Abstract
HCV is an important human pathogen that represents a model for chronic infection since the majority of infected individuals fail to clear the infection despite generation of virus-specific T cell responses during the period of acute infection. While viral sequence evolution at targeted MHC class I restricted epitopes represents one mechanism for immune escape in HCV, many targeted epitopes remain intact under circumstances of viral persistence. In order to explore alternative mechanisms of HCV immune evasion, we analyzed patterns of expression of a major inhibitory receptor on T cells, programmed death-1 (PD-1), from the time of initial infection and correlated these with HCV RNA levels, outcome of infection, and sequence escape within the targeted epitope. We show that the level of PD-1 expression in early HCV infection is significantly higher on HCV-specific T cells from those who progress to chronic HCV infection compared to those who clear infection. This correlation is independent of HCV RNA levels, compatible with the notion that high PD-1 expression on HCV-specific CD8 T cells during acute infection inhibits viral clearance. Viral escape during persistent infection is associated with reduction in PD-1 levels on the surface of HCV specific T cells, supporting the necessity of ongoing antigenic stimulation of T cells for maintenance of PD-1 expression. These results support the idea that PD-1 expression on T cells specific for nonescaped epitopes contributes to viral persistence and suggest that PD-1 blockade may alter the outcome of HCV infection.
Keywords: CD8+ T cells, HCV infection, Programmed Death-1, Viral Escape
Introduction
Chronic viral infections, such as those caused by human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV) are among the leading causes of death in the world (1). While most viral infections induce successful T cell responses that completely eliminate them, HCV, HBV, and HIV have developed mechanisms to evade immune elimination, allowing them to persist in many if not all infected individuals. HCV is found in virtually every region of the world with an estimated 170 million people and 1–2% of the general population of most countries infected (2). Chronic HCV may cause cirrhosis or liver cancer and, in the United States, is the most common indication for liver transplantation as well as the most common cause of hepatocellular carcinoma (3–7). In the United States, there are approximately four million people with persistent hepatitis C infection and more than ten thousand HCV-related deaths each year, with mortality from HCV expected to double in this decade and possibly surpass that caused by HIV (4, 8, 9).
HCV is not only an important cause of disease, but it is also an ideal infection in which to study viral evasion mechanisms because infection persists in the majority of infected individuals but not all since approximately 25% of those infected successfully clear the virus. This permits comparison of immune responses between those individuals who successfully control the infection and those who fail. The immune correlates that determine whether a patient resolves infection or proceeds to chronic infection are not completely defined, but viral escape is likely a contributing factor in HCV as well as in HBV and HIV infection. Because immune responses take weeks to develop, and pathogens replicate rapidly, it is well-recognized that immune escape mutations may blunt the effectiveness of the immune response (10). Indeed, evasion of the immune response via substitution within T cell epitopes has been demonstrated during HCV, HBV, SIV, and HIV infections (11–22). However, detectable levels of CD8 T cell recognition do not always result in escape substitutions and many CD8 T cell epitopes remain intact while others mutate within the same host (11, 23). Reversion of immune-escape mutations after transmission to a new host suggests that immune escape can be associated with a significant cost to the virus in terms of fitness (17). Therefore, it is possible that some substitutions result in virus with such significantly reduced fitness that substitution at that position is not observed. Reversion of immune escape mutations can even occur in the original host without viral elimination. This finding not only highlights the presumed fitness cost of certain immune escape mutations but also emphasizes the importance of additional immune evasion mechanisms that inhibit the immune system from eliminating HCV despite an expanded T cell repertoire capable of recognizing viral epitopes. Among others, cell surface receptors associated with T cell regulation have been shown to play important roles in regulating T cell function and responsiveness in chronic murine viral infections and may play a role in control of human viral infections.
One of the most important categories of candidate cell membrane receptors that could be participating in the hyporesponsiveness of human virus-specific T cells is the growing family of inhibitory (or regulatory) receptors. A number of inhibitory receptors that down-regulate T cell function have been recently characterized and their engagement appears to inhibit CD8 effector function. Programmed Death -1 (PD-1) is an ITIM-containing inhibitory receptor expressed on activated T cells that binds two known ligands: B7-H1/PD-L1, which is expressed at high levels in the liver, and B7-DC/PD-L2, which is predominantly expressed on dendritic cells (24–27). Although B7-DC/PD-L2 binds PD-1 with higher affinity than B7-H1/PD-L1, B7-H1/PD-L1 appears to be the major PD-1 ligand in vivo responsible for inhibition of T cell function. Antibody blockade of B7-H1/PD-L1 and PD-1 partially reversed the inactivity of exhausted lymphocytic choriomeningitis virus (LCMV)-specific T cells. PD-1 has recently been shown to be upregulated on HIV and HCV-specific T cells, suggesting that PD-1 upregulation may be an important mechanism for viral immune evasion in these chronic human viral diseases (28–32). However, the absence of analysis from the time of documented infection of PD-1 expression and of viral sequence evolution precluded determination of the role of antigen persistence in maintenance of PD-1 expression. A recent study examining PD-1 expression on CD8 T cells from SIV infected macaques demonstrated that PD-1 expression gradually declined on CD8 T cells specific for an SIV-derived epitope that had undergone mutational escape (33). The authors suggested that sustained antigen-specific T cell receptor (TCR) stimulation is the primary determinant of PD-1 expression, but the study examined just one mutated epitope so the decline could have been characteristic of some other feature of the host or the epitope rather than the mutation. In addition, the mutation studied completely abrogates recognition so it is not clear if a decline in PD-1 expression would be observed with mutations that decrease but decrease but do not completely abrogate recognition. Similar data have not yet been demonstrated with other epitopes, in other infections, or in humans.
In order to more clearly define variables associated with PD-1 expression and outcome of HCV infection, we performed a detailed analysis of PD-1 expression on HCV-specific T cells monitored longitudinally in a cohort of patients studied from the time of infection. PD-1 levels were ascertained at multiple time points post infection and correlated with HCV RNA levels, sequence variation within the cognate epitopes, and outcome of infection. We demonstrate that PD-1 upregulation precedes epitope mutation and viral escape is associated with reduction in PD-1 expression on the surface of HCV specific T cells, supporting the necessity of ongoing antigenic stimulation of T cells for upregulation of PD-1. We observed an inverse correlation between PD-1 expression during acute infection and subsequent viral clearance, independent of HCV RNA levels, indicating that PD-1 expression on T cells specific for nonescaped epitopes contributes to viral persistence.
Materials and Methods
Study Subjects
Blood samples were obtained from consenting HCV infected adults participating in a prospective study of young Intravenous Drug Users in Baltimore, MD, as previously described (36). Table I shows the demographics of subjects that were studied. At each visit, participants were provided counseling to reduce the risks of drug use. Blood was drawn for the isolation of serum, plasma, and peripheral blood mononuclear cells (PBMC) in a protocol designed for monthly follow-up. Serum and plasma were stored at −80°C, PBMC were isolated by Ficol-hypaque (GE Healthcare Bio-Sciences AB, Sweden) gradient centrifugation and cryopreserved in liquid nitrogen.
Table I.
Demographic characteristics of the 20 adult study subjects
| Characteristic | Value |
|---|---|
| Median Age at seroconversion (range) | 24 (20–46) |
| Race | |
| Caucasian | 100% |
| Other | 0% |
| % Male | 60% |
| Genotype | |
| 1a | 16 |
| 1b | 1 |
| 2b | 1 |
| 3a | 1 |
| Unknowna | 1 |
| Outcome of infection (%) | |
| Persistent | 14 (70) |
| Control | 4 (20) |
| Unknownb | 2 (10) |
| Mode of infection injection drug use (%)c | 19 (95) |
| Mean length of follow-up | 673 days (106–1521) |
| Mean number of time points studied | 3.5 (1–6) |
| HCV RNA levels during the first 180 days following | Median 95,700 IU/ml |
| infection | (0 – 6.66 × 106 IU/ml) |
| HCV RNA levels after more than 180 days following | Median 47,300 IU/ml |
| infection | (0 –7.56 × 106 IU/ml) |
| Mean number of epitopes tested per patient | 2.5 (1–5) |
For one subject who cleared HCV viremia rapidly and missed a monthly visit, HCV antibody seroconversion but no period of viremia was observed. Therefore, the genotype of that subject’s infection is unknown.
