Abstract
Homing endonuclease genes are “selfish” mobile genetic elements whose endonuclease promotes the spread of its own gene by creating a break at a specific target site and using the host machinery to repair the break by copying and inserting the gene at this site. Horizontal transfer across the boundary of a species or population within which mating takes place has been thought to be necessary for their evolutionary persistence. This is based on the assumption that they will become fixed in a host population, where opportunities of homing will disappear, and become susceptible to degeneration. To test this hypothesis, we modeled behavior of a homing endonuclease gene that moves during meiosis through double-strand break repair. We mathematically explored conditions for persistence of the homing endonuclease gene and elucidated their parameter dependence as phase diagrams. We found that, if the cost of the pseudogene is lower than that of the homing endonuclease gene, the 2 forms can persist in a population through autonomous periodic oscillation. If the cost of the pseudogene is higher, 2 types of dynamics appear that enable evolutionary persistence: bistability dependent on initial frequency or fixation irrespective of initial frequency. The prediction of long persistence in the absence of horizontal transfer was confirmed by stochastic simulations in finite populations. The average time to extinction of the endonuclease gene was found to be thousands of meiotic generations or more based on realistic parameter values. These results provide a solid theoretical basis for an understanding of these and other extremely selfish elements.
Keywords: horizontal gene transfer, selfish gene, VDE, meiosis, haploid-diploid cycle
Homing endonucleases are a large class of proteins that are common in simple eukaryotes and in prokaryotes (1). Their coding regions (called genes here for convenience), are inserted into a specific DNA sequence and spliced out of either the transcribed mRNA as an intron, or out of the translated protein as an intein. Homing endonuclease genes act as “selfish” mobile genetic elements, because their gene product introduces a break at an “empty” target site and forces the host machinery to repair the break by copying the endonuclease gene. These genes are not essential to their hosts. Currently, much is known about the molecular structure and action of homing endonucleases and their genes, but little information is available on their population biology (2).
From a population genetics standpoint, a homing endonuclease allele could easily become fixed in a host population and susceptible to degeneration because homing opportunities disappear. Therefore, the long-term survival of a homing endonuclease allele at a specific site is proposed to depend on horizontal transfer across the boundary of a species or population, within which mating takes place, to another species or population that do not yet contain the homing endonuclease allele at that particular site (3, 4). Specifically, they have been hypothesized to persist over a long evolutionary period only if they can transmit to other species before extinction. For this reason, active homing endonuclease genes are often used as an indicator of DNA transfer events. This interpretation is consistent with phylogenetic analysis of homing endonuclease genes such as ω and PI-SceI (VDE) of Saccharomyces cerevisiae (5), the group I intron of the mitochondrial cox1 gene (6), and the group I intron of the 23S rRNA genes of hyper-thermophilic bacteria.
Although these interpretations are widely accepted, a functional homing intein endonuclease in the PRP8 gene of euascomycetes (Pezizomycota) has been found that can survive within a species for several hundred million years, in the absence of interspecies horizontal transfer (4). Because the conditions that allow the evolutionary persistence of homing endonuclease genes without horizontal transfer are unknown, this process must be explored. In this work, we investigate if horizontal gene transfer across the boundary of a species or population within which mating takes place is necessary for the evolutionary maintenance of homing endonuclease genes. We built a model of a hypothetical homing endonuclease gene that spreads through meiotic double-strand break repair, similar to VDE (7). We analyzed its allele frequency over evolutionary periods in which an alternation of meiotic generations was interspersed with stages of haploid and diploid mitosis, followed by degenerate mutation, resulting in a pseudogene, which would be lost. We mathematically explored conditions that allowed persistence of the homing endonuclease gene, and numerically confirmed the evolutionary dynamics.
We discovered conditions for persistence of the homing endonuclease gene, elucidated their dependence on parameters such as its cost, and generated phase diagrams. Persistence was possible through autonomous periodic oscillation, or through 2 other types of evolutionary dynamics, even in the absence of horizontal transfer. These results were confirmed by stochastic simulations in finite populations.
Model.
We constructed a model of a haploid-diploid cycle in a Mendelian population of unicellular eukaryotes, in which the haploid cells correspond to gametes in higher eukaryotes. We focused on a homing endonuclease locus that moves by homing in the early phase of meiosis. At each round of meiosis, which was designated as a meiotic generation, the survival of each allele depended on the frequencies of the interacting haploid genotypes in the population. The model is illustrated in Fig. 1A, and symbols are explained in Table 1.
