Abstract
Despite intense investigation over the past century, the molecular mechanisms that regulate maintenance and adaptation of the heart during postnatal development are poorly understood. Myocardin is a remarkably potent transcriptional coactivator expressed exclusively in cardiac myocytes and smooth muscle cells during postnatal development. Here we show that myocardin is required for maintenance of cardiomyocyte structure and sarcomeric organization and that cell-autonomous loss of myocardin in cardiac myocytes triggers programmed cell death. Mice harboring a cardiomyocyte-restricted null mutation in the myocardin gene (Myocd) develop dilated cardiomyopathy and succumb from heart failure within a year. Remarkably, ablation of the Myocd gene in the adult heart leads to the rapid-onset of heart failure, dilated cardiomyopathy, and death within a week. Myocd gene ablation is accompanied by dissolution of sarcomeric organization, disruption of the intercalated disc, and cell-autonomous loss of cardiomyocytes via apoptosis. Expression of myocardin/serum response factor-regulated myofibrillar genes is extinguished, or profoundly attenuated, in myocardin-deficient hearts. Conversely, proapoptotic factors are induced and activated in myocardin-deficient hearts. We conclude that the transcriptional coactivator myocardin is required for maintenance of heart function and ultimately cardiomyocyte survival.
Keywords: apoptosis, heart failure, serum response factor, sarcomere, intercalated disc
Despite intense investigation over the past century, the molecular mechanisms that regulate maintenance and adaptation of the heart during postnatal development remain poorly understood. Transcriptional coactivators have evolved to modulate information encoded within the genome required for organismal homeostasis and survival. Myocardin is a remarkably potent transcriptional coactivator expressed exclusively in cardiomyocytes and smooth muscle cells (SMCs) during postnatal development (1–3). Myocardin physically associates with serum response factor (SRF) at CArG box motifs to synergistically activate transcription (1–3). In the SMC lineage, myocardin promotes the contractile SMC phenotype by activating a subset of SRF-dependent genes encoding SMC-restricted contractile and cytoskeletal proteins (4–7). By contrast, the function of myocardin in the heart during embryonic and postnatal development remains unclear. Myocardin-null mutant mice survive only through embryonic day (E)10.5, exhibiting a block in vascular SMC differentiation (8). At least through E10.5, the hearts of myocardin-null embryos appear morphologically normal (8).
Consistent with its role as a transcriptional coactivator, myocardin may regulate the adaptive response of the heart to growth factors and hemodynamic stress (9–12). Expression of dominant-negative myocardin-mutant protein blocks expression of genes encoding some cardiac-specific myofibrillar proteins and disrupts heart tube formation (9). Forced expression of myocardin in murine embryonic stem cells activates SRF-dependent cardiac- and SMC-restricted genes (4, 10). In addition, myocardin transactivates multiple SRF-dependent genes associated with the “fetal/hypertrophic program,” including ANF, α-skeletal actin, and SM22α (1, 11). Forced expression of myocardin in isolated cardiomyocytes induces cardiomyocyte hypertrophy, while expression of a dominant-negative myocardin-mutant protein blocks cardiomyocyte hypertrophic response (12).
In the studies described in this article, we generated and characterized mice in which the myocardin gene (Myocd) was conditionally ablated in cardiomyocytes. Ablation of the Myocd gene in the adult heart leads to the rapid progression of heart failure, dilated cardiomyopathy, and death within a week. Myocd gene ablation is accompanied by dissolution of sarcomeric organization, disruption of the intercalated disc, and the cell-autonomous loss of cardiomyocytes via programmed cell death. Consistent with this observation, SRF/myocardin-activated genes encoding cardiomyocyte-specific myofibrillar and structural proteins were profoundly repressed. These data provide conclusive evidence that the transcriptional coactivator myocardin is required for maintenance of heart function and, ultimately, cardiomyocyte survival.
Results
Cardiac-Restricted Ablation of the Myocd Gene.
