Abstract
The human MHC represents the strongest susceptibility locus for autoimmune diseases. However, the identification of the true predisposing gene(s) has been handicapped by the strong linkage disequilibrium across the region. Furthermore, most studies to date have been limited to the examination of a subset of the HLA and non-HLA genes with a marker density and sample size insufficient for mapping all independent association signals. We genotyped a panel of 1,472 SNPs to capture the common genomic variation across the 3.44 megabase (Mb) classic MHC region in 10,576 DNA samples derived from patients with systemic lupus erythematosus, Crohn's disease, ulcerative colitis, rheumatoid arthritis, myasthenia gravis, selective IgA deficiency, multiple sclerosis, and appropriate control samples. We identified the primary association signals for each disease and performed conditional regression to identify independent secondary signals. The data demonstrate that MHC associations with autoimmune diseases result from complex, multilocus effects that span the entire region.
Keywords: autoimmunity, genes, genetics
Following its discovery in mice in 1936 (1), the human MHC was mapped to the short arm of chromosome 6 and was studied extensively for both gene and variation content. The first full sequence of this region was completed and reported in 1999 by the MHC Sequencing Consortium (2). Gene density was greater than expected: from 224 identified loci, 128 were predicted to be expressed, and about 40% were predicted to have immunological functions. Among these loci, the classic HLA class I (HLA-A, -B, -C) and class II (HLA-DP, -DQ, -DR) gene clusters involved in antigen processing and presentation are the best characterized in terms of structure, diversity, and function.
The ability to respond to an antigen, whether foreign or self, and the nature of that response are determined to a large extent by the unique amino acid sequences of HLA alleles, an observation that followed the first association studies between HLA genotypes and susceptibility to diseases (3, 4). More than 100 diseases, many of which are autoimmune, have been associated with HLA genes. In some disorders, single HLA genes seem to be implicated in susceptibility (e.g., HLA-B27 and ankylosing spondylitis) (5); in others, specific heterodimers (e.g., DQB1*02 in celiac disease) (6) or complex interactions between alleles at multiple genes within the HLA (e.g., HLA-DR, -DQ and rheumatoid arthritis) have been described (7). In most cases, the MHC region is the strongest genetic component in autoimmune diseases (8–11). However, the extensive allelic variation and linkage disequilibrium (LD) across the MHC, together with the limited number of MHC genes examined in most association studies reported to date, have confounded attempts to resolve unequivocally the location of the primary signals responsible for disease susceptibility (12, 13).
To identify the genetic variants within the MHC that are disease specific and those that are shared across multiple inflammatory diseases, we examined a set of 10,576 DNA samples derived from patients with systemic lupus erythematosus (SLE), Crohn's disease (CD), ulcerative colitis (UC), rheumatoid arthritis (RA), myasthenia gravis (MG), selective IgA deficiency (IgAD), multiple sclerosis (MS), and appropriate control samples. Building on published high-density genetic maps of the extended MHC (14–16), we genotyped a panel of 1,472 SNPs to capture the common genetic variation across the 3.44 megabase (Mb) classic MHC region. We imputed classic HLA alleles in individuals, which we validated in a subset of samples for which classic HLA typing was available. Although the majority of causal variants in this region remain to be identified, the results demonstrate that susceptibility to these diseases results from complex multilocus effects that span the entire region, with evidence for shared loci.
Results
SNP-Based Screening for Testing both HLA and non-HLA Variation.
Based on a high-resolution haplotype map of HLA alleles and SNPs across the extended MHC (16), we designed and genotyped a panel of 1,472 SNPs in a cohort of 10,576 individuals (supporting information (SI) Text), aiming to capture common variation across the MHC region including classic HLA alleles. The study participants were recruited according to well-established diagnostic and inclusion criteria (see Materials and Methods).
To maximize the uniformity of the genotype data and the comparability of results across all the cohorts, sample handling, genotyping, allele calling, quality control, and association analyses were performed simultaneously on the entire set of DNA samples (see SI Text). To increase efficiency, 3 sets of shared controls were used, representing the general geographic regions from which the majority of patient samples were collected (the United States, the United Kingdom, and Sweden); this approach previously has been shown to be reliable (17). Overall, 83.8% of SNPs and 97.48% of all DNA samples passed quality-control thresholds (see Methods), with a final call rate of 99.0%.
We imputed the HLA alleles in all samples, using the high-resolution haplotype map of the region (including SNPs and HLA alleles) as the reference (16) (see SI Text). This study is the first to use imputation methodology for the association analysis of HLA alleles. Although imputation for SNP alleles now is well established, the high diversity and complex haplotype structure makes imputation of HLA alleles considerably more difficult (16, 18). Notably, the training data used here to make HLA allele predictions (16) is relatively small (range of 142–172 haplotypes with 4-digit resolution per locus), and low-frequency HLA alleles suffer from few observations in the training data (see ref. 16 for a list of classic alleles for which training data were available). We therefore measured the sensitivity, specificity, and positive predictive value of the imputed HLA alleles in a validation study using data available from the 1958 Birth Cohort (18) and data on samples from the current project for which classic HLA data were available (Fig. S1 and Table S1). For the 16 alleles for which there is strong or suggestive evidence of disease association (as described in later sections), the average sensitivity is 92%, the average specificity is 98%, and the average positive predictive value is 86%. These figures indicate that, at least for alleles for which we make a claim of disease association, imputation accuracy is sufficient to expect little loss of power relative to direct typing.