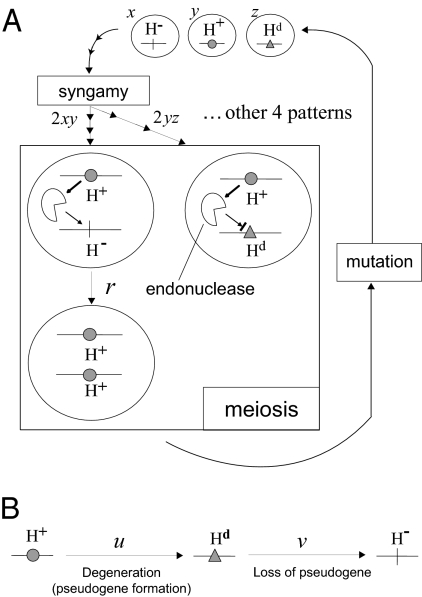
Fig. 1.
Model of homing endonucleases. (A) Life cycle of homing endonuclease alleles: allele H+ produces a homing endonuclease that cleaves the empty allele, H−, on its homologous chromosome, in a diploid cell during meiosis. The DNA double-strand break is repaired by the host with the H+ allele as the template. This process of homing results in gene conversion from H− to H+. The homing endonuclease does not cleave Hd, which represents a pseudogene degenerated from H+. Diploid cells are formed by syngamy from all pair-wise combinations of haploid cells, whose probabilities depend on the frequency of each haploid cell type. (B) Mutation: the H+ allele becomes pseudogene Hd by degenerate mutation at the rate u. Pseudogene Hd is occasionally lost from its target site with the rate v, which is smaller than u.
Table 1.
Definitions
| Variable and parameter | Symbol | Value |
|---|---|---|
| Homing endonuclease absent allele | H– | – |
| Homing endonuclease positive allele | H+ | – |
| Homing endonuclease defective (pseudogene) allele | Hd | – |
| Frequency of H– | x | – |
| Frequency of H+ | y | – |
| Frequency of Hd | z | – |
| Rate of homing (DNA double-strand breakage by the homing endonuclease and succeeding repair by a host cell) | r | – |
| Cost of carrying a homing endonuclease gene H+ on a host cell | c1 | – |
| Cost of carrying a homing endonuclease pseudogene Hd on a host cell | c2 | – |
| Relative fecundity of a cell carrying an H+ allele to that carrying only H– empty sites | α | e−c1 |
| Relative fecundity of a cell carrying an Hd allele to that carrying only H– empty sites | β | e−c2 |
| Rate of degeneration (pseudogene formation) | u | – |
| Rate of losing pseudogene (precise excision) | v | u/10 |
| Effective population size | N | – |
If α > β, then β = α 2(c2 = 2c1). In contrast, if β > α, then β = (c2 = c1/2).
Each haploid cell carried one of 3 homing endonuclease alleles: a gene encoding the homing endonuclease (H+), a pseudogene formed by its degenerate mutation (Hd), or its empty target site (H−). A zygote was formed by random mating of the haploid cells. All 6 combinations of haploid cells could be produced by syngamy, with probabilities given by the product of the frequency (x, y, or z) of each allele. For example, the probability of the formation of diploid H+/H− was 2xy, and that of H+/Hd was 2yz. We assumed that the haploid cells could freely switch their mating type (8) or that the homing endonuclease alleles were randomly associated with the mating types.
During meiosis of an H+/H− diploid cell, the homing endonuclease produced from the H+ allele cleaves the unique empty target site (H−) of the homologous chromosome. Homing endonucleases are known to have a single target site in a haploid genome (9). Although they can evolve new target specificities (10, 11), they also tolerate individual bp variation at their recognition sequences (12). We therefore ignored the possibility that mutations at their target prevented recognition. The DNA double-strand break generated by endonuclease cleavage is efficiently repaired by the recombination repair system of the host cell that is induced at the meiosis stage. Repair occurs with no deleterious effects (13) through the copying of the homologous H+ allele, which results in gene conversion from H− to H+. This homing process takes place at the rate r. The homing endonuclease will not cleave the H+ or Hd allele because of insertion at the recognition site. The pseudogene Hd and the empty target site H− do not produce the functional homing endonuclease, so the homing process and the accompanying change in allele frequency can occur only in H+/H− diploids.
