Abstract
During early heart development, Tbx2 gene expression is initiated in the cardiac crescent and then becomes restricted to the outflow tract and the atrioventricular region. We identified a Tbx2 regulatory region, enriched in multiple Smad sites, sufficient to reproduce Tbx2 expression patterns overlapping Bmp2 and Bmp4 gene activity in the heart. The role of Tbx2 in cardiogenesis was analyzed by using Cre-LoxP activated Tbx2 transgenic misexpression in chamber myocardium. Ventricular Tbx2 misexpression exhibited an abnormally narrow chamber lumen owing to the expansion of Hyaluronan synthase 2 expression in the ECM or cardiac jelly and the appearance of the endocardial cushions (ECs). Excessive Tbx2 also induced Tgfβ2, which coincided with the outgrowth epithelial-mesenchymal transformed cells in ventricular and atrial tissues modifying cardiomyocyte identity from chamber type to non-chamber type. Tbx2, a central intermediary of Bmp-Smad signaling, has a central part in directing Has2 and Tgfβ2 expression, facilitating EC formation.
Keywords: cardiac jelly, cardiomyocyte identity, epithelial-mesenchymal transformation, extracellular matrix, misexpression
The heart develops, as a modular organ, driven by distinct transcriptional regulatory programs that control each anatomical region (1). A member of the T-box factor family, Tbx2, which first appears in the cardiac crescent and then later restricted to non-chamber myocardium (My) [outflow tract (OFT), atrioventricular canal (AVC), inner curvature, and inflow tract) (2, 3)], is a valuable model of modular cardiac gene activity. Tbx2 is central for endocardial cushion (EC) formation and chamber specification, and may be a transcriptional repressor (4, 5). Expression of chamber-specific myocardial genes, which include Nppa (encoding atrial natriuretic factor, ANF), Gja5 (encoding connexin 40, Cx40), and Gja1 (encoding connexin 43, Cx43), were repressed by Tbx2 (3–5). Tbx2 null mutant embryos exhibited small AVC and defective OFT septation (3), whereas Tbx2 transgenic expression blocked chamber formation (4) and cell proliferation in the OFT and AVC (6).
The ECs form from localized expansion of the ECM also named cardiac jelly (7, 8) found in the cardiac OFT and AVC segments the simple heart tube into a complicated structure composed of the aortic sac, common ventricular chamber, and atrial chamber. Some endocardial cells invade into the ECM through epithelial-mesenchymal transformation (EMT) to remodel the cushion tissue into the mature valves. Several signaling pathways have been implicated in EC formation. The Bmp pathway is essential for both processes; expansion of ECM and EMT in the EC formation (9–14). Tgfβ2 performs crucial and sequential roles in EC formation and may also be regulated by Bmp2/4 during cardiogenesis (9–11). The hyaluronan (HA) synthase 2 (Has2) has been recently shown to have essential role in expansion of ECM and EMT (16). Also, Tbx2 may be a direct target of Bmp2/4 signaling pathway during EC formation (2).
Here, we delineated an 80-bp regulatory region within the Tbx2 5′ flanking sequences, which contain multiple Smad DNA binding sites that recapitulate expression of Tbx2 in the AVC and OFT. Previously, myocardial-specific inactivation of Bmp2 also inhibited the appearance of several factors, including Tbx2, Tgfβ2, and Has2, and blocked cushion formation (9). To define the regulatory hierarchy shared by Bmp2/4-dependent genes, we analyzed embryos in which murine Tbx2 was misexpressed in the developing chamber My, using a mouse genetic system based on Cre/loxP recombination. Tbx2 altered cardiogenic lineage specification by expanding the ECM and EMT to drive EC formation via the induction of Tgfβ2 and Has2 gene activity in embryonic hearts.
Results
Smad Signaling Drives Tbx2 Transgene Activity via a Distal Enhancer.