To elucidate the function of myocardin in the heart, genetically engineered mice harboring a homozygous conditional null mutation in the Myocd gene (MyocdF/F) (6, 13) were intercrossed to αMyHC-Cre mice (6, 13) that express Cre recombinase under the transcriptional control of the cardiomyocyte-specific α-myosin heavy chain (αMyHC) promoter to generate αMyHC-Cre/MyocdF/F-mutant mice. Quantitative RT-PCR (qRT-PCR) revealed an 80% decrease in myocardin mRNA in hearts harvested from 3-month-old αMyHC-Cre/MyocdF/F mutants [supporting information (SI) Fig. S1A]. Immunoblot analysis revealed a 90% reduction in myocardin protein in the hearts of αMyHC-Cre/MyocdF/F mutants compared to control littermates (Fig. S1B). Consistent with these findings, myocardin expression in the nucleus was markedly attenuated in hearts harvested from αMyHC-Cre/MyocdF/F-mutant mice (Fig. S1C). By contrast, qRT-PCR performed with mRNA prepared from the aorta of 3-month-old αMyHC-Cre/MyocdF/F-mutant mice and control MyocdF/F littermates demonstrated comparable levels of myocardin gene expression (Fig. S1D). These data demonstrate that αMyHC-Cre/MyocdF/F-mutant mice exhibit a strong hypomorphic phenotype, related to cardiomyocyte-restricted deletion of the Myocd gene, as opposed to a true null phenotype.
MyHC-Cre/MyocdF/F-Mutant Mice Develop Lethal Cardiomyopathy.
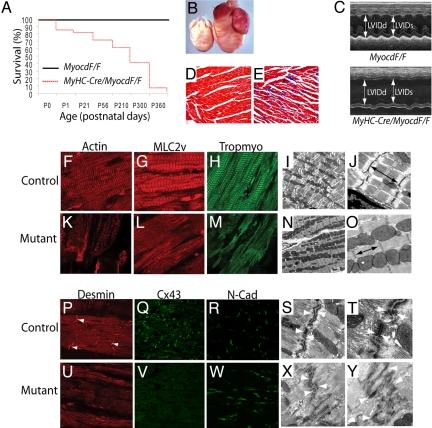
αMyHC-Cre/MyocdF/F-mutant mice were born in the anticipated Mendelian ratio. However, ≈20% of the αMyHC-Cre/MyocdF/F-mutant mice died before 3 weeks of age (P21) and 90% of the mutants died by 10 months of age (Fig. 1A). To date, no αMyHC-Cre/MyocdF/F-mutants (n = 62) have survived beyond 1 year. Hearts harvested from 10-month-old αMyHC-Cre/MyocdF/F-mutant mice exhibit marked four-chamber enlargement (Fig. 1B). Pericardial effusions and atrial or ventricular thromboses were observed in each αMyHC-Cre/MyocdF/F-mutant heart (n = 10). Echocardiography performed on 10-month-old αMyHC-Cre/MyocdF/F-mutant mice and controls revealed marked enlargement of the left atrium, left ventricle (LV), right atrium, and right ventricle compared to control littermates (Fig. 1C and Table S1). Indices of systolic function were severely depressed in αMyHC-Cre/MyocdF/F mutants (see SI Text, Table S1, Movies S1–S4). The mean ejection fraction was 57.8% ± 5.8% in control mice compared to 24.5% ± 7.3% in αMyHC-Cre/MyocdF/F mutants (P < 0.0001). αMyHC-Cre/MyocdF/F-mutant hearts exhibited extensive loss of cardiomyocytes and myocardial fibrosis (Fig. 1 D and E). Remarkably, 11.2% ± 1.2% of the LV myocardium demonstrated fibrosis (blue stain) in αMyHC-Cre/MyocdF/F-mutant hearts when compared to 0.46% ± 0.05% fibrosis in MyocdF/F-control hearts.
Fig. 1.