After the homing step, each diploid cell that results from production of a zygote was assumed to undergo meiosis to regenerate haploid cells. The frequency of each allele was calculated at this stage. We assumed a cost to the host cell of carrying a homing endonuclease gene, c1, manifested as a reduced growth rate. The relative fecundity of a cell carrying an H+ allele to one carrying only an H− empty target site depended on the cost expressed as α = e−c1. Also assumed was the cost of carrying a pseudogene, c2, which was also represented by a reduced growth rate. The relative fecundity of a cell carrying Hd allele to one carrying only H− alleles similarly depended on the cost expressed as β = e−c2. Assuming an independent contribution of H+ or Hd to the cost, the relative fecundity of a diploid cell was given by the product of the fecundities of 2 haploid cells Two contrasting cases for the relative amounts of costs c1and c2 (αand β) were assumed. If the cost of carrying an intact homing gene was more than the cost of carrying a pseudogene (β > α), we set β = (c2 = c1/2). In contrast, if carrying a pseudogene gene was more costly (α > β), then β = α2(c2 = 2c1). Because the cost to the host of carrying some homing endonuclease genes is very small (2), we assumed in our numerical analysis that c1 was not large, and therefore α was high (for example, c1 = 0.01,α = 0.99).
This calculation is summarized in Table 2 as a mating table containing all patterns of syngamy, with probabilities of occurrence and the number of progeny for each pattern. The relative number for a particular haploid progeny from one type of diploid is given by the probability of occurrence of syngamy, the rate of homing, and the relative fecundity of the diploid cell. Note that β can be replaced according to the definition of Table 1.
Table 2.
Mating table
| Zygote genotypes | Probability | Progeny |
||
|---|---|---|---|---|
| H– | H+ | Hd | ||
| H–/H– | x2 | 1 | 0 | 0 |
| H+/H– | 2xy | a(1−r)/2 | a2r+a(1−r)/2 | 0 |
| H+/H+ | y2 | 0 | a2 | 0 |
| Hd/H– | 2xz | β /2 | 0 | β /2 |
| Hd/Hd | z2 | 0 | 0 | β 2 |
| Hd/H+ | 2yz | 0 | α β /2 | α β /2 |
From the mating table (Table 2), we arrived at the following equations. The fitness W, which is the expected number of progeny per haploid individual, was calculated for each allele as follows:
The allele frequency in the next meiotic generation can be calculated as the expected number of haploid cells harboring each allele—for example, W(y)y—divided by the total number of haploid cells in the population, or the mean fitness W̄ = W(x)x+W(y)y+W(z)z:
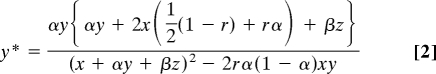 |
where ynext = y* and znext = z* are the frequencies of H+ and Hd in the next meiotic generation. Note that x can be replaced by x = 1−y−z. The frequency of H− in the next meiotic generation was calculated by using x* = 1−y*−z*.
When the effect of mutation was introduced, Eq. 2 changed as follows:
As illustrated in Fig. 1B, only a limited case of mutations was taken into account. That is, only degeneration of the H+ allele to Hd at rate u, and precise loss of the pseudogene creating an empty target site H− at rate v, was assumed. As v is conceivably smaller than u, we assumed v = u/10 in our numerical analysis. All other mechanisms with the potential to change allele frequency, such as gene conversion caused by mechanisms other than homing, horizontal gene transfer, or migration were not assumed here. We defined horizontal gene transfer as transfer of a gene across the boundary of a species or population within which mating takes place, i.e., a Mendelian population. An empty target site H− could be converted to H+ only through the homing process, resulting in an increase in the frequency of the homing endonuclease gene in the population.
We mathematically explored conditions for persistence of the H+ allele based on the above equations. Stability of 3 marginal equilibria in which each of the 3 alleles predominates in the population was analyzed. Stability of the internal equilibrium in which the 3 alleles could coexist was then analyzed, together with the conditions for its existence. We summarized the parameter dependence of the conditions that allowed the evolutionary maintenance of the homing endonuclease gene with phase diagrams, and numerically confirmed the evolutionary dynamics. [See SI Appendix for details on the stability analyses and derivation of the phase diagrams.]
We also carried out computer simulations in finite populations with an effective size N ≥106 (14) to consider the possibility of stochastic extinction, and examined the validity of our mathematical analysis. Because we did not assume that any of the mechanisms described earlier recovered the H+ allele, it would inevitably become extinct during a long evolutionary time span because of degeneration caused by accumulation of mutations. The average time until the extinction of the H+ allele was calculated by 50 runs of simulation for each parameter set.