We subsequently performed association tests for individual SNPs across the MHC region as well as for the (imputed) HLA alleles for each disease. For 3 of the diseases (SLE, RA, and MS), we exploited the availability of separate European and American cohorts, designating 1 cohort as the “screening cohort” and the other as the “replication cohort.” As shown in Table 1, the high degree of concordance for the top SNP and HLA alleles between cohorts indicates the robustness of the approach and the high quality of the sample collections. For example, the top 6 SLE markers (in both sides of the RCCX module and HLA-DQA1) are identical in both groups. Thus, for all subsequent analyses, samples were pooled to increase power. These composite results across all diseases are presented in Fig. S2, and the most highly associated SNPs and HLA alleles are summarized in Table 2. In fact, with the exception of MG, all the top associations observed in the separate cohorts are of genome-wide significance.
Table 1.
Top association SNP and HLA signals in screening and replication datasets
| Disease | ID | Genomic position | Screen dataset |
Replication dataset |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Site | OR | P-value | Rank | Site | OR | P-value | Rank | |||
| SLE | rs1269852 | 32188168 | UK | 2.6 | 6.26E-19 | 1 | US | 2.00 | 6.23E-09 | 1 |
| DRB1*0301 | 32600001 | UK | 2.3 | 2.15E-15 | 8 | US | 1.87 | 8.78E-08 | 11 | |
| RA | rs6457617 | 32771828 | SWE | 0.3 | 3.1E-27 | 7 | US | 0,4 | 5.1E-16 | 2 |
| DQA1*0301 | 32716004 | SWE | 3.13 | 9.17E-19 | 1 | US | 4.8 | 1.91E-49 | 1 | |
| MS | rs3135391 | 32518964 | UK | 3.66 | 6.86–29 | 2 | US | 3.2 | 7.03E-22 | 1 |
| DRB1*1501 | 32600020 | UK | 3.65 | 4.18E-29 | 1 | US | 3.1 | 1.22E-20 | 5 | |
Table 2.
Top disease specific association signals for the MHC in entire datasets
| Disease | Type | ID | Genomic position | Top association signals relative position | OR | P-value | Reciprocal conditional p-value | r2 in disease dataset |
|---|---|---|---|---|---|---|---|---|
| SLE | SNP | rs1269852 | 32151660 | Between TNXB and CREBL1 | 2.4 | 5.63E-29 | 1.64E-06 | 0.78 |
| SLE | HLA | DRB1*0301 | 32605000 | DRB1 | 2.1 | 1.06E-23 | 0.51 | |
| UC | SNP | rs4639334 | 32653366 | Between DRB1 and DQA1 | 4.0 | 1.30E-11 | 0.24 | 0.71 |
| UC | HLA | DRB1*1101 | 32605000 | DRB1 | 4.6 | 1.59E-11 | 0.32 | |
| CD | SNP | rs382259 | 32280470 | NOTCH4 region | 2.3 | 1.40E-09 | 7.19E-06 | 0.14 |
| CD | HLA | DRB1*1101 | 32605000 | DRB1 | 2.2 | 2.08E-05 | 0.025 | |
| RA | SNP | rs2395175 | 32513003 | Between BTNL2 and DRA | 3.7 | 1.36E-96 | 1.30E-05 | 0.78 |
| RA | HLA | DQA1*0301 | 32660500 | DQA1 | 4.0 | 1.78E-107 | 7.33E-15 | |
| MG | SNP | rs2523674 | 31541152 | 3' of HCP5 | 1.5 | 2.89E-04 | 0.011 | 0.16 |
| MG | HLA | HLA-C*0701 | 31346016 | HLA-C | 1.6 | 1.02E-03 | 0.047 | |
| IgAD | SNP | rs2187668 | 32713861 | Intronic DQA1 | 2.8 | 5.31E-14 | 0.69 | 0.66 |
| IgAD | HLA | DQB1*0201 | 32676000 | DQB1 | 2.8 | 3.04E-17 | 0.0013 | |
| MS | SNP | rs3135391 | 32482210 | DRA (synonymous coding) | 3.4 | 5.51E-49 | 1 | 0.98 |
| MS | HLA | DRB1*1501 | 32605000 | DRB1 | 3.3 | 6.62E-48 | 1 |
To determine whether the associations detected for the top SNPs and the top HLA alleles are related or independent, we performed reciprocal logistic regression tests and also examined the correlation (as measured by the coefficient of determination, r2) between these SNPs and the HLA alleles and generally found consistency between them (Table 2).