Two subjects were lost to follow-up before 300 days following infection, the minimum time of follow-up required to meet the definition of any outcome.
The only study subject not infected via injection drug use was infected in a common source outbreak.
HCV RNA levels
HCV RNA levels were determined using the quantitative RT-PCR assay (COBAS AMPLICOR HCV Monitor version 2.0 or COBAS TaqMan™ HCV Test (Roche Molecular Systems Inc., Branchburg, N.J., USA) according to the manufacturers instructions. The COBAS AMPLICOR HCV and COBAS TaqMan™ assays have a lower limit of quantitation of 2.8 log10 IU/ml and 1.7 log10 IU/ml, respectively. HCV clearance was defined as the presence of anti-HCV antibody with HCV RNA undetectable by the COBAS TaqMan™ HCV Test (Roche Molecular Systems Inc., Branchburg, N.J., USA) in serum or plasma specimens from ≥ 2 consecutive visits obtained at least 300 days after initial detection of viremia. Persistence was defined by detection of HCV RNA by either COBAS HCV assay in serum or plasma specimens obtained at least 300 days after initial viremia.
Multimers
APC-labeled multimers were obtained through ProImmune (Springfield, VA, USA) and Beckman Coulter Immunomics (San Diego, CA, USA). They comprised a panel of nine HCV multimers A2 (KLVALGINAVNS3:1406–1415, ALYDVVTKLNS5:2594–2602, RLWHYPCTVE23:614–622, CINGVCWTVNS3:1073–1081, VLSDFKTWLNS4:1987–1995), A01 (ATDALMTGYNS3:1435–1443) B7 (GPRLGVRATCore:41–49), B35 (HPALVFDITNS2:881–889, HPNIEEVALNS3:1359-1357). Eight CMV, EBV and Flu multimers were used as controls and were included in the evaluation. These consisted of A01-CMV (VTEHDTLLYpp50 245–253) A2-CMV (NLVPMVATVpp65:495–504) B7-CMV (TPRVTGGGAMpp65:417–426), A1-Flu (CTELKLSDYNP 44–52), A2-Flu (GILGFVFTLMP 58–66), A2-EBV (GLCTLVAMLBMLF-1 259–267, CLGGLLTMVLMP-2 426–434) and B8-EBV (RAKFKQLLBZLZ-1 190–197).
Phenotypic analysis
PBMC were thawed and rested at 37°C/5% CO2 for at least 5 hours. 1–2 × 106 cells were resuspended in 50µl/well of 10% human serum (Gemini Bio-Products, California, USA)/10% Fetal Calf Serum (FCS, Hyclone, Utah, USA). 50µg of hIgG (Jackson ImmunoResearch Labs Inc. PA, USA) and the cells were incubated at room temperature for 10 minutes, 5µl of antigen specific multimer was added and the incubation continued for a further 20 minutes. Either biotinylated PD-1 (anti-hPD-1.5, Medarex, CA) or IgG4 isotype control (Medarex, CA) were added at a final concentration of 5µg/ml and the cells incubated for an additional 20 minutes at room temperature (37). PBMC were washed twice with FACS buffer (1% BSA/0.1% NaN3/PBS) and resuspended in 50µl of 10% human serum (Gemini Bio-Products, California, USA)/10% FCS. To this Strepavidin-PE (10µg/ml, eBioscience, USA) and CD8-PerCP Cy5.5 (BD Pharmingen, USA) were added and PBMC were incubated at room temperature for 20 minutes. PBMC were washed twice with FACS buffer and resuspended 200µl of 2% paraformaldehyde (J.T. Baker, NJ, USA) and analyzed on a FACSCanto Flow Cytometer with 24 hours. At least 50,000–100,000 cells were collected in the lymphocyte gate, from which CD8+ cells were isolated. Subsequently, CD8+ multimer positive (multimer +) cells were selected and PD-1 expression was measured in the gated total CD8+ multimer+ T cells.
Viral sequencing
From 140–280 uL of serum or plasma, the 5.2 kb region from the 5’UTR to the NS3/NS4A junction was amplified as previously described (38). For each specimen, forty clones were isolated and stored, and the first twenty were selected for preliminary clonal analysis. Preliminary clonal analysis was performed by sequencing and phylogenetic analysis of a 450 nt region spanning the E1/E2 junction including hypervariable region 1 (HVR1). Sequence contigs were assembled and aligned using Aligner version 2 (CodonCode Corp., Dedham, MA) and trimmed to equal length using BioEdit (39). The GTR+I+G analytical model (parameters available on request from the authors) was selected using the AIC criterion as implemented in ModelTest version 3.7 (40) and PAUP* version 4b10 (Sinauer Associates, Sunderland, MA). Phylogenetic trees were estimated using the neighbor-joining algorithm implemented in PAUP*. For each specimen, the sequence closest to the phylogenetic center of the tree (COT) was identified as previously described (41) and sequenced. These 5.2 kb contigs were assembled, aligned, and edited as described above. Because longitudinal sequencing provided validation of conserved clonal sequences, only novel epitope substitutions required validation by sequencing additional stored cDNA clones.
Assessment of Potential Escape Mutations
Peptides originating from the HCV H77 genome (genotype 1a) were used to assess T cell function by the Elispot assay. Amino acid replacements in epitopes were tested as potential immune-escape substitutions by synthesizing each of the variant peptides and testing them in serial dilution as antigens for bulk PBMC and T cell lines raised to original and mutated versions of the epitope in an IFN-γ ELISpot assay. These methods have been described in detail previously (11). The observed amino acid replacements were classified as escape substitutions if they reduced autologous T cell recognition of the epitope in which it was located as measured by at least a two-fold reduction in SFC at two or more peptide dilutions.
Statistical analysis
Linear regression was used to examine the relationship between the covariates and the level of PD-1. Generalized estimating equations (GEE) methods were used to account for the multiple observations per participants (42). Analysis was performed using SAS version 9 (Cary, North Carolina). Differences were considered significant if p-values were <0.05.
Results
Characteristics of study populations
The demographic characteristics of the twenty subjects studied are listed in Table I. Subjects were selected on the basis of expression of human leukocyte antigen (HLA) alleles for which multimers were available as well as on the basis of having previously demonstrated peptide-specific responses by interferon-gamma (IFN-γ) ELISpot. Subjects with HIV infection were excluded. Using peptides representing T cell epitopes identified previously, HLA-matched major histocompatibility complex multimers with peptides recognized by the subject were used to assess PD-1 expression on HCV specific CD8 T cells obtained from the subjects at time points from initial detection of CD8 T cell responses through the first four years following infection. As controls, we performed multimer/antibody costaining for Epstein Barr virus (EBV) cytomegalovirus (CMV), and influenza (Flu) specific T cells. Statistical analysis of the relationship between expression of PD-1 with gender, age, HCV RNA levels, status of antigen (intact or mutated) and virologic outcome of infection was then performed. PD-1 expression on HCV-specific T cells was essentially uniformly positive and generally unimodal (Figure 1). Therefore, we used mean levels of PD-1 expression (mean fluorescence intensity, MFI) on the entire multimer-positive population of T cells rather than percentage positivity, as an unbiased quantitative estimate of PD-1 expression. Other studies of PD-1 have also demonstrated that the relative expression levels of PD-1 is probably the more useful marker than is the presence or absence of PD-1 as measured by percentage (43).