Results
The conditions for evolutionary maintenance of the homing endonuclease gene were classified by the magnitude of α and β. If β > α—i.e., the cost of carrying the homing endonuclease gene H+ is larger than the cost of carrying the homing endonuclease pseudogene Hd—the homing endonuclease gene can be maintained through periodic oscillation of the 3 alleles. In contrast, if β < α, the homing endonuclease gene can predominate in the population. Phase diagrams and the evolutionary dynamics are as follows.
Phase Diagrams and Evolutionary Dynamics for β > α.
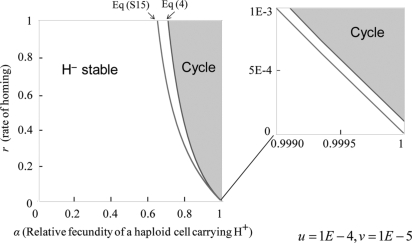
If β > α, which is the case when the pseudogene has lost its homing endonuclease activity but retains splicing activity, thus imposing only a minor additional cost on the host cell, the pseudogene Hd is advantageous over H+ if there is no H− allele in the equilibrium. In this case, the phase diagram shows 2 regions: the stable H− monomorphism and the limit cycle of all 3 homing alleles (Fig. 2).
Fig. 2.
Phase diagram for α < β indicating the region in which the H+ allele is maintained through periodic oscillation. The black line is obtained from the inequality shown in Eq. 4. In the gray region (Right), a limit or heteroclinic cycle occurs. H− is stable in the white region on the left side of the black boundary irrespective of its initial frequency. The gray line is obtained from Eq. S15 (SI Appendix), indicating that an internal equilibrium exists (Right). The enlargement (Upper Right) shows how these 2 boundaries shift with mutation. The black boundary line shifts upward by the mutation rate u. In contrast, the gray boundary line shifts downward. The 2 boundaries intersect at (α,r) = (1, 0). These diagrams are illustrated with mutation rates u = 10−4,v = 10−5, which are higher than expected for clarity.
The black line in the phase diagram shows the boundary of the area of H− predominance obtained as follows:
indicating that H− is stable if the rate of homing is smaller than a threshold. Specifically, the empty allele H− predominates if the cost of homing is considerably high compared with the rate of homing.
In the region to the right of the black line, where Eq. 4 is not satisfied, however, none of the 3 alleles can permanently predominate in the population. All equilibria, including internal equilibrium, are unstable, and limit or heteroclinic cycles occur, depending on the presence or absence of mutation. Accordingly, the H+ allele can be maintained through a limit cycle in the presence of mutation, if the homing rate r is considerably high compared with the cost of carrying a homing endonuclease gene.
The gray line in the phase diagram of Fig. 2 is the boundary for the area of the existence of the internal equilibrium. This is obtained from Eq. S15 in the SI Appendix, indicating that an internal equilibrium exists on its right. In the space between the black and gray lines, H− is stable, irrespective of its initial frequency, and its evolutionary dynamics move away from the internal equilibrium toward marginal equilibrium where H− dominates. The enlargement in Fig. 2 shows how these 2 boundaries shift with mutation. In the presence of mutation, the black boundary line shifts upward by the mutation rate u, while the gray boundary line shifts slightly downward.