Next, we identified the set of variants that are statistically equivalent to each top (primary) association, providing a list of potential causal variants and the genomic region in which the causal gene is likely to be located (Table S2). Finally, we conditioned each disease dataset on the most significant association to identify secondary association signals independent of the peak association (Table 3). For many of the diseases, there was evidence for multiple additional association signals (Fig. 1 and Tables S3 and S4).
Table 3.
Most significant secondary associations identified following conditioning on primary association signals
| Type | ID | Type | ID | Relative position | P-value | |
|---|---|---|---|---|---|---|
| SLE | SNP | rs1269852 | SNP | rs3135391 | Within HLA-DRA | 3.90E-06 |
| HLA | DRB1*1501 | 1.46E-05 | ||||
| UC | SNP | rs4639334 | SNP | rs382259 | NOTCH4 region | 4.82E-06 |
| HLA | DQB1*0502 | 4.82E-03 | ||||
| CD | SNP | rs382259 | SNP | rs4713436 | Within CDSN | 3.31E-04 |
| HLA | - | - | - | |||
| RA | HLA | DQA1*0301 | SNP | rs6457614 | Between HLA-DQA1 and HLA-DQA2 | 1.21E-17 |
| HLA | DQB1*0501 | 1.48E-21 | ||||
| MG | SNP | rs2523674 | SNP | - | - | - |
| HLA | - | - | - | |||
| IgAD | HLA | DQB1*0201 | SNP | rs3135352 | Between BTNL2 and HLA-DRA | 7.70E-07 |
| HLA | DRB1*1501 | 6.65E-07 | ||||
| MS | SNP | rs3135391 | SNP | rs2743951 | Between HLA-F and HLA-G | 1.88E-04 |
| HLA | HLA-B*4402 | 1.08E-08 |
Conditional logistic regression analysis was performed on each disease for the top HLA and top SNP. The top HLA and SNP signal following conditioning are shown for each of these alleles.
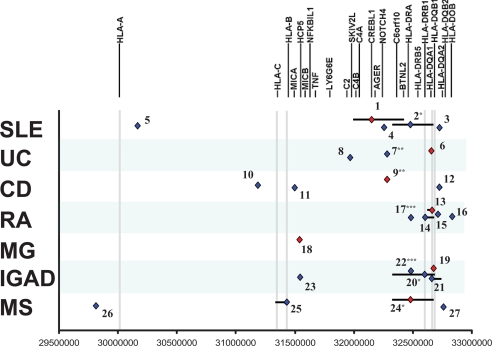
Fig. 1.
Summary of primary and secondary signals for all 7 diseases. Top primary association signals (red) and putative independent secondary signals (blue) are shown for each disease along with their location within the region. Independent secondary signals were defined as those with residual conditional association p-values < 0.001, following logistic regression analysis for the top primary association signal and showing pair-wise correlation (r2) lower than 0.2 with other neighboring signals. The correlation neighborhood or extent of correlation around each marker, at an r2 > 0.8, is illustrated by black lines. Numbers attached to each signal refer to additional information on span and location of the correlation neighborhoods (see Table S4).
Systemic Lupus Erythematosus.
In the combined United Kingdom and United States SLE dataset, the top signal is rs1269852, located between TNXB and CREBL1 (Table 2 and Fig. S2), in strong LD with HLA-DRB1*0301 (r2 = 0.78). Although the top imputed HLA allele is HLA-DRB1*0301, conditioning on the TNXB-CREBL1 association indicates that the HLA-DRB1*0301 signal may be dependent on this SNP, whereas the SNP itself shows association over and above that exhibited by HLA-DRB1*0301 (Table 2). However, the imperfect correlation between the imputed and classically typed HLA-DRB1*0301 allele in this dataset suggests that the impact of this class II association may be underestimated. Conditioning on the top signal (rs1269852) identifies a number of secondary signals; the best is rs3135391 within HLA-DRA (Table 3) and variants in LD with this allele including HLA-DRB1*1501 (r2 = 0.98) (Table 3). Other signals potentially independent of rs1269852 are seen in class I (between RNF39 and TRIM31), class III (NOTCH4), and class II (HLA-DQB1-DQA2) (Fig. 1 and Table S4). The analysis detected further associations in class III; hence there seem to be at least 3 separate signals in this region tagged by the SKIV2L gene, rs1269852, and NOTCH4 (19). Together, these data indicate the presence of multiple SLE risk alleles located across the class I, class II, and class III regions.
Ulcerative Colitis.