MyHC-Cre/MyocdF/F-mutant mice develop lethal dilated cardiomyopathy. (A) Kaplan-Meier survival curves for control MyocdF/F mice (black line) (n = 60) and MyHC-Cre/MyocdF/F-conditional mutant mice (red dashed line) (n = 62). (B) Hearts harvested from P300 MyHC-Cre/MyocdF/F-mutant (n = 12) mice exhibit four-chamber enlargement compared to MyocdF/F-control (n = 12) hearts. (C). An M-mode echocardiogram demonstrating that P300 MyHC-Cre/MyocdF/F-mutant mice (n = 10) exhibit increased LV systolic and diastolic dimensions and decreased systolic function compared to MyocdF/F-control mice (n = 10). (D, E) A Masson's trichrome-stained section of LV myocardium demonstrating that P300 MyHC-Cre/MyocdF/F-mutant (E) hearts (n = 10) exhibit cardiomyocyte disarray with fibrosis (blue stain) compared to MyocdF/F-control (D) hearts (n = 10). (F–H, K–M) LV myocardium harvested from P300 MyHC-Cre/MyocdF/F-mutant mice (n = 6) and MyocdF/F controls (n = 3) immunostained with antibodies that recognize α-cardiac actin (F, K), MLC2v (G, L), and tropomyosin (H, M). Note the loss of myofibrillar striations in the MyHC-Cre/MyocdF/F hearts (mutant) (K–M) compared to MyocdF/F (control) hearts (F–H). (I, J, N, O) Electron microscopy of hearts harvested from P300 MyHC-Cre/MyocdF/F-mutant mice (n = 4) and MyocdF/F controls (n = 4) revealed shortening and contracture of sarcomeres (arrows) in MyHC-Cre/MyocdF/F-mutant hearts (N, O) compared to MyocdF/F-control hearts (I, J). (P–R, U–W) Immunohistochemical analyses revealed that expression of Desmin (P, U) and Cx43 (Q, V) are markedly repressed in hearts harvested from MyHC-Cre/MyocdF/F-mutant mice compared to MyocdF/F controls, while comparable levels of N-Cadherin (R, W) immunostaining are observed. (S, T, X, Y) Electron microscopy of MyocdF/F-control hearts (S, T) revealed distinct adherens junctions and desmosomes (arrows). By contrast, lightly-stained serpiginous intercalated discs with indistinct desmosomes and adherens junctions (arrows) were observed in MyHC-Cre/MyocdF/F-mutant hearts (X, Y). Original magnification, ×200 (D, E); ×800 (F–H, K–M, P, U); ×600 (Q, R, V, W); ×10,000 (I, N); ×40,000 (J, O, S, X); ×100,000 (T, Y).
Disruption of Cardiomyocyte Structural Organization.
Confocal microscopic analyses of heart sections prepared from αMyHC-Cre/MyocdF/F-mutant and MyocdF/F-control hearts demonstrated the extensive loss of myofibrillar striations, including loss of α-cardiac actin-, MLC2v-, and α-tropomyosin-immunostaining (Fig. 1 F–H and K–M). This was observed in all four chambers of the heart, although the extent of myofibrillar loss varied somewhat within adjacent regions of the LV (see Fig. 1 K–M). Electron microscopy (EM) revealed shortened, hypercontracted sarcomeres, with a mean sarcomere length of 1.4 μM ± 0.1 μM in mutant hearts versus 2.0 μM ± 0.1 μM in control hearts (arrows, Fig. 1 I, J, N, and O). The intercalated discs of αMyHC-Cre/MyocdF/F-mutant hearts also appeared markedly abnormal (Fig. 1 P–Y). Expression of desmin- and connexin 43 (Cx43) was virtually extinguished in MyHC-Cre/MyocdF/F-mutant hearts (see Fig. 1 P, Q, U, and V). Immunoblot analysis revealed an 80% decrease in Cx43 protein in the mutant hearts compared to controls. By contrast, expression of N-cadherin and β-catenin, components of the adherens junction, was observed in the αMyHC-Cre/MyocdF/F-mutant hearts (see Fig. 1 R and W). EM revealed serpiginous intercalated discs with indistinct desmosomes and adherens junctions in MyHC-Cre/MyocdF/F-mutant hearts (see arrows, Fig. 1 S, T, X, and Y). Taken together, these data demonstrate that αMyHC-Cre/MyocdF/F-mutant mice develop dilated cardiomyopathy accompanied by disruption of the sarcomere and gap junction structure.
Myocardin Is Required for Maintenance of the Adult Heart.