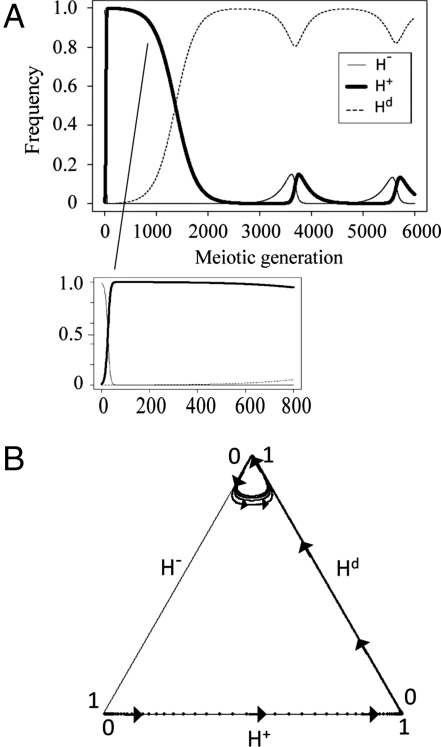
The evolutionary dynamics of the limit cycle of all 3 homing alleles is numerically illustrated in Fig. 3. The H+ allele is maintained through periodic oscillation of the 3 homing endonuclease alleles. At the beginning, the majority of the allele population is H−, providing ample opportunity for homing, which enables the spread of the H+ allele. After the apparent near-fixation, however, the H+ allele is susceptible to degeneration because there is little opportunity for homing. This, in turn, enables the spread of the Hd allele. The first 800 meiotic generations in Fig. 3 Upper Right show this fixation-to-degeneration process. Hd has a relative advantage over H+ while it still imposes cost c2 (assumed to be c1/2 and thus β = ) on the host, and is thus disadvantageous versus H− when the threat by H+ becomes small. This, in turn, increases H− frequency, which again generates the homing opportunity for the H+ allele. The above process is repeated, and the period is approximately 2,000 generations according to the parameter set of Fig. 3. The period decreases monotonically as the relative fecundity of a cell carrying H+ decreases, as is shown in Fig. S1. The limit cycle enables the H+ allele to be maintained over thousands of meiotic generations in the absence of horizontal transfer. A schematic diagram of the limit cycle of homing endonuclease alleles in a population is shown in Fig. S2. The result is consistent with cyclical models of the gain, degeneration and loss of homing endonuclease genes that are formulated based on the presence of horizontal transfer (3, 5). Results under different cost and mutation parameter settings are summarized in Fig. S3. In all of the parameter spaces explored, the limit cycle appeared and the homing endonuclease gene was maintained.
Fig. 3.
Evolutionary dynamics of periodic oscillation of homing endonuclease alleles for α < β. Results obtained numerically for a parameter set in the Cycle region of Fig. 2 with mutations: α = 0.99 (c1 = 0.01), r = 0.2, u = 6 × 10−6,v = 6 × 10−7. The initial frequencies are 99% for H− and 1% for H+.
Stochastic Simulations for β > α.
To consider stochastic extinction, and examine the validity of our mathematical analysis, we also carried out simulations in finite populations. Because we did not assume any mechanism for recovery of the H+ allele, such as horizontal transfer, migration, or reverse mutation, the homing endonuclease allele would inevitably become extinct at some point over a long evolutionary period. The average time until extinction is expected to depend on the cost and mutation rate. The results summarized in Table 3 indicate that the average time is more than thousands of meiotic generations, unless the cost is very large. An example of the long-term trajectory under the same parameter set as in Fig. 3, with N≥106 illustrated in Fig. S4.
Table 3.
Average logarithmic time to extinction of H+ allele in stochastic simulations for α < β
| u | N = 106 | N = 107 | N = 108 |
|---|---|---|---|
| c1 = 0.01 (α ≈ 0.990) | |||
| 6 × 10−6 | 3.8±0.4 | >5 | >5 |
| 6 × 10−5 | >5 | >5 | >5 |
| 6 × 10−4 | >5 | >5 | >5 |
| c1 = 0.03 (α ≈ 0.970) | |||
| 6 × 10−6 | 3.4±0.3 | >5 | >5 |
| 6 × 10−5 | >5 | >5 | >5 |
| 6 × 10−4 | >5 | >5 | >5 |
| c1 = 0.05 (α ≈ 0.951) | |||
| 6 × 10−6 | 3.1±0.2 | >5 | >5 |
| 6 × 10−5 | >5 | >5 | >5 |
| 6 × 10−4 | >5 | >5 | >5 |
The average logarithmic time (± SD) in terms of meiotic generation was calculated from 50 runs of simulation. The rate of homing, r, was 0.2. All logarithms are to base 10.
Phase Diagrams and Evolutionary Dynamics when β < α.
If β < α, e.g., if the intron or intein has lost the ability to be spliced out and consequently has a deleterious effect on its host, the pseudogene Hd is disadvantageous compared with the H+ allele. In this case, the phase diagram shows 3 regions: H− stable, bistable, and H+ stable (Fig. S5). In this phase diagram, the black line, obtained from Eq. 4, represents the boundary of the area of H− predominance as in Fig. 2. As shown earlier, H− is stable on its left. In contrast, the gray line in the phase diagram is the boundary of the area of H+ predominance in a population. In the absence of mutation, this is obtained from Eq. 5:
which indicates that H+ is stable if the rate of homing r is larger than the threshold. In the presence of mutation, an H+-dominating marginal equilibrium exists if β < α:
where higher terms of u are disregarded. The condition for the local stability of H+-predominant equilibrium (Eq. 6) is again approximated by Eq. 5. Therefore, in the deep gray “H+ stable” region where Eq. 5 is satisfied, H+ predominates irrespective of its initial frequency. In the light gray “bistable” region, where both Eq. 4 and Eq. 5 are satisfied, the result depends on the initial frequency. In the white “H− stable” region, H− predominates irrespective of its initial frequency. The enlargement in Fig. S5 shows how these 2 boundaries shift with mutation. Similar to Fig. 2, the black boundary line shifts upward by the mutation rate u, while the gray boundary line shifts slightly downward. In contrast to the case of α < β, no internal equilibrium exists when α > β.