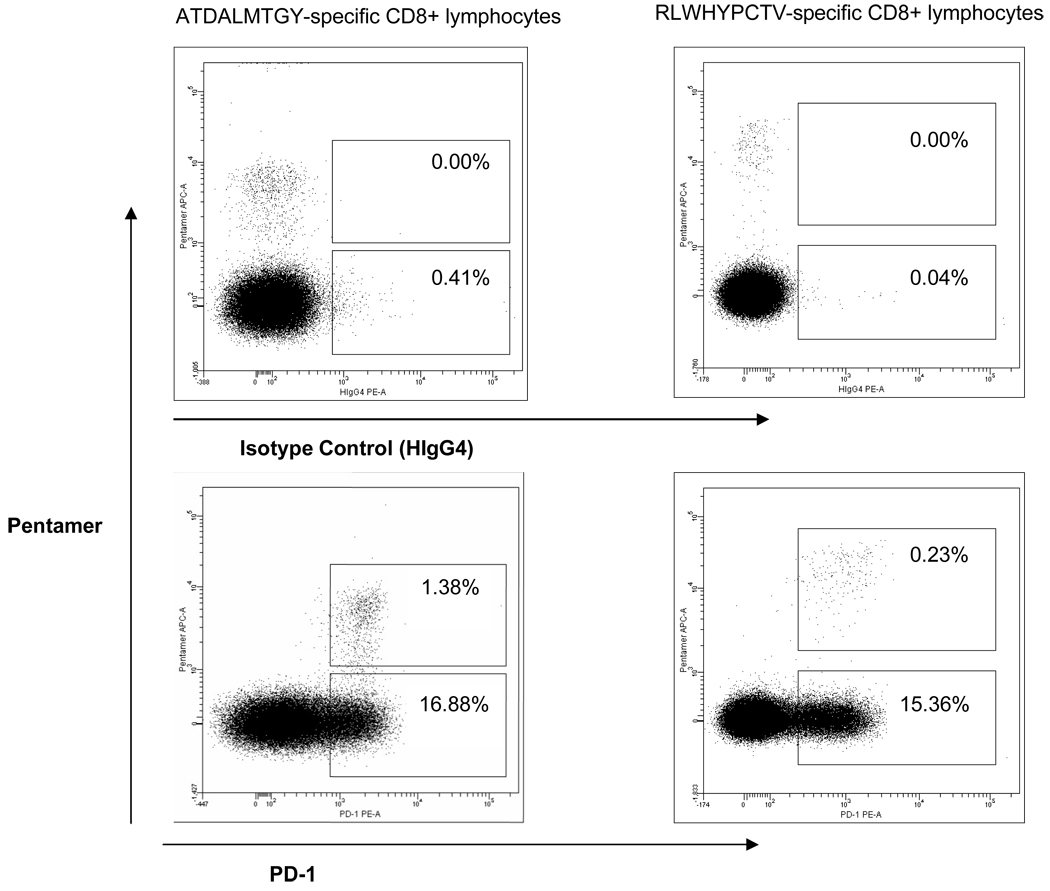
Figure 1. PD-1 staining patterns for two representative subjects.
PD-1 expression on HCV-specific T cells was essentially uniformly positive and generally unimodal. Therefore, the mean levels of PD-1 expression (mean fluorescence intensity, MFI) on the entire multimers positive population of T cells rather than percentage positivity was used as an unbiased estimate of PD-1 expression. The numbers in the top boxes indicate the percentage of isotype control (top row) or PD-1 positive (bottom row) CD8 positive HCV-specific T cells.
Statistical Analysis for Correlations with PD-1 Level
Given that we had different numbers of subjects in each outcome group and they were sampled at varying numbers of time points following infection, we chose the generalized estimating equations (GEE) as our statistical method. Using this method, which accounts for different numbers of subjects sampled at different numbers of time points, we determined that PD-1 levels, as measured by the MFI on HCV specific T cells, correlate positively with HCV RNA levels but not with subject gender or age (Table II). Race could not be considered since all the study subjects were Caucasian. When the levels of PD-1 were compared between subjects based on outcome of infection, they were significantly higher on HCV specific T cells in those with persistent infection compared to the levels on T cells from subjects who cleared infection, both in the first 180 days of infection and using all time points assessed. In a multivariate model, both log10 HCV RNA and clearance were significantly associated with the PD1-level. When the model was changed so that clearance was outcome, PD-1 levels were again associated with clearance (p=0.0007). Due to the limited variability of HCV-RNA in the clearance group a model which included HCV RNA could not be tested. Earlier studies on PD-1 expression in chronic viral infections have not assessed whether the difference in T cell PD-1 expression between subjects who control HIV or HCV and those with progressive HIV or chronic HCV infection is based on the level of virus even though our study and others have shown that levels of virus (HIV or HCV) and PD-1 expression on virus-specific T cells are positively correlated. Given that HCV RNA levels are higher in those with persistent infection than in those who have cleared infection, we analyzed this association controlling for HCV RNA levels and found that PD-1 levels are independently associated with outcome of infection. In fact, multivariate analysis revealed that in the first 180 days of infection, PD-1 levels were correlated only with outcome of infection, not with HCV RNA levels. These data are most compatible with the notion that PD-1 expression or a factor which regulates it has causal effects on outcome of infection rather than that PD-1 expression simply represents a marker for presence and levels of antigen.
Table II.
Factors Associated with PD-1 Levela on HCV Specific T Cells
| Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Estimate (95% CI) | p value | Estimate (95% CI) | p value | |||
| Total | Female | 0.15 | −0.12 – 0.42 | 0.2916 | |||
| follow-up | Age by 10 yrs | −0.12 | −0.28 – 0.05 | 0.1639 | |||
| periodb | Log10 HCV RNA | 0.06 | 0.03 – 0.09 | 0.0003 | 0.05 | 0.02 – 0.09 | 0.0006 |
| HCV clearance | −0.25 | −0.43 – −0.07 | 0.0075 | −0.14 | −0.26 – −0.01 | 0.0382 | |
| First 180 | Female | 0.00 | −0.28 – 0.28 | 0.9979 | |||
| days of | Age by 10 yrs | 0.39 | −0.08 – 0.86 | 0.1000 | |||
| infectionc | Log10 HCV RNA | 0.07 | 0.03 – 0.10 | 0.0005 | 0.03 | −0.01 – 0.07 | 0.1537 |
| HCV clearance | −0.41 | −0.50 – −0.32 | <0.0001 | −0.35 | −0.51 – −0.19 | <0.0001 | |
measured by mean fluorescence intensity
129 observations used from 17 participants
47 observations used from 13 participants
PD-1 Levels on HCV Specific T cells Versus the Total CD8 Population T cells
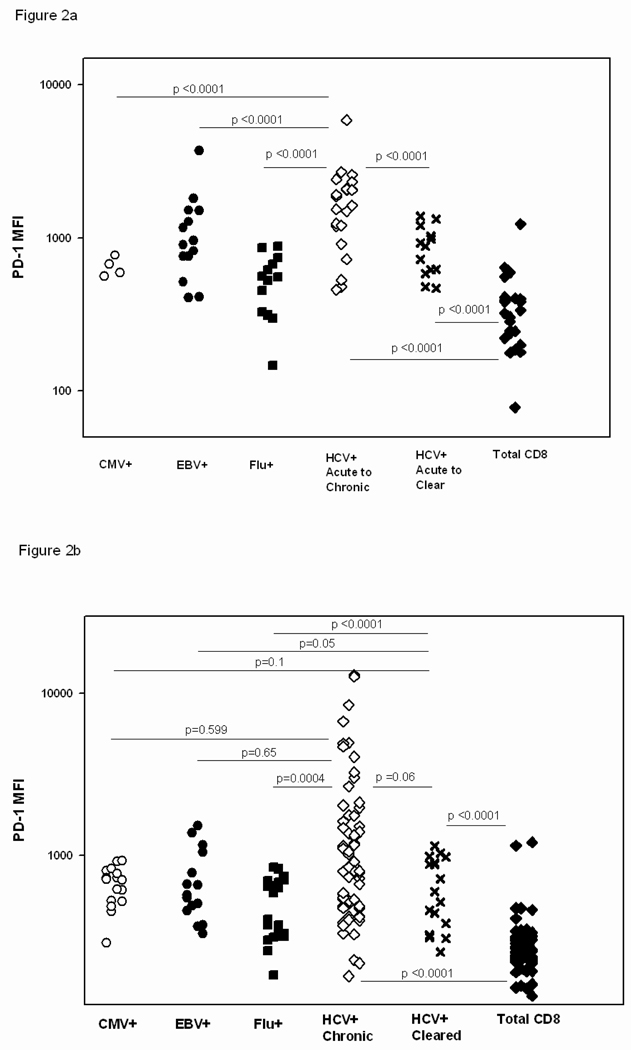
PD-1 is significantly upregulated on HCV-specific T cells versus the total CD8 T cell population at all stages of infection regardless of the outcome of HCV infection (Figure 2). Given that the level of PD-1 remains high on the cell surface of T cells specific for recognized HCV antigens relative to the level on the general CD8 T cell population long after control of HCV infection, continued recognition of antigen does not seem to be required to maintain PD-1 levels above that of the general CD8 population (Figure 2b).