Both the SNP and the HLA data convincingly show that the main signal (Table 2 and Fig. S2) is located in a narrow genomic window containing the HLA-DRB1 gene (although < 10 kb from the 5′ end of HLA-DQA1) and strongly suggest that the more common HLA-DRB1*1101 allele (≈10%) plays a primary role in UC susceptibility. An association with this allele also was detected by a recent meta-analysis of published data (4). This primary association to the DRB1 locus is consistent with 3 recently published genome-wide association studies in UC (20–22). Interestingly, upon conditional analyses, the top secondary signal is localized around the NOTCH4 gene (Table 3), a region also implicated in our analyses of SLE and CD. Independent of this signal is a cluster of associated alleles in the BAT8-C2-RDBP-SKIV2L region including intronic variants in each of the first 3 genes (Fig. 1 and Table S4). A role for this class III segment has not been characterized previously for UC but overlaps with 1 of the secondary signals in SLE, albeit to an independent set of alleles.
Crohn's Disease.
Cross-disease risk factors in the MHC also seem to exist between CD and UC, because the current CD data reveal a significant association with the HLA-DRB1*1101 allele (Table 2 and Fig. S2). Testing of the individual SNPs also provides evidence of association at rs382259, which is independent of HLA-DRB1*1101, shows greater statistical significance, and is located in the adjacent NOTCH4 region. Association with this SNP is supported by the significant association of an adjacent SNP (rs419132, P = 1.0 × 10−8), at a distance of less than 2 kb (pairwise r2 between these 2 SNPs is 0.94). As noted earlier, this signal also is observed as the major secondary signal in UC. Furthermore, this same region was identified in the recent meta-analysis of CD genome-wide association studies (23), further supporting the importance of this region in CD risk. Searching the entire dataset for association signals independent of rs382259, we identified independent association signals in the class I region near the CDSN gene, in the region between HLA-B and MICA, and in the DQA1-DQB1-DQA2-DQB2 region (Fig. 1, Table 3, and Table S4). Of interest, the DQA1-DQB1-DQA2-DQB2 region contains 3 intronic SNPs in HLA-DQA2 (rs9276431, rs2213567, and rs2213568) and a synonymous coding SNP in HLA-DQB2 that are associated, albeit with p-values just shy of the significant threshold set for these conditional analyses. These results are consistent with previous reports of a modest association with MICA and HLA-DRB1 alleles (HLA-DRB1*0103, HLA-DRB1*04, HLA-DR7, and HLA-DRB3*0301). These results also suggest an important role for the HLA-DQA/DQB region, an observation that is consistent with a recent meta-analysis of CD genome-wide association studies (23). These results also suggest that previous reports of an association with TNF alleles (24) actually may represent a residual signal from the MICA association.
Rheumatoid Arthritis.
Early studies in RA noted significant association with multiple alleles at the HLA-DRB1 locus (*0101, *0401, and *0404), and Gregersen et al. noted that this allelic heterogeneity could be explained by a shared amino acid sequence or “shared epitope” at positions 70–74 of the HLA-DRβ1 protein (7). More recently, it has become evident that the RA association with the MHC is restricted to RA individuals positive for antibodies to citrullinated protein antigens (ACPA) (25). Therefore, the current study was restricted to the ACPA-positive form of RA. The top signal maps to DQA1*0301, and the top SNP is rs2395175, located ≈ 2.5 kb upstream from HLA-DRA. Although these 2 signals show high correlation (r2 = 0.78), reciprocal conditional analysis reveals them to be independent (Fig. S2 and Table 2). DQA1*0301 is reported to be in LD with DRB1*0401 and *0404, and although these shared-epitope alleles were not the top primary or secondary association signals, they were associated (P = 4.07 × 10−77 and 1.17 × 10−12, respectively) and showed residual signal upon conditioning on DQA1*0301 (P = 0.0025 and 2.28 × 10−5, respectively).
Conditioning on either the top SNP or DQA1*0301 identifies additional independent effects, the strongest of which is to DQB1*0501 (Fig. 1 and Table 3). DRB1*0101, another shared-epitope allele, is part of this secondary signal, although with slightly lower (almost equivalent) association signal (see Table S4).
The data also suggest the presence of additional independent signals located within the class II region: rs6457617 (located between HLA-DQA1 and -DQA2), which was also independently reported by the Wellcome Trust Case Control Consortium study (17), rs2621326 (located within the HLA-DOB locus) in a region previously reported by Lee and colleagues (26), and rs3129878 (located in an intron of the HLA-DRA locus), a signal that also is shared with IGAD (see later sections). Additional significant signals can be observed beyond the ones described here, including the HLA-DRB1*0404 allele (1 of the shared-epitope alleles) and a signal in the HLA-DPB1-Col11A2 region, which have been reported previously (25, 26).
Myasthenia Gravis.