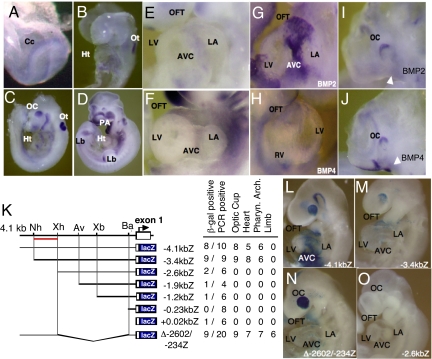
Tbx2 expression was first detected in the cardiac crescent and notochordal plate (Fig. 1A). At E8.5, Tbx2 expression was maintained only in the posterior portion of the looping heart (Fig. 1B). As the heart matured, the expression of Tbx2 became further limited to the AVC and OFT region (Fig. 1 E and F). Tbx2 mRNA was also detected in the optic cups, otic vesicles, pharyngeal arches, and limb buds of embryonic day (E)9.5 and E10.5 mouse embryos (Fig. 1 C and D). Transient F0 transgenic mice harboring 4.1 kilobase (kb) of Tbx2 5′ flanking sequences linked to the lacZ reporter gene (Fig. 1K) revealed β-galactosidase activity in the OFT, AVC, and a portion of the left atrium, whereas LacZ activity was absent from the right atrium and the ventricles (Fig. 1L). Also, Tbx2, a downstream target of murine Bmp2/4 signaling (2, 9), was colocalized to the AVC and OFT (Fig. 1 G–J). After serial and gap deletion mutagenesis strategy, recapitulated Tbx2 expression was delineated to a region between −3.4 and −2.6 kb in transgenic mouse embryos (Fig. 1 L–O).
Fig. 1.
Mouse Tbx2 gene activity in early cardiac development was recapitulated by distal cis-acting regulatory regions coincided with Bmp2 and Bmp4 expression. Tbx2 expression was first detected in the cardiac crescent at E7.5 (A), and restricted to the posterior part of looping heart at E8.5 (B). The expression of Tbx2 was also observed in the developing eye, otic cup, pharyngeal arches, and limb buds (B–D) At E9.5 and E10.5, Tbx2 transcripts were localized in the OFT and AVC (E and F). . Expression of Bmp2 and Bmp4 (G and H) was observed in the OFT and AVC (I and J). Schematic representation of serial deleted 5′ flanking sequences of the Tbx2 gene, which were analyzed for transgene expression patterns in E9.5–E10.5 F0 founder embryos (K). An upstream regulatory region that directs Tbx2 gene expression was indicated by a red bar. LacZ was expressed in the optic cup, heart, and pharyngeal arches using 4.1 kb (L), 3.4 kb (M), and Δ−2602/−234 (N) 5′ flanking fragments. No LacZ staining was observed in −2.6-kbZ transgenic embryos (O). Cc, cardiac crescent; Ht, heart; LA, left atrium; Lb, limb bud; LV, left ventricle; OC; optic cup; Ot, otic cup; PA, pharyngeal arch. Restriction sites shown above are: Nh, NheI; Xh, XhoI; Av, AvrII; Xb, XbaI; and Ba, BamHI.
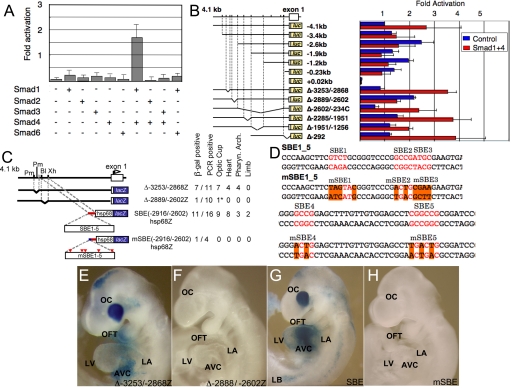
Paired Smad 1/4 proteins, primary intracellular mediators of Bmp signals (12), activated transcription of the −4.1-kb reporter construct (Fig. 2A). Other Smad factor combinations did not strongly activate the Tbx2 reporter, whereas the inhibitory Smad 6 (13, 14) blocked Tbx2 gene activity. Tbx2 regulatory region responsive to Bmp signaling was localized to a 290-bp region expression in the developing heart (Fig. 2 B and C), which contains at least five conserved Smad sites, two of which, SBE1 and SBE5 sequences, were potent Smad1/4 cofactor binding sites (Fig. S1). Schematic representation of Tbx2 transgenes analyzed in E9.5 F0 embryos and a summary of the tissue restricted expression activity is shown in Fig. 2C, whereas five SMAD sites in SBE1-5 were mutated by site directed mutagenesis as shown in mSBE1-5 (Fig. 2D). The gap deletion mutant Δ−2899/−2602 hsp68lacZ construction, in which the five Smad sites were removed from the Tbx2 5′ flanking sequence, showed a complete loss of lacZ expression activity in the hearts of F0 transgenic embryos (Fig. 2 C and F). The Smad site enriched region (SBE; −2916/−2602) linked to a minimal hsp68 promoter lacZ transgene, revealed robust expression in the OFT and AVC, sufficient to recapitulate the restricted Tbx2 expression pattern in the heart (Fig. 2 C and G), whereas site directed Smad site mutations eliminated gene activity in transgenic mice (Fig. 2 C and H).