Figure 2. PD-1 expression on HCV-specific T cells is higher than on influenza-specific T cells and the general CD8 T cell population regardless of outcome of infection or the phase of infection.
The MFI of PD-1 on the T cell surface was compared between HCV specific T cells from subjects with either outcome of infection, control specific T cells and the general CD8 population in the first 180 days of HCV infection (HCV+ acute to chronic or cleared, 2a) and at time points following 180 days of HCV infection (HCV+ chronic or cleared, 2b). The level of PD-1 expression is also higher in acute infection on HCV specific T cells from subjects who remain persistently infected than those who clear HCV infection. Each data point represents the MFI of the PD-1 level on a specific tetramer positive population within an individual.
PD-1 Levels on HCV Specific T cells in Subjects Who Progress to Chronic Infection Versus Those Who Clear Virus
PD-1 is significantly upregulated on HCV-specific T cells versus T cells specific for influenza from the acute phase through chronic infection in individuals who progress to chronic HCV infection (Figure 2). PD-1 levels are also higher on HCV-specific T cells than on T cells specific for EBV and CMV in the first 6 months of HCV infection but not in later stages of HCV infection in subjects who progress to chronic infection (Figure 2). This suggests a change in PD-1 modulation on HCV-specific T cells in the later stages of infection relative to EBV- or CMV- specific T cells and that regulation of PD-1 might differ in the acute and chronic phases of infection.
There were not enough data points in the first six months of infection for control (EBV, CMV or influenza) antigen-specific T cells from subjects who cleared HCV infection to compare them to HCV-specific responses in the early phase of infection. However, the level of PD-1 on HCV-specific T cells for subjects who cleared infection was significantly higher than on influenza-specific but not other control antigen-specific T cells after the first six months of infection (Figure 2b). HCV infection is characterized by a longer duration of viremia than is influenza, and PD-1 may be regulated in part by duration of viremia or another difference between influenza and cleared HCV infection. Significantly, the level of PD-1 on HCV-specific T cells in the first six months of infection for subjects who cleared infection was lower than on HCV-specific T cells from subjects who failed to clear HCV infection (Figure 2a, p<0.0001). Smaller differences were observed between these two groups when PD-1 levels were assessed beyond 6 months after initial infection (Figure 2b, p= 0.06). The range of PD-1 expression on HCV specific T cells in the chronic phase of HCV infection is broad with some levels less than 100 and others exceeding 2000. However, the bulk of the data points for the HCV+ Chronic population in Figure 2b fall in the lower half of PD-1 MFI’s measured, resulting in an insignificant difference between the two groups more than six months after infection. This further suggests that regulation of PD-1 may differ between the acute versus chronic phases of infection. We therefore further analyzed the association between PD-1 levels, infection outcome, and HCV RNA levels in early and later infection.
PD-1 Levels on HCV-Specific T cells Stratified by HCV RNA Levels and Outcome of Infection
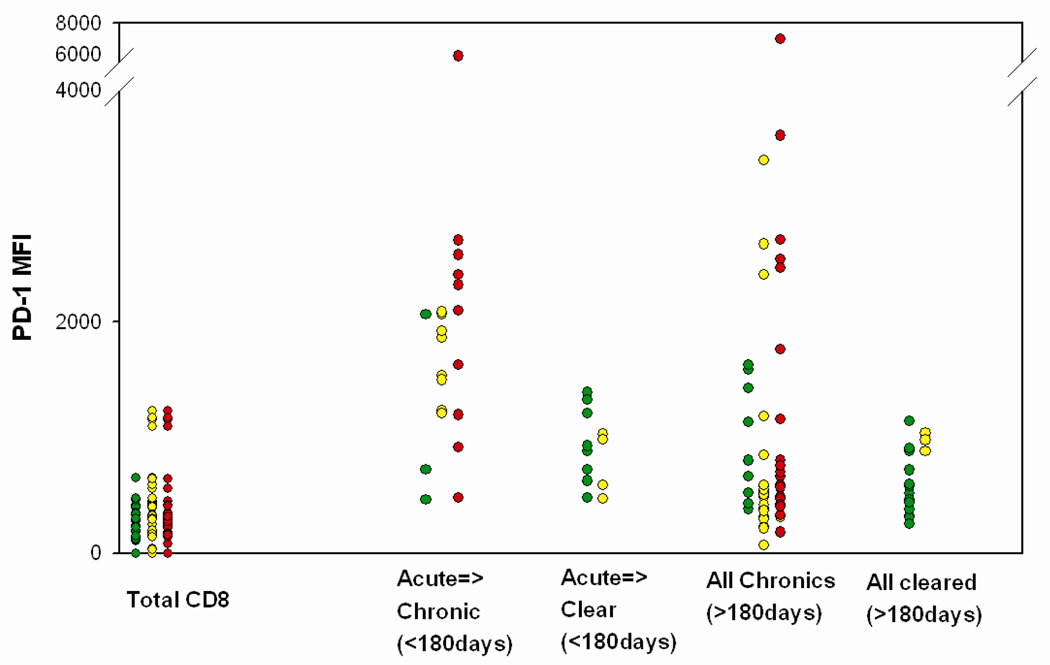
As shown in Table II and Figure 2, the level of PD-1 is significantly higher in early infection on HCV specific T cells from those who fail to control infection versus T cells from those who clear HCV infection. Despite high HCV RNA levels being generally associated with high PD-1 levels, the levels of PD-1 on HCV specific T cells ranged widely at all HCV RNA levels both early and late in infection (Figure 3). We observed that some HCV-specific T cells from subjects with chronic infection expressed low PD-1 levels in the setting of high circulating HCV RNA levels (Figure 3). Low levels of PD-1 have also been observed on some HIV-specific T cells with high HIV RNA levels (28). We hypothesized that T cells with low levels of PD-1 expression in the setting of high viral RNA levels recognize epitopes that have mutated in order to escape the immune response. Viral amino acid substitutions in an epitope that abrogate the generation of peptide-MHC complexes on the cell surface result in functional antigen loss even in the setting of high HCV RNA levels. If intact antigen is required for a T cell to receive the signals that induce PD-1 upregulation, T cells specific for HCV epitopes that have escaped via impaired presentation on antigen presenting cells would be expected to have low levels of PD-1 expression.
Figure 3. PD-1 expression on HCV specific T cells stratified by HCV RNA level during early and late infection.
HCV RNA levels at the time the T cells were acquired are indicated by colored symbols. Red: >100,000 copies/mL Yellow: 600–100,000 copies/mL Green: <600 copies/mL. Acute infection was defined as time points less than 180 days from initial viremia. PD-1 levels are highly variable in chronic infection. Some HCV-specific T cells from subjects with chronic infection expressed low PD-1 levels in the setting of high circulating HCV RNA levels (Red dots at low PD-1 MFI values).