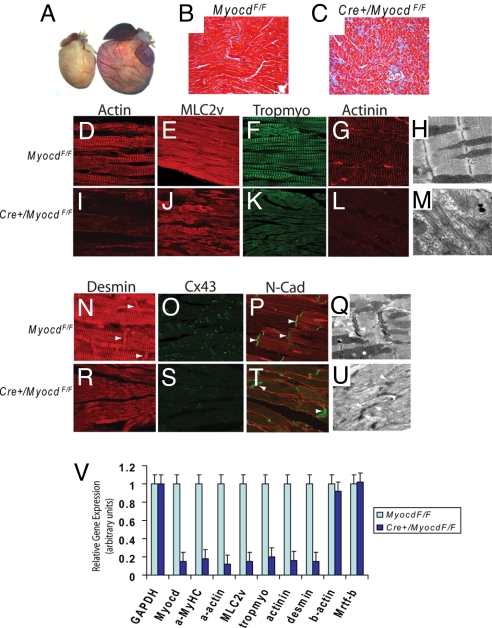
To determine whether myocardin is required for maintenance of the adult heart, MerCreMer/MyocdF/F-conditional mutant mice were generated in which expression of tamoxifen-inducible Cre was placed under the transcriptional control of the cardiomyocyte-specific αMyHC promoter (14). Four days following the initiation of tamoxifen treatment, qRT-PCR revealed an 85% decrease in myocardin mRNA in hearts harvested from MerCreMer/MyocdF/F-mutant mice compared to control MyocdF/F hearts (Fig. 2V). Four-month-old tamoxifen-treated MerCreMer/MyocdF/F mice (n = 24) became notably lethargic within 3 to 4 days of tamoxifen exposure and died within 7 days. By contrast, tamoxifen-treated MyocdF/F-control mice (n = 24) appeared healthy throughout the course of tamoxifen-treatment. Six days after tamoxifen exposure, echocardiography revealed severe dilated cardiomyopathy involving all four chambers in MerCreMer/MyocdF/F-mutant mice, but not in tamoxifen-treated control mice (Table S2 and Movies S5–S8). Remarkably, the mean ejection fraction was 56.0% ± 4.9% in tamoxifen-treated MyocdF/F mice compared to 14.9% ± 4.7% in tamoxifen-treated MerCreMer/MyocdF/F mice (P < 0.001). Of note, no significant difference in ejection fraction was observed between tamoxifen-treated MyocdF/F- and MerCerMer+-control mice.
Fig. 2.
Myocardin is required for maintenance of cardiac structure and function. (A) Hearts harvested 6 days following the initiation of tamoxifen treatment from MerCreMer/MyocdF/F mutants (n = 6) exhibited massive enlargement compared to hearts of tamoxifen-treated MyocdF/F controls (n = 6). (B, C) Masson's trichrome-stained section of LV myocardium harvested 6 days after tamoxifen exposure of MyocdF/F-control mice (n = 6) (B) and MerCreMer/MyocdF/F-mutant mice (n = 6) (C) revealed myocyte disarray and fibrosis (blue stain) in the tamoxifen-treated mutant hearts. (D–G, I–L) Sections of LV myocardium immunostained with antibodies that recognize α-cardiac actin (D, I), MLC2v (E, J), tropomyosin (F, K), and α-actinin (G, L) demonstrated the loss of myofibrillar striations in tamoxifen-treated MerCreMer/MyocdF/F (Cre+/MyocdF/F)-mutant hearts (I–L) compared to MyocdF/F-control hearts (D–G). EM revealed the loss of sarcomeres with loosely organized and randomly-oriented myofibers in the hearts of tamoxifen-treated MerCreMer/MyocdF/F mutants (M) (n = 3) compared to MyocdF/F-control mice (H) (n = 3). (N–P and R–T) Disruption of intercalated disc structure including loss of desmin (arrows) and Cx43-immunostaining was observed in tamoxifen-treated MerCreMer/MyocdF/F-mutant hearts (R–T) compared to MyocdF/F-control hearts (N–P). By contrast, comparable levels of N-Cadherin (N-Cad) are observed. (Q, U) EM revealed abnormal intercalated disc structure in tamoxifen-treated MerCreMer/MyocdF/F-mutant (U) compared to MyocdF/F-control hearts (Q). Original magnification, ×200 (B, C); ×800 (D–G, I–L, N, P, R, T); ×600 (O, S); ×40,000 (H, M, Q, U). (V) A graphic representation of cardiac gene expression as assessed by qRT-PCR 4 days after tamoxifen exposure in MyocdF/F-control (n = 3) and MerCreMer/MyocdF/F-mutant (n = 3) mice. The light blue bars (controls) and dark blue bars (mutants) show the relative level of GAPDH, myocardin (Myocd), α-MyHC (a-MyHC), α-cardiac actin (a-actin), MLC2v, tropomyosin (tropmyo), α-actinin, desmin, β-actin (b-actin), and Mrtf-b mRNA. Data are expressed as mean gene expression (arbitrary units) ± SEM.