Fig. 2.
Delineation of the distal Smad factor dependent Tbx2 enhancer. (A) Multiple Smad expression vectors tranfected into CV1 cells revealed activation caused by Smad1/4 of the 4.1 kb of 5′ flanking sequence of Tbx2 fused in-frame to the luciferase. The bars represent the average of three independent transfections, and the error bars represent the SE of corrected luciferase activity relative to the pCMV5 control vector. (B) Activation by Smad1/4 required the −2889/−2602 region. (C) Schematic representation of Tbx2 transgenes analyzed in E9.5 F0 embryos and a summary of the tissue restricted expression activity. (D) Five SMAD sites in SBE1-5 were mutated by site directed mutagenesis in mSBE1-5. (E–J) LacZ expression patterns in E9.5 F0 embryos. (E) Δ−3253/−28682hsp68Z transgene; a gap deletion mutant, of a region immediately upstream of the multiSmad sites. (F) Δ−2899/−2602hsp68lacZ; a gap deletion mutant in which the multiple Smad sites were removed from the 5′ flanking sequence. (G) SBE(−2916/−2602)hsp68lacZ; the region from −2916 to −2602bp, containing five Smad sites, was linked to the hsp68lacZ reporter gene. (H) mSBE(−2916/−2602)hsp68lacZ; mutations were inserted at the multiple Smad sites in the −2916/−2602 fragment. LacZ expression were recapitulated by the DNA fragments containing Smad binding sites (E and G), whereas gap deletion and point mutagenesis of Smad binding sites eliminated gene activity in transgenic mice (F and H). Restriction sites are: BlpI; Xh, XhoI.
Abnormal Deposition of ECM in mTbx2-Misexpressing Embryonic Hearts.
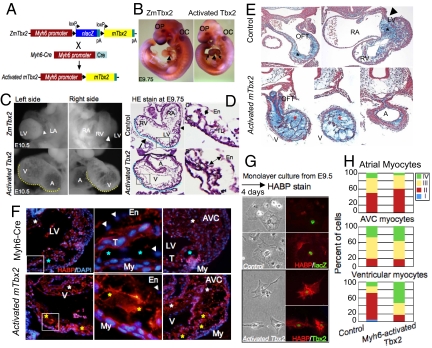
To study the role of Tbx2 in the cardiac morphogenesis, we generated transgenic mice that conditionally misexpressed murine Tbx2 gene in embryonic chamber-cardiomyocytes (Fig. 3 A and B; Fig. S2). Activated Tbx2 embryos exhibited enlarged hearts, with marked myocardial hypoplasia associated with rich deposition of ECM in the compact and trabecular My (Fig. 3 C and D). Expansion of the EC (cardiac jelly) between the endocardium (En) and My (Fig. 3E) stained with alcian blue for acidic glycosaminoglycans (15) caused a narrow ventricular lumen. In control littermates, acidic glycosaminoglycans were deposited mainly in the ECM of OFT and AVC regions at E10.5 compared with the expanded ECM induced by activated Tbx2.
Fig. 3.