In the MG dataset there is a paucity of significant signals in the class II region; the strongest associations arise from the class I region. Specifically, we observed that the top association was to rs2523674, which is located 3.5 kb away from the 3′ end of the HLA complex protein 5 (HCP5) gene, between MICA and MICB. The strongest imputed associated HLA allele is HLA-C*0701. There is strong LD between HLA-C*0701 and HLA-B8, but the latter shows a slightly weaker association than HLA-C*0701. The SNP rs2523674 and these HLA alleles are independent of each other (Table 2 and Fig. S2). The conditional analyses performed with rs2523674 and HLA-C*0701 did not reveal any statistically significant additional signals, although this result may be a reflection of the modest sample size and/or disease heterogeneity rather than a feature of the genetic architecture of this disease. Taken together, the data confirm the strong influence of HLA-C and suggest an independent effect of rs2523674 in the HCP5 region.
Selective IgA Deficiency.
The association of IgAD with the MHC is clearly documented (27), but the precise location of the genetic effect within the region has remained elusive. The peak association in the current study is with the HLA-DQB1*0201 allele (P < 10−16) (Table 2 and Fig. S2). This association is ≈1,000-fold more significant than that of the imputed HLA-DRB1*0301 and the top SNP (rs2187668) that tags the DRB1*0301 allele. Conditioning upon HLA-DQB1*0201 demonstrates that the DRB1*0301 signal is not independent of the primary signal at HLA-DQB1. The analyses also reveal an independent secondary association with HLA-DRB1*1501 that is protective (Table 3) as well as other suggestive risk alleles in the class II and class III regions located between BTNL2 and HLA-DRA and in LD with the protective DRB1*1501 allele (Fig. 1 and Table S4).
Multiple Sclerosis.
The association of MS with HLA genes, specifically the DRB1*1501 allele, has been a consistent finding across nearly all studies, including this study, in which the top HLA signal is DRB1*1501, detected both with a tagging SNP (rs3135391) and by imputed HLA (Table 2 and Fig. S2). Trends for association with DRB1*0301 and DRB1*0401 also were detected, with the DRB1*0401 association nearly reaching a threshold for significance (P < 10−5). Conditioning for rs3135391, putative independent clusters in the class III and class I regions emerge, with HLA-B*4402 being the top signal (Fig. 1, Table 3, and Table S4). This allele is in LD with HLA-C*0501 (r2 = 0.58), an allele that has been associated with MS in the absence of DRB1*1501 (28). When the MS cases were compared with unrelated controls, the results remained unchanged, and a case-control analysis demonstrated a similar under-representation of HLA-B*4402 on DRB1*1501 haplotypes from MS patients compared with controls. The significance of the residual signals in the HLA-A region after conditioning for rs3135391 (Fig. S2 and Table S3) remains to be determined.
Discussion
In the multisystem autoimmune disease SLE, the MHC represents the strongest risk locus genomewide (10, 29, 30), and case-control studies in predominantly European-derived populations have revealed a consistent association with the HLA-DRB1*0301 and HLA-DRB1*1501 class II alleles and their respective haplotypes (31, 32). The particularly strong LD across these haplotypes has confounded attempts to identify clearly additional independent signals, such as those previously reported for complement C4-null alleles, as well as TNF and SKIV2L variants, all encoded within the class III region (19, 32, 33). The influence of copy number variation at the complement C4/RCCX locus in relation to the association signals demonstrated in this study remains to be established. The current high-density SNP analysis confirms that the predominant signals in SLE map to the class II and class III regions. Also, there seem to be at least 2 class III associations, with peaks on either side of the RCCX module, in addition to a further signal centered around NOTCH4, a gene involved in development and cell fate; another independent signal is located in the class I region near TRIM31.
UC and CD are related inflammatory diseases of the gastrointestinal tract, classified together as inflammatory bowel diseases. Several independent genome-wide scans in inflammatory bowel disease have shown evidence of linkage to the MHC region (34–36), and recent genome-wide association studies of nonsynonymous SNPs in UC confirmed the association with the MHC, specifically within the BTNL2 and HLA-DRB1 genes (17, 37). In European-derived populations, HLA-DRB1*0103 represents the most reproducible association observed to date in UC, albeit at a low prevalence of less than 2% (38). Previous studies of the HLA loci in CD have identified association with 4 independent HLA alleles, DRB1*07, DRB1*0103, DRB1*04, and DRB3*0301. One of 4 recent genome-wide association studies of CD, as well as the meta-analysis of these studies, also found association with this region, specifically with the nearby region delimited by the HLA-DQA1 to DQA2 genes (23, 39). The results from the current study, however, strongly implicate the more common HLA-DRB1*1101 allele (≈10% allele frequency) in both UC and CD susceptibility. Upon conditional analyses, the top secondary signal is localized around NOTCH4, a gene also implicated in our analyses of SLE and CD.