The evolutionary dynamics of the bistable and H+ stable regions are numerically illustrated in Fig. S6A and B, respectively. The former dynamics were confirmed by randomly assigning 100 patterns of initial frequencies to the 3 alleles, showing that the homing endonuclease gene H+ predominates if its initial frequency is relatively high. The latter shows that the homing endonuclease gene H+ predominates irrespective of its initial frequency.
Stochastic Simulations for β < α.
Under N≥106, H+ does not become extinct within 105 meiotic generations for any parameter set used in Table 3. An example of unrealistically disadvantageous parameter sets for the H+ allele, and its trajectory to extinction, is illustrated in Fig. S7.
Discussion
Our mathematical analyses revealed that evolutionary persistence is indeed possible for a homing endonuclease gene in the absence of gene transfer across the boundary of a species or population within which mating takes place. The dynamics enabling the persistence were classified by the cost to the host cell of carrying the homing endonuclease gene H+ or its pseudogene Hd, and 2 phase diagrams were constructed.
If the cost of the pseudogene was lower than that of the homing endonuclease gene (β > α), persistence of the homing endonuclease gene was possible through autonomous periodic oscillation in the population, in the presence of mutation (Figs. 2 and 3). An example of this case would be if the pseudogene lost homing endonuclease activity but retained splicing activity. The endonuclease domain of VDE is known to degenerate after becoming fixed in a population. This type of pseudogene is common in Saccharomyces (7).
Conversely, if the cost of the pseudogene was higher than that of the intact homing endonuclease gene (β < α), persistence of the homing endonuclease gene was possible through the evolutionary dynamics of bistability. Furthermore, if the homing rate became sufficiently high, persistence of the homing endonuclease gene was possible irrespective of its initial frequency (Fig. S5 and Fig. S6B). An example of this case would be the presence of a pseudogene whose intron or intein has a reduced ability to be spliced out (15), which could have a deleterious effect on the host cell.
In the former case, opportunities of homing must be regularly created, even after fixation of the homing endonuclease gene, for the evolutionary maintenance of homing endonuclease genes. This is because the pseudogene is advantageous over the homing endonuclease gene and can always invade the population. Horizontal gene transfer is believed to be the most effective mechanism for this. Although we do not deny the effectiveness of horizontal gene transfer, our results suggest that horizontal gene transfer is not the only mechanism that enables persistence of homing endonuclease genes over a long evolutionary period. This was confirmed by stochastic simulations on finite populations with effective population sizes ≥106, which was consistent with estimates for yeast and other small eukaryotes (14, 16). The average time to extinction of allele H+ is thousands of meiotic generations, unless its cost is high. Because meiosis in S. cerevisiae is initiated only upon nutritional starvation of diploid cells, its frequency is expected to be much smaller than that of mitosis of either haploid cells or diploid cells. Although the exact ratio of meiosis to mitosis in natural environments has not been examined for S. cerevisiae (17), the ratio in S. paradoxus, the closest wild relative of S. cerevisiae, is estimated to be 1:1,000 in Europe and 1:3,000 in the Far East (14). Therefore, the ratio could range from 1:100 to 1:10,000 for S. cerevisiae. The thousands of meiotic generations would then correspond to 105 to 107 mitotic generations. In addition, the doubling time for unicellular organisms in natural environments is likely to be much longer than in laboratory conditions. Escherichia coli reproduces at a doubling time of 40 h in the human intestine (18), but exhibits a doubling time of 0.5 h under laboratory conditions. We could not find comparable estimates for S. cerevisiae, which can be regarded as an opportunistic pathogen on fruits and other plants (17), and might assume a similar 102-fold difference. In addition, microbes are likely to be in a resting stage more often than a dividing stage. We found no estimate of how often yeast organisms are in a dividing stage in natural environments, but it could be 1 to 100. Taken together, the average time for extinction estimated above appears to be, in practice, very long. Any mechanism increasing H+ frequency such as reverse mutation or migration would further promote its persistence.