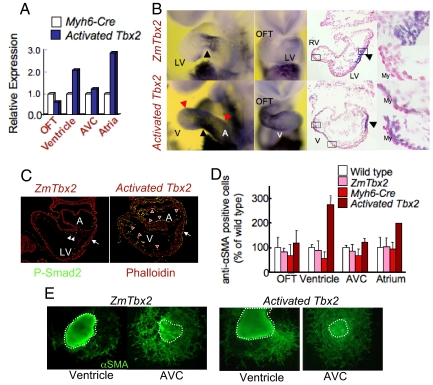
Abnormal cardiac morphogenesis induced by activated mTbx2 coincided with excessive HA deposition. (A) Schematic representation of Myh6-Cre induced activation of mTbx2 by breeding ZmTbx2 mice to the Cre deleter mouse line, Myh6-Cre. (B) Cardiomyocyte-specific activated mTbx2 was detected by immunoperoxidase staining in the atria and ventricles at E9.75. (C) Enlarged images highlight embryonic hearts at E10.5. Relative to control littermates, activated Tbx2 embryos exhibited an enlarged heart, which appeared as swollen single ventricular and atrial chambers, and a dilated AVC (yellow dotted line in C) at E10.5. (D) Histological sections of activated mTbx2 in embryonic hearts at E9.75 stained with HE staining showed rounded ventricular chamber (blue dotted line) and an abnormally narrow ventricular lumen due to expansion of EC like structure between the En and My (two-headed arrow in D), and arrow indicate intraventricular sulcus (IVS) and arrowheads indicate AVC. (E) Histological sections of embryonic hearts stained at E10.5 with alcian blue revealed acidic glycosaminoglycans (blue stain) not only deposited in the OFT and AVC regions of control littermates (black asterisks), but throughout the chambers of activated Tbx2 hearts (red asterisks). (F) Visualization of HA on sections of E9.75 embryos with HABP (red). HA was deposited between En and My in the inner curvature and AVC of activated mTbx2 embryos and control littermates (white asterisks) and not in the outer curvature of control littermates. Extra deposition of HA was observed between En and My in the outer curvature of mTbx2-misexpressing ventricles (yellow asterisks). DAPI stain was used to visualize nuclei (blue). (G) Visualization of HA on cultured ventricular myocytes from embryos of E9.5 or E9.75 with HABP (red). Immunocytochemical detection of lacZ and Tbx2 (green). (H) Percentage of classified cardiomyocytes with HA secretion. Type I cardiomyocytes, no secretion of HA; Type II, little amount of HA around the cells; Type III, much HA is secreted around the cells; Type IV, much HA is secreted around the cells and deposited in the cells (Fig. S4). Majority of ventricular cardiomyocytes secreted excessive HA in the activated mTbx2 embryos. T, trabeculae; CL, compact layer; and TL, trabecular layer.
Glycosaminoglycan HA, a major constituent of the cardiac jelly, may be required for the expansion of EC (16,17). Excessive acidic glycosaminoglycans deposited through the cardiac tube in mTbx2-misexpressing embryos were also revealed by deposition of HA with the biotinylated-HA binding protein (BP; Fig. 3F). The specific binding of HABP to HA was eliminated by hyaluronidase treatment (Fig. S3). HA deposition was observed between the En and My in the inner curvature and the AVC of both control littermates and mTbx2-misexpressed mice, and in between the En and My of the outer curvature of mTbx2-misexpressed ventricles (Fig. 3F). Also, cultured cardiomyocytes from quartered hearts identified by lacZ and or by Myh6 induced mTbx2 expression (Fig. 3G; Fig. S4) were classified by the amount of HA secretion (Fig. 3H). Tbx2-misexpressing ventricular myocytes secreted greater amount of HA compared with ventricular myocytes from control littermates.
Has2 and Tgfβ2 are Tbx2 Downstream Gene Targets.
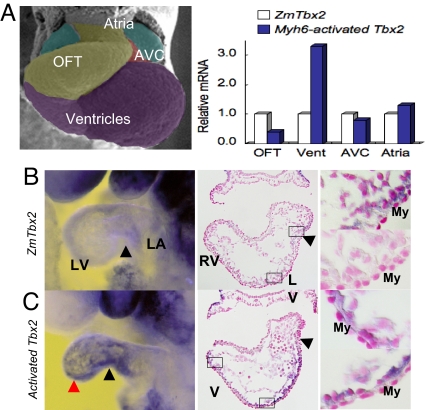
Is there a hierarchical relationship between Tbx2, Has2, and Tgfβ2? HA synthatase (Has)2, the major enzyme responsible for HA synthesis in the AVC and atria, was strongly up-regulated in the ventricular My by mTbx2 misexpression (Fig. 4). In comparison, other components of the ECM, including Col1a1 (encoding type I collagen), Cspg2 (encoding chondroitin sulfate proteoglycan 2, versican), Fn1 (encoding fibronectin 1), and Tnc (encoding tenascin C) did not show obvious differences (data not shown). Has2 is a direct Tbx2 target, because T-box binding sites were conserved between Has2 promoter regions, which recruited endogenous Tbx2 from cardiac nuclear chromatin extracts and was transactivated by Tbx2 (SI Materials and Methods and Fig. S5).