In RA, the top independent signals map to HLA-DQA1*0301 and -DRB1*0101, both shared-epitope alleles (7). Because different DRB1 genotypes are known to modulate risk for RA in a hierarchical fashion, genotypic combinations for conditioning may be necessary to expose the full complexity of the underlying genetic association. For example, DRB1*0401/0401 has been associated with very high risk, often in the range of an odds ratio between 20 and 30, whereas the combination of DRB1*0101/*0401 is associated consistently with lower risk (40). Hierarchical risk profiles of different HLA alleles also are likely to be present in MS (41) and in other autoimmune disorders as well. Also, in RA, at least 2 other independent signals were observed, in the DQ region for DQB1–0501 and in the DPB1-CollIA2 region. These data thus indicate that multiple class II-related loci and alleles are associated only with ACPA-positive RA (25). This finding, in turn, may suggest that MHC class II alleles linked to this subset of RA may be involved in determining the magnitude of response to different citrullinated proteins/peptides.
MG is characterized by autoimmunity at the postsynaptic neuromuscular junction. Prior studies had suggested that the class I region has a role in susceptibility (42, 43). The current analysis reveals that, compared with the other autoimmune disorders, the genetic role of the MHC in MG is distinctive, consisting of a single signal in the class I region, specifically in the vicinity of the HLA complex protein 5 gene (HCP5) located between MICA and MICB. The strongest associated imputed HLA allele is HLA-C*0701. An underlying biologic heterogeneity of MG is likely; thus it will be of great interest to determine whether endophenotypes such as the identity of the autoantibody (acetylcholine receptor vs. muscle-specific kinase vs. titin), the presence of thymoma, or the occurrence of associated autoimmune diseases are associated with distinctive HLA-region signals. In this regard, an association with MHC class II genes has been reported in patients with muscle-specific kinase or titin antibodies (44).
Multiple HLA haplotypes are known to be positively associated with IgAD (serum IgA < 0.07 g/L), the most common form of primary immunodeficiency in the Western world (45, 46). The peak primary association in the current study is to the HLA-DQB1*0201 allele, with a significant secondary association with HLA-DRB1*1501 that is protective, and other putative risk alleles in the class II and class III regions. Recently, mutations in MSH5, encoded within the class III region, affecting class-switch recombination and associated with the HLA-B14-DR1 haplotype, have been suggested to constitute a primary risk factor for IgAD (47). However, no mutations in MSH5 were identified on the HLA-B8-DR3 susceptibility haplotype, suggesting a more complex MHC etiology in this disease.
Genetic studies of the HLA region in MS consistently yielded convincing evidence for the presence of a major susceptibility gene or genes (48). This signal maps primarily to the 1-Mb segment spanning the class II genes. There is debate, however, as to whether the HLA-DRB1*1501 association explains the entire MHC genetic signal and whether other independent genes of interest exist within the class III, class I, and/or telomeric to the class I regions (28, 49). The results of the present study highlight the role of HLA class I gene products and are consistent with the known dose effect of DRB1*1501 haplotypes on MS susceptibility suggesting that a second disease gene exists within the MHC (50) and also consistent with the current working model of MS pathogenesis which includes a prominent role for CD8+ T cells (51, 52). Further, MHC class I molecules may act as a molecular address for CD8+ T cells at the blood−brain barrier, facilitating the transendothelial migration of antigen-specific T-cells into the brain (53). This situation might have evolved as a result of selective pressure to promote antiviral immune surveillance of the central nervous system.
Because the conditional analysis used here is based generally on the assumption that a single primary association signal allows conditioning on a relatively homogeneous set of cases and controls that exhibit this primary association, additional layers of complexity are likely to exist. Nevertheless, the data clearly demonstrate that, in contrast to the prevailing single-locus model, the MHC associations with chronic inflammatory diseases result from complex, multilocus effects that span the entire MHC region. Furthermore, because this MHC-specific panel of SNPs was typed and analyzed uniformly across multiple diseases, it was possible to begin to identify a set of shared genetic variants (Fig. 1) that should, with additional mapping efforts, lead to a better understanding of common pathogenic mechanisms. For example, consistent independent signals across class I genes may indicate a possible role for killer cell immunoglobulin-like receptor-HLA class I combinations, perhaps by promoting innate immune responses to viral infections. In addition, although we highlight associations to specific HLA or non-HLA variants for each disease, we have shown that for any association signal there are many equivalent genetic variants, demonstrating that narrowing the associations and identification of the causal genes will require even larger disease cohorts, potentially in different populations of distinct ancestry. Finally, no single approach will be sufficient to dissect fully the complex set of HLA and non-HLA genetic factors, and thus a combination of the approach described herein with classic typing and deep resequencing will reveal the disease-specific and shared risk factors involved in chronic immune-mediated diseases.
Materials and Methods
Cohorts.
The complete data set studied consisted of 10,576 individuals (SI Text). Diagnostic criteria, ascertainment protocols, and clinical and demographic characteristics for cases and controls are summarized in previously published reports: SLE (30), CD (54), UC (54), RA (55, 56), MG (57), IGAD (58), MS (48), and the 1958 birth cohort (59). Appropriate institutional review boards approved all studies, and written informed consent was obtained from all participants.
Genotyping Assay Design.