Hearts of tamoxifen-treated MerCreMer/MyocdF/F-mutant mice were markedly enlarged with large pericardial effusions (Fig. 2A). Histological assessment of heart sections obtained from tamoxifen-treated MerCreMer/MyocdF/F-mutant mice and MyocdF/F-control littermates revealed cardiomyocyte disarray with myocyte loss and fibrosis in the mutant hearts (Fig. 2 B and C). Striking derangements in cardiomyocyte structure and myofibrillar organization were observed throughout the heart (compare Fig. 2 D–G and I–L). Myofibrillar striations corresponding to α-cardiac actin, MLC2v, tropomyosin, and α-actinin, were virtually absent in tamoxifen-treated hearts (see Fig. 2 I–L). A dramatic loss of sarcomeres was observed in the hearts of tamoxifen-treated MerCreMer/MyocdF/F-mutant mice, with sparse, randomly oriented myofibers observed throughout the sarcoplasm (Fig. 2 H and M). Once again, the structure of the intercalated disc was disrupted in tamoxifen-treated MerCreMer/MyocdF/F-mutant mice, with dramatic loss of desmin and Cx43 immunostaining (Fig. 2 N, O, R, and S). Cardiomyocytes of tamoxifen-treated MerCreMer/MyocdF/F-mutant mice exhibited indistinct desmosomes and adherens junctions (Fig. 2 Q and U). qRT-PCR performed with cardiac mRNA harvested 4 days following the initiation of tamoxifen exposure revealed that the expression of SRF/myocardin-regulated genes encoding myofibrillar proteins was profoundly attenuated in MerCreMer/MyocdF/F-mutants compared to control MyocdF/F mice. The expression of α-cardiac actin, α-myosin heavy chain, myosin light chain 2v, tropomyosin, α-actinin, and desmin genes were reduced by 80 to 90% in tamoxifen-treated MerCreMer/MyocdF/F hearts compared to controls (see Fig. 2V). By contrast, comparable levels of GAPDH, β-actin, and Mrtf-b mRNA were observed (see Fig. 2V). Taken together, these data demonstrate that myocardin is required for maintenance of cardiac contractile function and structural organization of the cardiomyocyte during postnatal development.
Induction of Programmed Cell Death.
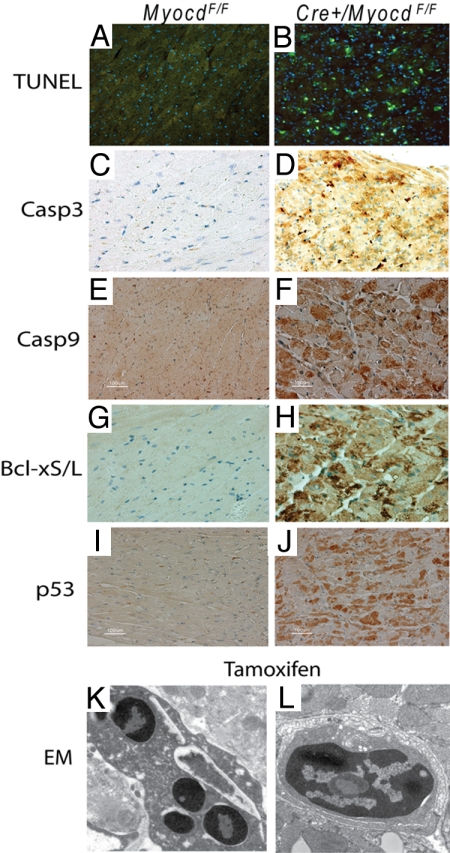
To determine whether the loss of cardiac myocytes observed in myocardin-deficient hearts resulted from cardiomyocyte apoptosis, TUNEL staining was performed on heart sections prepared from tamoxifen-treated MerCreMer/MyocdF/F-mutant and MyocdF/F-control mice. Remarkably, 12% ± 2% of LV cardiomyocytes stained TUNEL-positive (green stain) in MerCreMer/MyocdF/F mice compared to 0.1% ± 0.01% in control mice (Fig. 3 A and B). Consistent with these findings, abundant caspase 3, caspase 9, and Bcl-xS/L were observed in cardiomyocytes of tamoxifen-treated MerCreMer/MyocdF/F mice, demonstrating activation of the intrinsic/mitochondrial apoptotic pathway (Fig. 3 C–H). Moreover, p53, a major orchestrator of cellular response to stress that has been implicated in programmed cell death (15), was induced in the hearts of tamoxifen-treated MerCreMer/MyocdF/F mice (Fig. 3 I and J). EM confirmed widespread cardiomyocyte apoptosis manifest as nuclear chromatin aggregation, nuclear fragmentation, and cytoplasmic apoptotic body formation in the cardiomyocytes of tamoxifen-treated MerCreMer/MyocdF/F-mutant mice (Fig. 3 K and L).