Fig. 4.
HA synthetase, Has2, activated by Tbx2 in cardiac chamber myocytes. (A) Real-time PCR analysis of dissected embryonic hearts at E9.5 or E9.75. Has2 transcripts were tripled in the ventricles of activated mTbx2 embryos. (B) Whole-mount in situ hybridization analysis for Has2 expression was detected in the AVC (black arrowheads) of control littermates whereas expanded to ventricles (red arrowheads) of activated Tbx2 embryos. The middle and right columns show sections after WISH. Has2 expression was activated in ventricular myocardial cells of activated Tbx2 embryos (right column). Nuclear were stained by nuclear fast red.
Tgfβ2 is normally expressed in the myocardial cells in the OFT and AVC, but was induced by Myh6-Cre activated mTbx2 in ventricular and atrial My (Fig. 5 A and B). Also, phosphorylated Smad2 appeared in the endocardial and myocardial cells of mTbx2-activated ventricles and atria (Fig. 5C), which coincided with robust in vitro EMT assays (Fig. 5 D and E); thus, suggesting that the Tgfβ2 signaling pathway was activated by mTbx2. Tgfβ2 may also be a direct target of Tbx2, because the Tgfβ2 promoter has conserved T-box binding sites, bound endogenous Tbx2, and was activated with a Tbx2-expression vector (SI Materials and Methods and Fig. S6).
Fig. 5.
Activated mTbx2 induced Tgfβ2 and enhanced EMT. (A) Real-time PCR analysis of quartered embryonic hearts at E9.5. Tgfβ2 was increased in the ventricles and atria of activated mTbx2 heart. (B) Whole-mount in situ hybridization analysis detected Tgfβ2 expression in the AVC (black arrowheads in controls) and OFT of control littermates which expanded to ventricles and atria of activated Tbx2 hearts (red arrowheads in misexpressed Tbx2). The middle and right columns show sections after WISH. Tgfβ2 expression was activated in ventricular myocardial cells of activated Tbx2 embryos (right column). Nuclear were stained by nuclear fast red. (C) Immunohistochemical detection of phospho-Smad2, an effector of Tgfβ signaling in the AVC (white arrows) En of control littermates (white arrowheads), which was expanded to the En of the ventricles and atria in activated mTbx2 hearts (red arrowheads). (D and E) Immunohistochemical detection of α-smooth muscle actin (SMA) showed increased EMT in in vitro collagen gel assays of ventricular and atrial explants. The percentage of anti-αSMA positive cells formed in explants from mTbx2-misexpressing ventricles and atria was approximately doubled compared with controls.
Discussion
Smad Factor Signaling Directed Tbx2 Expression Through a Distal Enhancer During Early Murine Cardiogenesis.
Tbx2 transcripts first appear in the cardiac crescent and then later became restricted to the OFT and AVC during early avian (2) and mouse embryogenesis (Fig. 1). Tbx2 is also expressed in the embryonic limb, optic cup, otic cup, and pharyngeal arches. Approximately 4 kb of 5′ flanking sequence of the murine Tbx2 gene was sufficient to reproduce much of these mouse embryonic expression patterns, as a lacZ reporter transgene. LacZ expression patterns closely overlapped with those of Bmp2 and Bmp4 in the heart and eye, and was delineated within the Tbx2 5′ flanking sequences, which contained Bmp directed multiple high-affinity binding sites for Smad transcription factors. Removal of these multiple Smad sites by gap deletion mutagenesis from the 4-kb flanking sequences blocked reporter gene activity in early embryos. In contrast, this short enhancer region linked to a minimal hsp68 promoter was sufficient for steering the restricted expression of lacZ in a pattern, highly similar to the endogenous Tbx2 gene activity. Previously, application of Bmp2 selectively induced cTbx2 expression in noncardiogenic embryonic tissue, and the Bmp antagonist Noggin down-regulated cTbx2 activity (2). Also, the appearance of murine Tbx2 was blocked in Bmp2 null mouse embryos (2). Thus, Tbx2 expression depended on Bmp signaling through Smad factors, a regulatory paradigm that also guides other modular-restricted genes in the developing heart.