Correlations between Viral Sequence Evolution and PD-1 Levels on HCV Specific T cells
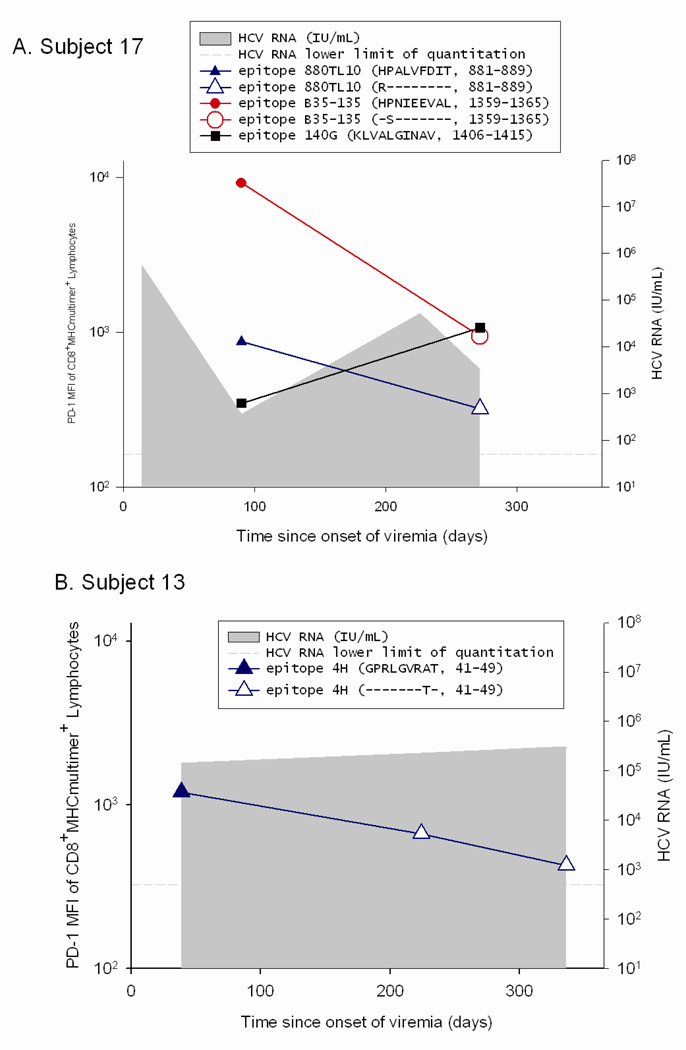
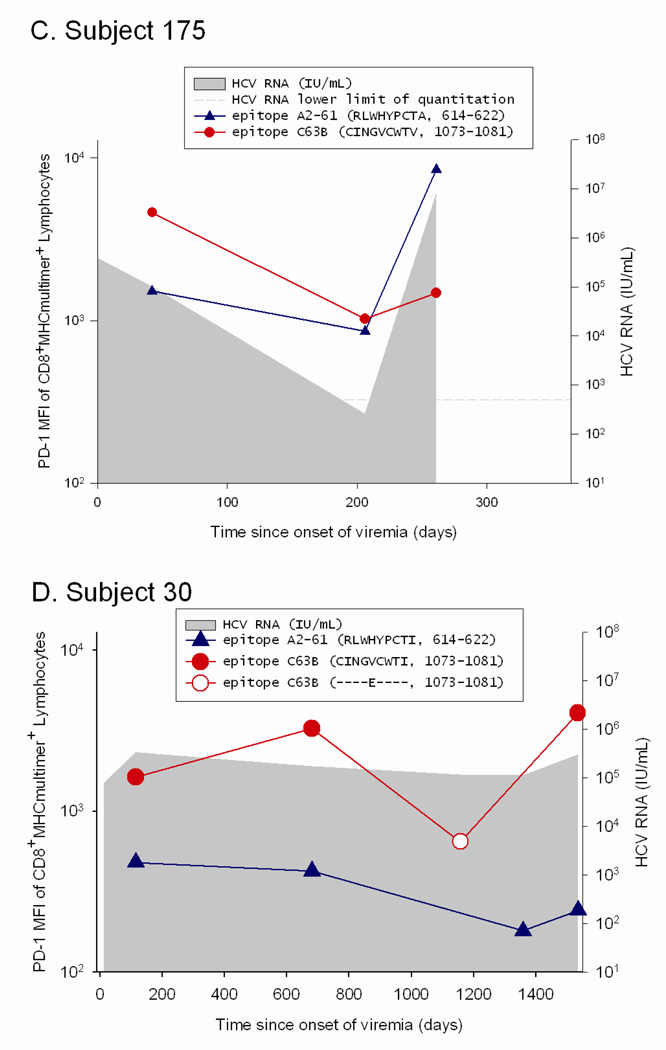
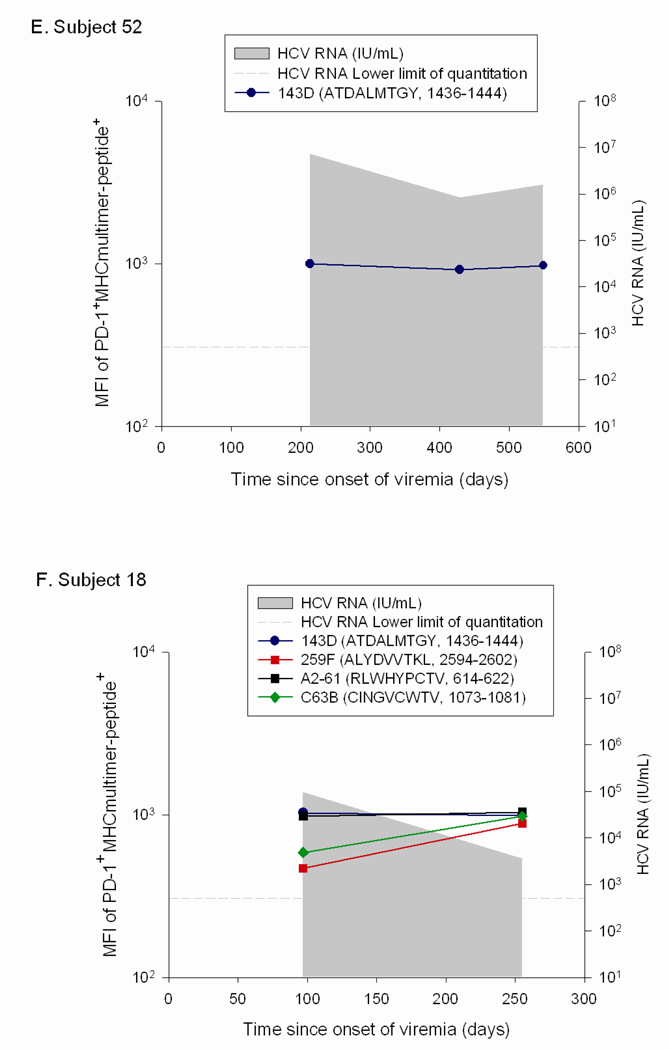
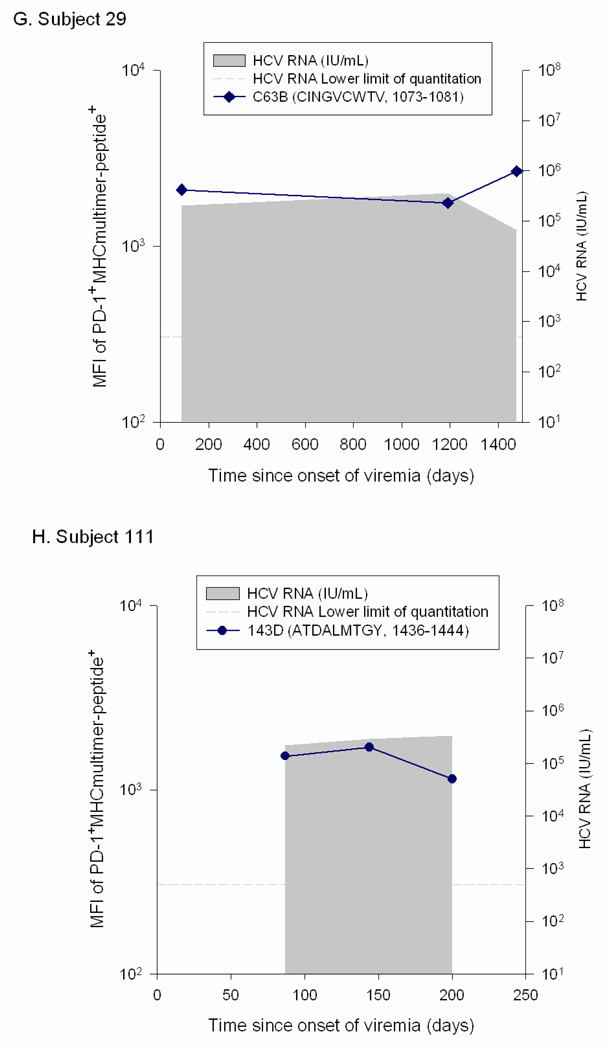
In order to determine if the low PD-1 levels seen on T cells in patients with high circulating HCV RNA levels were due to escape mutations, the viral sequence was assessed at initial infection and surrounding the time at which the PD-1 levels were found to be low relative to initial PD-1 levels. We compared the levels of PD-1 on T cells specific for epitopes that underwent substitutions that impaired T cell recognition to that on T cells specific for epitopes that remained constant over the same time period in the same subjects. For two subjects with some epitopes that underwent substitution while other epitopes were maintained constant (Subjects 17 and 30), the decline in PD-1 levels was only observed on T cells specific for the epitopes that underwent substitution (Figure 4). In contrast, PD-1 levels remained stable or increased for epitopes in the same host that did not undergo substitution. Subject 17 recognized three epitopes, two of which underwent substitution in the first six months of infection (880TL10 and B35-135) and the other of which remained intact (140G) (Figure 4a). T cells specific for 880TL10 and B35-135 showed three and ten fold decreases in PD-1 levels following substitution, respectively. In contrast, Subject 17’s T cells recognizing 140G, which remained constant over that time period, demonstrated a three fold increase in PD-1 expression. Subject 17’s 880TL10 and B35-135 HCV epitopes are both HLA B*3501 restricted. The substitutions resulted in 365 and 1690 fold reductions in HLA B*3501 binding, respectively. We have previously demonstrated that these mutations significantly impair T cell recognition in vitro but do not completely abrogate recognition.11 With these substitutions, T cell recognition of those epitopes is likely significantly impaired in vivo despite high levels of circulating virus. Similarly, in subject 13, who had one HCV specific T cell response that could be assessed with multimers, a substitution in that T cell epitope that resulted in impaired but not complete loss of recognition (Figure 5), was associated with nearly a three fold drop in PD-1 expression (Figure 4b). Two additional findings further highlight the role of antigen in modulating PD-1 levels on HCV specific T cells. In Subject 175, HCV levels transiently fell to below the limit of quantitation at Day 206 post infection followed thereafter by reemergence of high viral titers. PD-1 levels drop precipitously on HCV-specific T cells at the time when virus becomes nearly undetectable and are re-induced after viral reemergence (Figure 4c). Interestingly, the kinetics and magnitude of PD-1 re-expression is quite different for the two distinct epitopes, suggesting that additional factors may contribute to regulation of PD-1 expression besides simply antigen dosage. In Subject 30, sequence escape at epitope C63B is observed at day 1144 following infection and is associated with a five-fold drop in PD-1 levels.