In experiments with VDE of S. cerevisiae, the cost of carrying a homing endonuclease gene to the host cell seems very small because effect of VDE on mitotic replication rates is less than 1% in inbred populations (19). The observation corresponds to the regions in our phase diagrams in which the H+ allele is maintained under the relatively low cost of carrying a homing endonuclease gene.
Although cyclical models of the gain, degeneration, and loss of homing endonuclease genes based on their horizontal transfer have been formulated (3, 5), the present work is, to our knowledge, the first published theoretical analysis to successfully summarize the parameter dependence of the conditions for the cycle in the absence of horizontal gene transfer. Whether the evolutionary maintenance of homing endonuclease genes through periodic oscillation without horizontal gene transfer occurs in nature is worth exploring.
Other than mutation, migration, and horizontal gene transfer, other mechanisms might affect endonuclease gene frequency. One possible mechanism is gene conversion of Hd/H− during meiosis, producing Hd/Hd or H−/H−, and resulting in a haploid cell with an empty site allele. Mitotic gene conversion in H+/H− diploids would be very low for VDE, because its meiosis-specific, karyopherin-mediated nuclear import is required for its action on the chromosomal site (13, 20). Another possible mechanism is reverse transcription of spliced mRNAs that do not contain introns, followed by homologous recombination of the cDNA with the genomic DNA, resulting in intron loss (21). These mechanisms would help create homing opportunities, and would be advantageous for the homing endonuclease allele.
In each meiotic generation, the survival of each homing endonuclease allele depends on the frequencies of the interacting haploid cells. Our haploid-diploid cycle model was constructed to analyze the population dynamics of homing endonuclease alleles that involved interdependence among several alleles. The same framework could be applied to other homing endonuclease genes that spread through comparable chromosome cycles. Homing endonucleases of bacteriophage T4 (I-Tev I, I-Tev II, F-Tev I, and F-Tev II) are one example (1). Bacteriophage double-strand break repair mechanisms involving homologous recombination (22) would provide an opportunity for homing during co-infection of a single host cell. Homing endonuclease ω of yeast mitochondria are another example. Mitochondria are biparentally inherited in S. cerevisiae and recombine through a haploid-diploid-like cycle in which a double-strand break induces homologous recombination and gene conversion (5).
Another model of the evolutionary trajectory of a selfish invasion, followed by slow decay to complete loss, has been proposed for vertically transmitted bacteria of the genus Wolbachia (23). In this case, crosses between a male infected with Wolbachia and an uninfected female produce only few progeny because of cytoplasmic incompatibility. The infected males thus sacrifice themselves to reduce the frequency of uninfected females. Invasion and temporary predominance of the infected cytotype is followed by invasion of the resistant cytotype, allowing the eventual spread of the uninfected cytotype. However, the dynamics are frequency-dependent and do not allow the uninfected cytotype to spread irrespective of its initial frequency. This characteristic of dynamics, and the asymmetric relationship between the males and females, distinguishes their behavior from that of the homing endonuclease genes.
Other selfish genetic elements that cause a meiotic drive, such as Segregation Distorter in Drosophila melanogaster (24) or the spore killer in ascomycetes (25), also have a 3-allele polymorphism, i.e., killer, sensitive, and resistant. To be precise, their “alleles” are a combination of a killer/non-killer and another allele. Segregation Distorter is different from the homing endonuclease genes, in that the polymorphism is maintained by a strong directional selection against the killer alleles in diploid organisms. In contrast, the life cycle and mechanism for maintaining polymorphism in spore killer seems similar to those of the homing endonuclease genes. The published model (23), however, allowed sensitive-to-killer mutations, and did not analytically find conditions for the limit cycle and bistability, as were found here. None of the aforementioned models, including the one presented here, consider spatial structure, meaning the spatial localization of interactions between organisms. Because spatial structure can have a significant effect on the qualitative differences in the direction of evolution (26), inclusion of this effect into the models is worth exploring. Another study on a selfish genetic element with 3 alleles (toxin-producing, sensitive, resistant) revealed that spatial structure promotes coexistence of the 3 alleles in a population (27).
To our knowledge, this study is the first theoretical analysis of the evolutionary persistence of homing endonuclease genes in the absence of horizontal transfer to successfully summarize parameter dependence as phase diagrams. The results will provide a solid theoretical basis for our understanding of the evolution of homing endonuclease genes and other selfish genetic elements that spread through a comparable cycle.
Supplementary Material
Acknowledgments.