Tbx2 Directs EC Formation.
Tbx2 is generally considered to be a transcriptional repressor. For example, Tbx2-null embryos have expanded chamber-specific gene expression into the AVC My (3). Also, Tbx2-overexpressing transgenic mice and Tbx20-null mice, in which Tbx2 is up-regulated, exhibited decreased expression of several chamber-specific genes and caused hypoplasia of ventricular chamber (4–6, 18, 19). However, Tbx2 also contains an activation domain (20), and is important for EC formation, because Tbx2 null mutant embryos exhibited small AVC and defective OFT septation (3). As shown here, Tbx2 enhanced ECM synthesis from myocardial cells and EMT from endocardial cells. Tbx2 directly bound Has2 and Tgfβ2 promoters and increased their transcriptional activities. Tgfβ signaling is crucial for EMT in the AV cushions (20–22). Accelerated appearance of EMT from the ventricular and atrial tissues, in vitro EMT assays and increased Smad2 phosphorylation in mTbx2-activated embryos supports activation of Tgfβ2 pathway by Tbx2.
Tbx2 also induced Has2 myocardial expression and increased HA deposition. HA is known as an essential factor for EC formation (8, 16, 23), and interacts with other molecules such as versican and fibrillin, which expands cardiac jelly providing extracellular space for cell migration (8). In addition to organizing extracellular environment, HA stimulates EMT of several types of epithelial cells (24) and endocardial cells dependent on Ras-activation (15) via ErbB2 receptor (23). In Tbx2-misexpressing embryos, extradeposition of HA was observed in the dilated chamber My; thus, Tbx2 has an important role in EC formation by increasing synthesis of HA. During normal heart development, chamber-cardiomyocytes undergo a critical maturation step that is manifested by a transition from production to degradation of ECM between E8.0 and E9.5 (25). Whereas HA is required for cushion formation, excess HA deposition may cause hemodynamic alteration and prevent cardiomyocyte differentiation necessary for chamber maturation.
Hierarchical Relationship Between Bmp2/4, Tbx2, Has2, and TGFβ2.
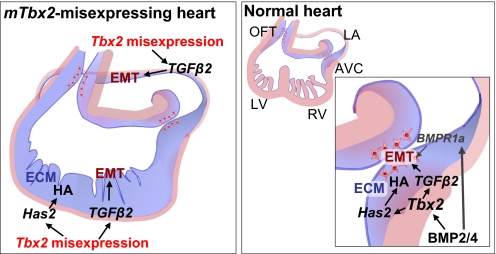
We propose a model in which Bmp-Smad responsive Tbx2 is stimulated to perform a central role in promoting EC formation by inducing expansion of ECM and EMT by directing Has2 and Tgfβ2 gene activity (Fig. 6). Several signaling pathways have been implicated in EC formation. The Bmp pathway is essential for expansion of ECM and EMT in EC formation. Myocardial-specific inactivation of Bmp2, Bmp4, and a Bmp type I receptor gene, Alk3, respectively, failed to form the EC (9, 10, 26). Sugi et al. (11) demonstrated that Bmp2 could substitute for the My to induce EMT. Also, noggin treatment of explants efficiently inhibited EMT. In both chicken and mouse EMT assays, Tgfβ2 is able to replace the overlying My to activate EMT in Tgfβ2 null mice (11, 16, 21, 22). Analysis of Tgfβ2-deficient mice also indicated that Tgfβ2 is important for valvulogenesis (27). Recent studies have shown that several factors, including Tbx2, Tgfβ2, and Has2, are downstream targets of Bmp2/4 pathway (9, 10). In addition to myocardial-derived Bmp function, Bmp signals directly to the cushion En through the Bmpr1a to induce EMT (9). In our experiments, Bmp2/4 were not up-regulated in Tbx2-misexpressing embryos (Fig. S7). Recently, Singh et al. (28) showed that Tbx20 directly interfered with Bmp/Smad signaling to suppress Tbx2 expression in the chambers; thereby, confining Tbx2 expression to the prospective AVC region. They also confirmed our observation that Tbx2 distal enhancer directs Tbx2 expression to the AVC and OFT. Here, we showed Bmp-Smad signaling dependent Tbx2 expression directed Has2 and Tgfβ2 gene activity to coordinately regulate EC formation.