We designed an Illumina GoldenGate panel of 1,536 SNPs consisting of 16 fingerprinting quality-control SNPs, 48 genomic control SNPs (60), 135 SNPs to tag the classic HLA types, and 1,337 SNPs to tag common SNP variations within the 3.44-Mb classic MHC region based on a high-density haplotype map using the Tagger algorithm (16, 61). All coordinates for SNPs and HLA markers are given relative to the National Center for Biotechnology Information Build 34 human genome assembly. Overall this set of SNPs captured variation of common (≥ 5%) HLA markers, less-common (<5%) HLA markers, common non-HLA markers, and less-common non-HLA markers, with an average maximum r2 of 0.80, 0.64, 0.90, and 0.62, respectively. We imputed classic HLA alleles in all individuals based on the available SNP data (18).
Supplementary Material
Acknowledgments.
This work was supported primarily by Grant AI067152 from the National Institutes of Allergy and Infectious Diseases. Additional support was supplied by Grants DK064869 and DK062432 from the National Institute of Diabetes and Digestive and Kidney Diseases (to J.D.R.), by Grant NS21799 from the National Institute of Neurological Disease and Stroke (to S.L.H.), and by the Swedish National Research Council. We also thank the disease-specific societies and foundations for support in DNA collections. We acknowledge the use of DNA from the British 1958 Birth Cohort collection (D. Strachan, S. Ring, W. McArdle, and M. Pembrey), funded by the Medical Research Council Grant G0000934 and Wellcome Trust Grant 068545/Z/02. The Broad Institute Center for Genotyping and Analysis is supported by Grant U54 RR020278 from the National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909307106/DCSupplemental.
References
- 1.Gorer PA. The detection of a hereditary antigenic difference in the blood of mice by means of human group A serum. Journal of Genetics. 1936;32:17–31. [Google Scholar]
- 2.The MHC Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 3.McDevitt HO. Discovering the role of the major histocompatibility complex in the immune response. Annu Rev Immunol. 2000;18:1–17. doi: 10.1146/annurev.immunol.18.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Fernando MM, et al. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genetics. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan MA, Mathieu A, Sorrentino R, Akkoc N. The pathogenetic role of HLA-B27 and its subtypes. Autoimmunity Reviews. 2007;6:183–189. doi: 10.1016/j.autrev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Vader W, et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci USA. 2003;100:12390–12395. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 8.Sawcer S, et al. A whole genome screen for linkage disequilibrium in multiple sclerosis confirms disease associations with regions previously linked to susceptibility. Brain. 2002;125:1337–1347. doi: 10.1093/brain/awf143. [DOI] [PubMed] [Google Scholar]
- 9.Aly TA, et al. Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci USA. 2006;103:14074–14079. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genetics. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klitz W, Stephens JC, Grote M, Carrington M. Discordant patterns of linkage disequilibrium of the peptide-transporter loci within the HLA class II region. Am J Hum Genet. 1995;57:1436–1444. [PMC free article] [PubMed] [Google Scholar]
- 13.Stenzel A, et al. Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet. 2004;114:377–385. doi: 10.1007/s00439-003-1075-5. [DOI] [PubMed] [Google Scholar]
- 14.Walsh EC, et al. An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet. 2003;73:580–590. doi: 10.1086/378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miretti MM, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bakker PI, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernando MM, et al. Identification of 2 independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genetics. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher SA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and 5 loci implicated in Crohn's disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franke A, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 22.Silverberg MS, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki A, Tsuchiya N, Hagiwara K, Takazoe M, Tokunaga K. Independent contribution of HLA-DRB1 and TNF alpha promoter polymorphisms to the susceptibility to Crohn's disease. Genes and Immunity. 2000;1:351–357. doi: 10.1038/sj.gene.6363689. [DOI] [PubMed] [Google Scholar]
- 25.Ding B, et al. Different patterns of associations with ACPA-positive and ACPA-negative rheumatoid arthritis in the extended MHC region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HS, et al. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Molecular Medicine (Cambridge, Mass.) 2008;14:293–300. doi: 10.2119/2007-00123.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vorechovsky I, Webster AD, Plebani A, Hammarstrom L. Genetic linkage of IgA deficiency to the major histocompatibility complex: Evidence for allele segregation distortion, parent-of-origin penetrance differences, and the role of anti-IgA antibodies in disease predisposition. Am J Hum Genet. 1999;64:1096–1109. doi: 10.1086/302326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeo TW, et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol. 2007;61:228–236. doi: 10.1002/ana.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 30.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao BP. Update on human systemic lupus erythematosus genetics. Current Opinion in Rheumatology. 2004;16:513–521. doi: 10.1097/01.bor.0000132648.62680.81. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickering MC, Walport MJ. Links between complement abnormalities and systemic lupus erythematosus. Rheumatology (Oxford) 2000;39:133–141. doi: 10.1093/rheumatology/39.2.133. [DOI] [PubMed] [Google Scholar]