(Figure 5b, Figure 4d) On Day 1521 following initial infection, the epitope reverted back to the original sequence. This reversion is associated with a six-fold increase in PD-1 expression on the cognate T cells. The A2-61 epitope recognized by Subject 30 does not undergo substitution and the level of PD-1 expression over the same time frame varies less than two fold. In contrast to levels on T cells specific for epitopes that underwent substitution, PD-1 levels remained stable (declined less than two fold) or increased over time for epitopes in four additional subjects with no viral escape (Figure 4e–h). Seven of the eight subjects studied had detectable T cell responses to at least one control (EBV, CMV, or influenza) antigen and PD-1 levels differed by less than two-fold over multiple time points on control epitope-specific T cells in the same time periods. (data not shown) In summary, epitope escape (or as in subject 175, decrease in viral titer to nearly undetectable levels) consistently resulted in decreases in PD-1 levels of 3-10 fold whereas decreases in PD-1 levels were never greater than 2 fold and usually increased over time when the cognate epitope sequence was maintained.
Figure 4. PD-1 upregulation requires maintenance of intact antigen and restoration of intact antigen following escape is associated with an increase in PD-1 levels.
Viral sequence was assessed at initial infection and at multiple subsequent time points at which T cells specific for known antigens were detectable, including those surrounding the time at which the PD-1 levels were found to be low relative to initial PD-1 levels. Levels of PD-1 on T cells specific for epitopes that either underwent substitutions that impaired T cell recognition or remained constant are shown. Time points at which the substituted epitopes were present are indicated with open circles and points at which the initial viral sequence was present are indicated with closed circles. A. Subject 17 recognized three epitopes: 880TL10 and B35-135 underwent substitution in the first six months of infection and 140G remained intact. PD-1 levels decline three and ten fold following substitution of 880TL10 and B35-135, respectively. T cells recognizing 140G demonstrated a three fold increase in PD-1 expression. B. Subject 13 had one HCV specific T cell response that underwent substitution with an associated drop of nearly three fold in PD-1 expression. C. In Subject 175, HCV levels transiently fell to below the limit of quantitation with a decline in PD-1 levels followed by reemergence of high viral titers and an increase in PD-1 levels on T cells specific for the epitope. D. Subject 30. Sequence escape at epitope C63B is associated with a five-fold decline in PD-1 levels. C63B reverted back to the original sequence in association with a six-fold increase in PD-1 expression on the cognate T cells. A2-61 does not undergo substitution and the level of PD-1 expression varies less than two fold. PD-1 levels are maintained constant or increase over time in the absence of viral sequence variation. (E–H) Levels of PD-1 on T cells specific for epitopes that remained constant are shown for four different HCV infected subjects who recognized at least one T cell epitope for which multimers were available. For all four subjects, PD-1 expression on HCV-specific T cells decreases less than two-fold or increases over time.
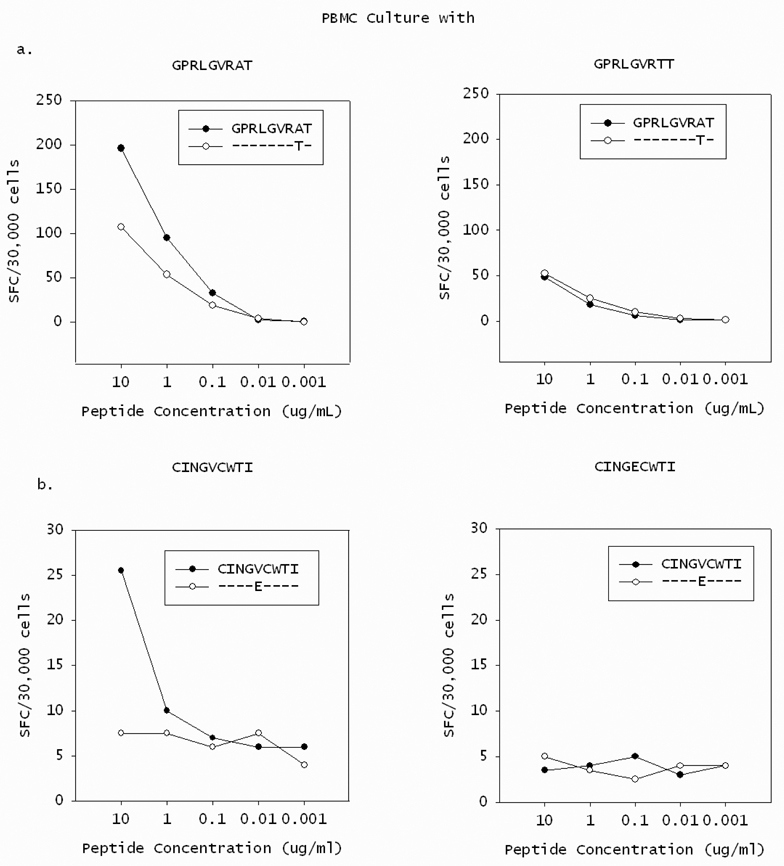
Figure 5. Amino acid substitutions in T cell epitopes reduce recognition by cognate T cells, validating them as escape mutations.