Fig. 6.
Model of Tbx2 function for the EC formation in the AVC. The mTbx2 misexpression induces expression of Has2; thereby, driving the synthesis and deposition of HA and/or cardiac jelly through the heart. Misexpressed Tbx2 signals also promoted expression of Tgfβ2 gene that supports the induction of EMT in the ventricles and atria. In the normal heart development, Tbx2 works as one of the important factor to induce expansion of ECM and EMT under the Bmp2/4-Smad signaling pathway. Bmp signals also directly to the En through the Bmpr1a.
Experimental Procedures
Generation of mTbx2 Reporter Gene Constructions.
A genomic fragment that contained the Tbx2 locus was isolated from a 129SVJ mouse genomic library. A 4.5-kb NotI fragment flanking the 5′ transcription start site and overlapping the first coding exon was cloned into the NotI site of pBluescript-KS for sequencing. The mTbx2 reporter construct were generated from 4.1 kb of mTbx2 flanking sequence 5′ was linked in-frame in front of the lacZ and luciferase cDNA from pPD46.21 and pGL3-Basic (Promega). Deletion constructs were generated by restriction endonuclease digestion. The region from −2916 to −2602bp, containing multiple Smad sites, was linked to the hsp68lacZ reporter gene (29). Mutations at multiple Smad sites in −2916/−2602 fragment were inserted using In-Fusion PCR cloning kit (Clontech) (Fig. 2D; Fig. S1C).
Whole-Mount in Situ Hybridization.
Staged mouse embryos were obtained after timed mating of mice with the morning of the copulation plug being E0.5. Embryos were fixed in MEMFA (0.1 M Mops/2 mM EGTA/1 mM MgSO4/3.7% formaldehyde) and stored in 90% methanol at −20 °C until use for hybridization. Whole-mount in situ hybridization was performed as described by Yamada et al. (2), except that polyvinyl alcohol was included to increase signal intensity. A full-length of mTbx2 cDNA (kindly provided by Roni Bollag) was cloned into the EcoRI site of pBluescript-KS to synthesize digoxigenin-labeled RNA probes. After restriction endonuclease digestion with SacI, antisense probes were transcribed with RNA labeling kit (Stratagene).
Transient Transfection Assays.
Monkey CV-1 fibroblasts were grown in DMEM with 10% FBS. Cells were plated at 1 × 105 cells per well in a 24-well plate and transfected 24 h later with DNA mixture containing a total of 2 μg of total DNA, which included 500 ng of luciferase reporter vector, 500 ng of β-galactosidase vector, and a total of 1 μg of pCMV5-derived vectors. Transfections were performed using Lipofectamine (Invitrogen) as described (30). Luciferase activity were measured using a luminometer to detect activates substrates, then normalized by β-galactosidase activity (29).
Histology.
Embryos were dissected in Dulbeco's PBS and fixed overnight at 4 °C in 4% PFA in PBS for histological analysis. After fixation, embryos were rinsed in PBS, then dehydrated through graded ethanol or methanol and embedded in paraffin wax. Sections were cut and stained with hematoxylin-eosin or alcian blue according to standard methods. The HA was detected with 2 μg/mL biotinylated HABP isolated from bovine nasal cartilage (Seikagaku). The HABP was detected using streptavidin-conjugated AlexaFluor (Invitrogen). For immunohistochemistry, sections were incubated with rabbit anti-TBX2 IgG (Upstate Biotechnology) or rabbit anti-β-galactosidase (Biogenesis). Primary antibody was detected with goat anti-rabbit IgG labeled with Alexa Fluor (Invitrogen).
Real-Time PCR.
Embryonic hearts at E9.5–E9.75 were divided to 4-parts, OFT, ventricles, AVC, atria at the posterior boundary of EC in OFT, anterior and posterior boundaries of EC. Dissected tissues were immediately frozen in liquid nitrogen and stored at −80 °C until embryo and yolk sac DNA was genotyped. Total RNA isolation and first strand cDNA synthesis were performed with TRIzol reagent, SuperScript III (Invitrogen) and random primer, as per the manufacturer's instructions.