We thank Drs. Satoru Nogami and Yoshikazu Ohya (University of Tokyo) for communication and helpful comments on the manuscript, and Drs. Masayuki Yamamoto, Akiko Sakai (University of Tokyo), and Yoshinobu Kaneko (Osaka University) for advice on yeast. The simulation was run on a computer at the Human Genome Center at the Institute of Medical Science, University of Tokyo. This work was supported by a grant from the Science and Technology Foundation of Japan (to K.Y.). The work of I.K. was supported by the 21st Century Center of Excellence Project, a grant from Foundation for the Fusion of Science and Technology, and Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science (21370001). This work was supported in part by The Graduate University for Advanced Studies (Sokendai).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908404106/DCSupplemental.
References
- 1.Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Harvard Univ Press; 2006. [Google Scholar]
- 3.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Butler MI, Gray J, Goodwin TJ, Poulter RT. The distribution and evolutionary history of the PRP8 intein. BMC Evol Biol. 2006;6:42. doi: 10.1186/1471-2148-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Y, Palmer JD. Multiple acquisitions via horizontal transfer of a group I intron in the mitochondrial cox1 gene during evolution of the Araceae family. Mol Biol Evol. 1999;16:1155–1165. doi: 10.1093/oxfordjournals.molbev.a026206. [DOI] [PubMed] [Google Scholar]
- 7.Okuda Y, Sasaki D, Nogami S, Kaneko Y, Ohya Y, Anraku Y. Occurrence, horizontal transfer and degeneration of VDE intein family in Saccharomycete yeasts. Yeast. 2003;20:563–573. doi: 10.1002/yea.984. [DOI] [PubMed] [Google Scholar]
- 8.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 9.Posey KL, Koufopanou V, Burt A, Gimble FS. Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species. Nucleic Acids Res. 2004;32:3947–3956. doi: 10.1093/nar/gkh734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosen LE, et al. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J Mol Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda T, Nogami S, Ohya Y. VDE-initiated intein homing in Saccharomyces cerevisiae proceeds in a meiotic recombination-like manner. Genes Cells. 2003;8:587–602. doi: 10.1046/j.1365-2443.2003.00659.x. [DOI] [PubMed] [Google Scholar]
- 14.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc Natl Acad Sci USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawasaki M, Nogami S, Satow Y, Ohya Y, Anraku Y. Identification of three core regions essential for protein splicing of the yeast Vma1 protozyme. A random mutagenesis study of the entire Vma1-derived endonuclease sequence. J Biol Chem. 1997;272:15668–15674. doi: 10.1074/jbc.272.25.15668. [DOI] [PubMed] [Google Scholar]
- 16.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 17.Landry CR, Townsend JP, Hartl DL, Cavalieri D. Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol. 2006;15:575–591. doi: 10.1111/j.1365-294X.2006.02778.x. [DOI] [PubMed] [Google Scholar]
- 18.Hartl DL, Dykhuizen DE. The population genetics of Escherichia coli. Annu Rev Genet. 1984;18:31–68. doi: 10.1146/annurev.ge.18.120184.000335. [DOI] [PubMed] [Google Scholar]
- 19.Goddard MR, Greig D, Burt A. Outcrossed sex allows a selfish gene to invade yeast populations. Proc R Soc Lond Ser B. 2001;268:2537–2542. doi: 10.1098/rspb.2001.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai Y, Nogami S, Kumagai-Sano F, Ohya Y. Karyopherin-mediated nuclear import of the homing endonuclease VMA1-derived endonuclease is required for self-propagation of the coding region. Mol Cell Biol. 2003;23:1726–1736. doi: 10.1128/MCB.23.5.1726-1736.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi N, Kobayashi I. Evidence for the double-strand break repair model of bacteriophage lambda recombination. Proc Natl Acad Sci USA. 1990;87:2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst LD, McVean GT. Clade selection, reversible evolution and the persistence of selfish elements: the evolutionary dynamics of cytoplasmic incompatibility. Proc R Soc Lond Ser B. 1996;263:97–104. [Google Scholar]
- 24.Charlesworth B, Hartl DL. Population dynamics of the segregation distorter polymorphism of Drosophila melanogaster. Genetics. 1978;89:171–192. doi: 10.1093/genetics/89.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nauta MJ, Hoekstra RF. Evolutionary dynamics of spore killers. Genetics. 1993;135:923–930. doi: 10.1093/genetics/135.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006;172:1309–1323. doi: 10.1534/genetics.105.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.