PBMC were cultivated in the presence of the peptide present at initial infection (closed circle) or at the point where PD-1 levels decreased (open circle) in Subject 17 (A) or Subject 30 (B). After 20 days of stimulation with the peptide present in initial infection (left panels) or the variant peptide (right panels), cells were tested for recognition of both peptides in serial dilutions in an IFN-γ Elispot assay. The number of spot forming colonies (SFC) per 30,000 cells is shown at each peptide dilution. In both cases, the variant peptide was less well recognized than the initial peptide by T cells expanded with the initial peptide (left panels). Stimulation with either of the variant peptides also resulted in reduced recognition of either the initial or variant peptides (right panels) and in reduced expansion of T cells as measured by T cell numbers (data not shown) compared to stimulation with the initial peptide. These substitutions were thus deemed escape mutations.
Discussion
We present the first detailed longitudinal analysis of PD-1 expression on HCV-specific T cells from the time of acute infection, correlating levels of surface expression with viral load, clearance versus persistence, and viral sequence evolution. Our data from early infection demonstrate that PD-1 upregulation precedes epitope mutation and correlates inversely with viral clearance independent of viral load. Analysis of patients with persistent infection indicates that maintenance of high PD-1 levels as a mechanism of immune evasion may be necessary only when the virus fails to escape via epitope mutation. Taken together, these results demonstrate that although intact antigen is not the sole factor determining levels of PD-1 expression, maintenance of PD-1 levels on an HCV-specific T cell requires persistence of intact antigen and that restoration of intact antigen (either by reemergence of virus or reversion back to the cognate epitope) following escape is associated with an increase in PD-1 levels. It is important to note that while changes in PD-1 levels on CD8 T cells specific for a given epitope are closely correlated with presence versus absence of mutation or major changes in viral load (i.e. Subject 175), the initial set points for PD-1 level on cognate T cells for each epitope varies widely even within the same patient. This could be due to any of a number of variables, including epitope density or mean affinity within the cognate TCR repertoire for each epitope.
There are conflicting data on whether the level of PD-1 on HCV specific T cells differs significantly between those who control HCV infection and those with persistent infection (31–32). All studies to date agree that PD-1 levels are high on HCV specific T cells versus levels on the general CD8 T cell population or on T cells specific for at least some control antigens in the acute phase of infection regardless of the outcome of infection. Our finding of higher PD-1 levels in the acute phase of infection on T cells from those who fail to control infection versus levels seen in those who clear matches the results of Urbani et al, but we did not see differences in the later stages of infection. A study by Kasprowicz et al found that PD-1 levels do not differ significantly between those who control infection and those who do not in the acute or chronic phase of infection. It is not clear why their results differ from ours, but sexual transmission is a more common route of infection in the cohort studied in Kasprowicz et al and the majority of subjects in both of the other studies presented with symptomatic infection. Our subjects almost all acquired the infection via injection drug use and all but one were asymptomatic, which is more common for acute HCV infection. There are known differences between patients who present with symptoms and those who do not, including the rate of clearance, but the reasons for those differences are not understood. It is therefore possible that some of the biologic differences that account for the development of symptoms also affect PD-1 expression. In addition, the duration of infection of those defined as acutely infected may be different between studies. The time from infection to manifestation of symptoms is highly variable, making it harder to pinpoint the duration of infection in studies that identify subjects on the basis of symptoms. Each of the three groups also used a different antibody against PD-1, which could also be responsible for the differences observed.
HCV and HIV RNA levels are known to be associated with outcome of infection, with persistence of HCV RNA with chronic HCV infection and higher HIV RNA levels associated with more rapid disease progression. Although previous studies have shown PD-1 levels to be positively correlated with viral RNA levels, those studies have not controlled for viral RNA levels in the assessment of the correlation between PD-1 levels and outcome of infection (28–31). We find that the level of HCV RNA is positively correlated with expression of PD-1 on HCV specific T cells, but also that the level of PD-1 expression is significantly higher in early infection on HCV-specific T cells from those who progress to chronic HCV infection compared to those who clear infection independent of HCV RNA levels. It is not possible from the current data to conclude that PD-1 levels during acute infection determine ultimate outcome, but the correlation between PD-1 levels on HCV-specific T cells during acute infection and outcome, independent of HCV RNA levels, certainly supports a contributory role. These data also suggest that another as yet undefined factor regulating PD-1 expression is associated with clearance of HCV infection. It is possible that initial innate responses to acute HCV infection differ between the two outcomes of infection and may affect PD-1 expression independently of viral RNA levels.
While there has been much focus on persistent antigen in the induction of chronically high PD-1 expression, it has recently been shown that PD-1 levels can also be dramatically affected by the presence versus absence of proinflammatory signals at the time of initial TCR engagement (34–35). A recent chimpanzee study also suggests that outcome of infection and loss of PD-1 expression may not be absolute and that PD-1 levels may not always be a marker of exhaustion (43). Consistent with data in HIV indicating that PD-1 may be differentially expressed on epitope-specific CD8 T cells within a single individual at the same time, we saw differential expression of PD-1 on contemporaneous HCV epitope-specific CD8 T cells from single individuals (28). HCV RNA levels and the cytokine milieu are constant in a patient at a given time point. Therefore, while distinct innate responses could create different cytokine milieus that affect PD-1 expression, HCV RNA levels and the cytokine milieu can not be the sole determinants of PD-1 expression based on our data and there must be epitope-specific differences.
Generation of escape mutations within T cell epitopes is a well described mechanism for viral escape from CD8 T cell responses. We sought to determine whether escape affected the level of PD-1 expression and therefore represented an epitope specific determinant of cell membrane levels of PD-1. Our data demonstrate that viral escape is associated with a reduction in PD-1 expression on the surface of HCV specific T cells and that reversion is associated with an increase in the level of PD-1, supporting the necessity of ongoing antigenic stimulation of T cells for upregulation of PD-1. However, maintenance of PD-1 on levels on the surface of HCV-specific T cells following control of HCV infection in those who cleared HCV above the levels seen on the general CD8 T cell population suggests that antigen need not be present to maintain some PD-1 on the surface of T cells after antigen exposure. Our data also suggest that maintenance of high levels of PD-1 as a mechanism of immune evasion is only necessary when the virus fails to escape via epitope mutation since PD-1 does not persist at high levels on the cell surface in the setting of escape.
Given that we observed lower levels of PD-1 on the cell surface of T cells specific for mutated epitopes than those specific for epitopes in the same individual who did not undergo substitution, variable PD-1 levels on T cells specific for different epitopes in the same subject at the same time point may in part be explained by the presence or absence of escape mutations in specific epitopes. However, the levels of PD-1 on the surface of T cells that did not undergo substitution also varied as much as five fold and so there must be alternative explanations for the variability of epitope specific PD-1 expression that are not due to escape, HCV RNA levels, and the cytokine milieu. Varying strength of TCR engagement, host TCR repertoires, antigen presentation, and other factors should be investigated as additional mechanisms of PD-1 expression modulation.
In summary, we demonstrated that high HCV RNA levels and maintenance of intact HCV epitopes are associated with high levels of PD-1 on the surface of HCV specific T cells. However, these factors did not fully explain the variability of PD-1 on the surface of HCV-specific T cells or the association of high PD-1 levels on HCV specific T cells in the acute phase with persistence of HCV infection. While additional research is needed to identify other factors affecting PD-1 expression and the outcome of HCV infection, these results suggest that PD-1 expression on T cells specific for non-escaped viral epitopes contributes to viral persistence and that PD-1 blockade may alter the outcome of HCV infection.
Acknowledgements
We gratefully acknowledge Minhua Han of Medarex Corporation and The Johns Hopkins University Immunology Core Laboratory for technical assistance. Shalyn Clute, Changyu Wang, and Alan Korman work for Medarex Corporation. The authors otherwise have no conflicting financial agreements.
Footnotes
This work was supported by grants from The Damon Runyon Foundation, The Dana Foundation, and US Public Health Service grants K08 DA11880, U19 AI040035 and R01 DA024565.