Primary Culture of Embryonic Cardiac Cells and Classification of Cardiomyocytes with HA Synthesis.
Quartered embryonic cardiac tissues were separated as described above. After trypsinization, isolated cardiac cells were cultured on gelatinized dishes by using culture medium containing DMEM (D5796; Sigma–Aldrich), 10% heat-inactivated FCS (HyClone), 0.1 g/mL penicillin, and 0.1 mg/mL streptomycin. After 4 days culture, cells were fixed in 4% PFA in PBS and detected HA, Tbx2 and β-galactosidase as described above. Classification of cardiomyocytes with HA secretion was followed as described in Fig. S4.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900635106/DCSupplemental.
References
- 1.Schwartz RJ, Olson EN. Building the heart piece by piece: Modularity of cis-elements regulating to Nkx2–5 transcription. Development. 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- 2.Yamada M, et al. Expression Of Chick Tbx-2, Tbx-3 and Tbx-5 genes during early heart development: Evidence for BMP2 induction of Tbx2. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- 3.Harrelson Z, et al. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 4.Christoffels VM, et al. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Devl Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 5.Habets PE, et al. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: Implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai CL, et al. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierpont ME, Markwald RR, Lin AE. Genetic aspects of atrioventricular septal defects. Am J Med Genet. 2001;297:289–296. [PubMed] [Google Scholar]
- 8.Schroeder JA, Jackson LF, Lee DC, Camenisch TD. Form and function of developing heart valves: Coordination by extracellular matrix and growth factor signaling. J Mol Med. 2003;81:392–403. doi: 10.1007/s00109-003-0456-5. [DOI] [PubMed] [Google Scholar]
- 9.Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 10.Gaussin V, et al. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc Natl Acad Sci USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol. 2004;269:505–518. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 12.Itoh S, Itoh F, Goumans MJ, Ten Dijke P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- 13.Galvin KM, et al. A role for smad6 in development and homeostasis of the cardiovascular system. Nat Genet. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 14.Yamada M, et al. Evidence for a role of Smad6 in chick cardiac development. Dev Biol. 1999;215:48–61. doi: 10.1006/dbio.1999.9419. [DOI] [PubMed] [Google Scholar]
- 15.Whiteman P. The quantitative measurement of Alcian Blue glycosaminoglycan complexes. Biochem J. 1973;131:343–350. doi: 10.1042/bj1310343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camenisch TD, et al. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin HS, Lloyd TR, Solursh M. Hyaluronate degradation affects ventricular function of the early postlooped embryonic rat heart in situ. Circ Res. 1994;74:244–252. doi: 10.1161/01.res.74.2.244. [DOI] [PubMed] [Google Scholar]
- 18.Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi JK, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 20.Paxton C, Zhao H, Chin Y, Langner K, Reecy J. Murine Tbx2 contains domains that activate and repress gene transcription. Gene. 2002;283:117–124. doi: 10.1016/s0378-1119(01)00878-2. [DOI] [PubMed] [Google Scholar]
- 21.Brown CB, Boyer AS, Runyan RB, Barnett JV. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: Roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP) Anat Rec. 2000;258:119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8:850–855. doi: 10.1038/nm742. [DOI] [PubMed] [Google Scholar]
- 24.Zoltan-Jones A, Huang L, Ghatak S, Toole BP. Elevated hyaluronan production induces mesenchymal and transformed properties in epithelial cells. J Biol Chem. 2003;278:45801–45810. doi: 10.1074/jbc.M308168200. [DOI] [PubMed] [Google Scholar]
- 25.Bernanke DH, Orkin RW. Hyaluronidase activity in embryonic chick heart muscle and cushion tissue and cells. Dev Biology. 1984;106:351–359. doi: 10.1016/0012-1606(84)90233-1. [DOI] [PubMed] [Google Scholar]
- 26.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartram U, et al. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta (2)-knockout mice. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- 28.Singh R, et al. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- 29.Chi X, et al. Complex cardiac Nkx2–5 gene expression activated by noggin-sensitive enhancers followed by chamber-specific modules. Proc Natl Acad Sci USA. 2005;102:13490–13495. doi: 10.1073/pnas.0504295102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown CO, III, et al. The cardiac determination factor, Nkx2–5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.