A. M., Olsen J. V., Mann M. (2004) Tyrosine phosphoproteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J. Biol. Chem. 279, 46438–46447 [DOI] [PubMed] [Google Scholar]
- 72.García J. M., Peña C., García V., Domínguez G., Muñoz C., Silva J., Millán I., Diaz R., Lorenzo Y., Rodriguez R., Bonilla F. (2007) Prognostic value of LISCH7 mRNA in plasma and tumor of colon cancer patients. Clin. Cancer Res. 13, 6351–6358 [DOI] [PubMed] [Google Scholar]
- 73.Herbsleb M., Birkenkamp-Demtroder K., Thykjaer T., Wiuf C., Hein A. M., Orntoft T. F., Dyrskjøt L.(2008) Increased cell motility and invasion upon knockdown of lipolysis stimulated lipoprotein receptor (LSR) in SW780 bladder cancer cells. BMC Med. Genomics. 1, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bihain B. E., Delplanque B., Khallou J., Chevreuil O., Troussard A. A., Michel L., Mann C. J., Yen F. T. (1995) Lipolysis-stimulated receptor: a newcomer on the lipoprotein research scene. Diabete Metab. 21, 121–126 [PubMed] [Google Scholar]
- 75.Yen F. T., Masson M., Clossais-Besnard N., André P., Grosset J. M., Bougueleret L., Dumas J. B., Guerassimenko O., Bihain B. E. (1999) Molecular cloning of a lipolysis-stimulated remnant receptor expressed in the liver. J. Biol. Chem. 274, 13390–13398 [DOI] [PubMed] [Google Scholar]
- 76.Yen F. T., Roitel O., Bonnard L., Notet V., Pratte D., Stenger C., Magueur E., Bihain B. E. (2008) Lipolysis stimulated lipoprotein receptor. A novel molecular link between hyperlipidemia, weight gain and atherosclerosis in mice. J. Biol. Chem. 283, 25650–25659 [DOI] [PubMed] [Google Scholar]
- 77.Mesli S., Javorschi S., Bérard A. M., Landry M., Priddle H., Kivlichan D., Smith A. J., Yen F. T., Bihain B. E., Darmon M. (2004) Distribution of the lipolysis stimulated receptor in adult and embryonic murine tissues and lethality of LSR−/− embryos at 12.5 to 14.5 days of gestation. Eur. J. Biochem. 271, 3103–3114 [DOI] [PubMed] [Google Scholar]
- 78.Dokmanovic-Chouinard M., Chung W. K., Chevre J. C., Watson E., Yonan J., Wiegand B., Bromberg Y., Wakae N., Wright C. V., Overton J., Ghosh S., Sathe G. M., Ammala C. E., Brown K. K., Ito R., LeDuc C., Solomon K., Fischer S. G., Leibel R. L. (2008)Positional cloning of “Lisch-Like”, a candidate modifier of susceptibility to type 2 diabetes in mice. PLoS Genet. 4,e1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shoelson S. E., Herrero L., Naaz A. (2007) Obesity, inflammation, and insulin resistance. Gastroenterology 132, 2169–2180 [DOI] [PubMed] [Google Scholar]
- 80.Steinberg G. R. (2007) Inflammation in obesity is the common link between defects in fatty acid metabolism and insulin resistance. Cell Cycle 6, 888–894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.