- 34.Satsangi J, et al. Contribution of genes of the major histocompatibility complex to susceptibility and disease phenotype in inflammatory bowel disease. Lancet. 1996;347:1212–1217. doi: 10.1016/s0140-6736(96)90734-5. [DOI] [PubMed] [Google Scholar]
- 35.Duerr RH, Barmada MM, Zhang L, Pfutzer R, Weeks DE. High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11–12. Am J Hum Genet. 2000;66:1857–1862. doi: 10.1086/302947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rioux JD, et al. Genomewide search in Canadian families with inflammatory bowel disease reveals 2 novel susceptibility loci. Am J Hum Genet. 2000;66:1863–1870. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnett HA, et al. BTNL2, a butyrophilin/B7-like molecule, is a negative costimulatory molecule modulated in intestinal inflammation. J Immunol. 2007;178:1523–1533. doi: 10.4049/jimmunol.178.3.1523. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad T, et al. The contribution of human leucocyte antigen complex genes to disease phenotype in ulcerative colitis. Tissue Antigens. 2003;62:527–535. doi: 10.1046/j.1399-0039.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 39.Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nature Reviews Genetics. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 40.Gregersen PK. Teasing apart the complex genetics of human autoimmunity: Lessons from rheumatoid arthritis. Clinical Immunology. 2003;107:1–9. doi: 10.1016/s1521-6616(02)00045-1. [DOI] [PubMed] [Google Scholar]
- 41.Barcellos LF, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 42.Janer M, et al. A susceptibility region for myasthenia gravis extending into the HLA- class I sector telomeric to HLA-C. Hum Immunol. 1999;60:909–917. doi: 10.1016/s0198-8859(99)00062-2. [DOI] [PubMed] [Google Scholar]
- 43.Vandiedonck C, et al. Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci USA. 2004;101:15464–15469. doi: 10.1073/pnas.0406756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraud M, et al. Linkage of HLA to myasthenia gravis and genetic heterogeneity depending on anti-titin antibodies. Neurology. 2001;57:1555–1560. doi: 10.1212/wnl.57.9.1555. [DOI] [PubMed] [Google Scholar]
- 45.Cobain TJ, French MA, Christiansen FT, Dawkins RL. Association of IgA deficiency with HLA A28 and B14. Tissue Antigens. 1983;22:151–154. doi: 10.1111/j.1399-0039.1983.tb01181.x. [DOI] [PubMed] [Google Scholar]
- 46.Hammarstrom L, et al. HLA antigens in selective IgA deficiency: Distribution in healthy donors and patients with recurrent respiratory tract infections. Tissue Antigens. 1984;24:35–39. doi: 10.1111/j.1399-0039.1984.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 47.Sekine H, et al. Role for Msh5 in the regulation of Ig class switch recombination. Proc Natl Acad Sci USA. 2007;104:7193–7198. doi: 10.1073/pnas.0700815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.International Multiple Sclerosis Genetics Consortium. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 49.Lincoln MR, et al. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat Genet. 2005;37:1108–1112. doi: 10.1038/ng1647. [DOI] [PubMed] [Google Scholar]
- 50.Barcellos LF, et al. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet. 2003;72:710–716. doi: 10.1086/367781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babbe H, et al. Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J Exp Med. 2000;192:393–404. doi: 10.1084/jem.192.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bitsch A, Schuchardt J, Bunkowski S, Kuhlmann T, Bruck W. Acute axonal injury in multiple sclerosis. Correlation with demyelination and inflammation. Brain. 2000;123:1174–1183. doi: 10.1093/brain/123.6.1174. [DOI] [PubMed] [Google Scholar]
- 53.Galea I, et al. An antigen-specific pathway for CD8 T cells across the blood-brain barrier. J Exp Med. 2007;204:2023–2030. doi: 10.1084/jem.20070064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Jager PL, et al. The role of the Toll receptor pathway in susceptibility to inflammatory bowel diseases. Genes and Immunity. 2007;8:387–397. doi: 10.1038/sj.gene.6364398. [DOI] [PubMed] [Google Scholar]
- 55.Klareskog L, et al. A new model for an etiology of rheumatoid arthritis: Smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 56.Plenge RM, et al. TRAF1–C5 as a risk locus for rheumatoid arthritis—a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsinzerling N, Lefvert AK, Matell G, Pirskanen-Matell R. Myasthenia gravis: A long term follow-up study of Swedish patients with specific reference to thymic histology. J Neurol Neurosurg Psychiatry. 2007;78:1109–1112. doi: 10.1136/jnnp.2006.109488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan-Hammarstrom Q, et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat Genet. 2007;39:429–430. doi: 10.1038/ng0407-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall IP, et al. Beta2-adrenoceptor polymorphisms and asthma from childhood to middle age in the British 1958 birth cohort: A genetic association study. Lancet. 2006;368:771–779. doi: 10.1016/S0140-6736(06)69287-8. [DOI] [PubMed] [Google Scholar]
- 60.Freedman ML, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 61.de Bakker PI, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.