This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nat.Med. 2004;10:S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Hepatitis C: global prevalence. Weekly Epidemiological Record. 1997:341–348. [Google Scholar]
- 3.Alter MJ. Epidemiology of hepatitis C in the West. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. MMWR. 1998;47:1–39. (No. RR-19) [PubMed] [Google Scholar]
- 5.Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- 6.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J.Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong MJ, El-Farra NS, Reikes AR, Co RL. Clinical outcomes after transfusion-associated hepatitis C. N.Engl.J.Med. 1995;332:1463–1466. doi: 10.1056/NEJM199506013322202. [DOI] [PubMed] [Google Scholar]
- 8.Wong JB, McQuillan GM, McHutchison JG, Poynard T. Estimating future hepatitis C morbidity, mortality, and costs in the United States. Am.J.Public Health. 2000;90:1562–1569. doi: 10.2105/ajph.90.10.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N.Engl.J.Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 10.Weiner A, Erickson AL, Kansopon J, Crawford K, Muchmore E, Hughes AL, Houghton M, Walker CM. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc.Natl.Acad.Sci.USA. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox AL, Mosbruger T, Mao Q, Liu Z, Wang XH, Yang HC, Sidney J, Sette A, Pardoll D, Thomas DL, Ray SC. Cellular immune selection with hepatitis C virus persistence in humans. J Exp.Med. 2005;201:1741–1752. doi: 10.1084/jem.20050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, Pillay T, Ouchi K, Reyor LL, Zur Wiesch JS, Gandhi RT, Chung RT, Bhardwaj N, Klenerman P, Walker BD, Allen TM. CD8 epitope escape and reversion in acute HCV infection. J.Exp.Med. 2004;200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura Y, Gushima T, Rawale S, Kaumaya P, Walker CM. Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection. J Virol. 2005;79:4870–4876. doi: 10.1128/JVI.79.8.4870-4876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seifert U, Liermann H, Racanelli V, Halenius A, Wiese M, Wedemeyer H, Ruppert T, Rispeter K, Henklein P, Sijts A, Hengel H, Kloetzel PM, Rehermann B. Hepatitis C virus mutation affects proteasomal epitope processing. J.Clin.Invest. 2004;114:250–259. doi: 10.1172/JCI20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang KM, Rehermann B, McHutchison JG, Pasquinelli C, Southwood S, Sette A, Chisari FV. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J.Clin.Invest. 1997;100:2376–2385. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen TM, O'Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 17.Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJ, Brander C, Walker BD. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J.Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J.Exp.Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, Ruiz L, Ramduth D, Jeena P, Altfeld M, Thomas S, Tang Y, Verrill CL, Dixon C, Prado JG, Kiepiela P, Martinez-Picado J, Walker BD, Goulder PJ. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp.Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokomaku Y, Miura H, Tomiyama H, Kawana-Tachikawa A, Takiguchi M, Kojima A, Nagai Y, Iwamoto A, Matsuda Z, Ariyoshi K. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J Virol. 2004;78:1324–1332. doi: 10.1128/JVI.78.3.1324-1332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu.Rev.Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 22.Ray SC, Fanning L, Wang XH, Netski DM, Kenny-Walsh E, Thomas DL. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J Exp.Med. 2005;201:1753–1759. doi: 10.1084/jem.20050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koibuchi T, Allen TM, Lichterfeld M, Mui SK, O'Sullivan KM, Trocha A, Kalams SA, Johnson RP, Walker BD. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J.Virol. 2005;79:8171–8181. doi: 10.1128/JVI.79.13.8171-8181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat.Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 25.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp.Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tseng SY, Otsuji M, Gorski K, Huang X, Slansky JE, Pai SI, Shalabi A, Shin T, Pardoll DM, Tsuchiya H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp.Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, Greenfield EA, Bourque K, Boussiotis VA, Carter LL, Carreno BM, Malenkovich N, Nishimura H, Okazaki T, Honjo T, Sharpe AH, Freeman GJ. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat.Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 28.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, Depierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006 doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 29.Radziewicz H, Ibegbu CC, Fernandez ML, Workowski KA, Obideen K, Wehbi M, Hanson HL, Steinberg JP, Masopust D, Wherry EJ, Altman JD, Rouse BT, Freeman GJ, Ahmed R, Grakoui A. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J.Virol. 2007;81:2545–2553. doi: 10.1128/JVI.02021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penna A, Pilli M, Zerbini A, Orlandini A, Mezzadri S, Sacchelli L, Missale G, Ferrari C. Dysfunction and functional restoration of HCV-specific CD8 responses in chronic hepatitis C virus infection. Hepatology. 2007;45:588–601. doi: 10.1002/hep.21541. [DOI] [PubMed] [Google Scholar]
- 31.Urbani S, Amadei B, Tola D, Cavallo MC, Mezzadri S, Missale G, Ferrari C. PD-1 expression in acute hepatitis C virus infection. Hepatology. 2006;44:292A–293A. [Google Scholar]
- 32.Kasprowicz V, Schulze zur WJ, Kuntzen T, Nolan BE, Longworth S, Berical A, Blum J, McMahon C, Reyor LL, Elias N, Kwok WW, McGovern BG, Freeman G, Chung RT, Klenerman P, Lewis-Ximenez L, Walker BD, Allen TM, Kim AY, Lauer GM. High PD-1 expression on HCV-specific CD8+ and CD4+ T cells during acute Hepatitis C irrespective of clinical outcome. J Virol. 2007 doi: 10.1128/JVI.02474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrovas C, Price DA, Mattapallil J, Ambrozak DR, Geldmacher C, Cecchinato V, Vaccari M, Tryniszewska E, Gostick E, Roederer M, Douek DC, Morgan SH, Davis SJ, Franchini G, Koup RA. SIV-specific CD8+T-cells express high levels of PD1 and cytokines but have impaired proliferative capacity in acute and chronic SIVmac251 infection. Blood. 2007 doi: 10.1182/blood-2007-01-069112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg MV, Maris CH, Hipkiss EL, Flies AS, Zhen L, Tuder RM, Grosso JF, Harris TJ, Getnet D, Whartenby KA, Brockstedt DG, Dubensky TW, Jr., Chen L, Pardoll DM, Drake CG. Role of PD-1 and its ligand, B7-H1, in early fate decisions of CD8 T cells. Blood. 2007 doi: 10.1182/blood-2006-12-062422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox AL, Netski DM, Mosbruger T, Sherman SG, Strathdee S, Ompad D, Vlahov D, Chien D, Shyamala V, Ray SC, Thomas DL. Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin.Infect.Dis. 2005;40:951–958. doi: 10.1086/428578. [DOI] [PubMed] [Google Scholar]
- 37.Wong RM, Scotland RR, Lau RL, Wang C, Korman AJ, Kast WM, Weber JS. Programmed death-1 blockade enhances expansion and functional capacity of human melanoma antigen-specific CTLs. Int.Immunol. 2007;19:1223–1234. doi: 10.1093/intimm/dxm091. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Netski DM, Mao Q, Laeyendecker O, Ticehurst JR, Wang XH, Thomas DL, Ray SC. duplicate use 7239. J.Clin.Microbiol. 2004;42:4223–4229. doi: 10.1128/JCM.42.9.4223-4229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall TA. BioEdit: Biological sequence alignment editor for Windows 95/98/NT version 7.0.4. [accessed 20 Feb 2005];software. 2001 Distributed by author http://www.mbio.ncsu.edu/RNaseP/info/programs/BIOEDIT/bioedit.html. [Google Scholar]
- 40.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 41.Rolland M, Jensen MA, Nickle DC, Yan J, Learn GH, Heath L, Weiner D, Mullins JI. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J.Virol. 2007;81:8507–8514. doi: 10.1128/JVI.02683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–1060. [PubMed] [Google Scholar]
- 43.Bowen DG, Shoukry NH, Grakoui A, Fuller MJ, Cawthon AG, Dong C, Hassleschwert DL, Brasky KM, Freeman GJ, Seth NP, Wucherpfennig KW, Houghton M, Walker CM. Variable Patterns of Programmed Death-1 expression on fully functional memory T cells after spontaneous resolution of Hepatitis C virus infection. J.Virol. 2008;82:5109–5114. doi: 10.1128/JVI.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]