Fig. 3.
Loss of myocardin triggers programmed cell death in the adult heart. Six days following the initiation of tamoxifen-treatment, hearts harvested from MyocdF/F-control (n = 6) and MerCreMer/MyocdF/F (Cre+/MyocdF/F)-mutant (n = 6) mice were fixed, sectioned, and immunostained. (A, B) This representative photomicrograph of the LV freewall of a tamoxifen-treated MerCreMer/MyocdF/F-mutant mouse (B) reveals widespread TUNEL-staining (green dots). By contrast, only rare TUNEL-positive myocytes are observed in this tamoxifen-treated MyocdF/F-control mouse (A). (C–J) Dramatic induction of activated caspase 3 (brown stain), caspase 9 (brown stain), and Bcl-xS/L (brown stain) is observed in tamoxifen-treated MerCreMer/MyocdF/F-mutant hearts compared to tamoxifen-treated MyocdF/F controls. (K, L) EM of tamoxifen-treated MerCreMer/MyocdF/F-mutant hearts reveals evidence of apoptosis, including nuclear chromatin aggregation, nuclear fragmentation, and cytoplasmic apoptotic body formation. Original magnification, ×400 (A–J); ×25,000 (K, L).
Identification of Cell-Autonomous Functions of the Myocd Gene.
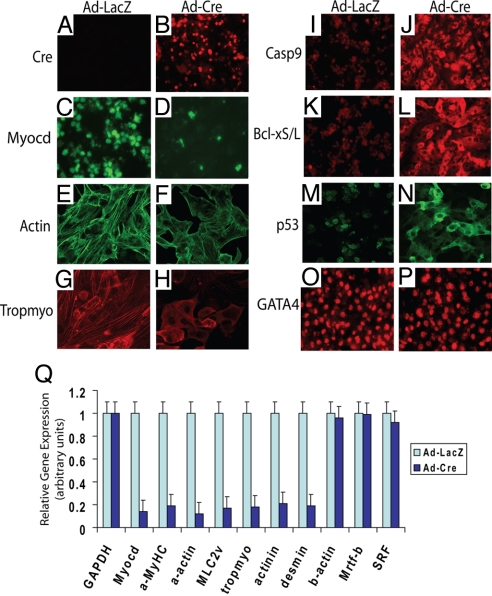
To determine whether myocardin acts in a cell-autonomous fashion to modulate cardiomyocyte structure and myofibrillar organization and to oppose programmed cell death, neonatal primary cardiomyocytes isolated from MyocdF/F-conditional mutant mice were transduced with the Ad-Cre replication-defective adenovirus (RdAV) encoding Cre recombinase, or as control Ad-LacZ encoding β-galactosidase. Seventy-two hours after transduction, greater than 90% of MyocdF/F cardiomyocytes were transduced with Ad-Cre (Fig. 4 A and B). In Ad-Cre transduced cultures, only rare myocardin-stained (green) nuclei were observed, consistent with a mean reduction of 86% in myocardin mRNA compared to Ad-LacZ-transduced cells (Fig. 4 C, D, and Q). Remarkably, within 72 h of Ad-Cre infection, the loosely arranged myofibrils observed in neonatal cardiomyocytes virtually disappeared in Ad-Cre-transduced MyocdF/F cardiomyocytes (Fig. 4 E–H). Moreover, widespread apoptosis was observed in Ad-Cre-transduced MyocdF/F myocytes, demonstrated by enhanced expression of cleaved caspase 9 and Bcl-xS/L (Fig. 4 I–L). Once again, p53 was dramatically up-regulated and translocated from the nucleus to the cytoplasm in Ad-Cre transduced cardiomyocytes (Fig. 4 M and N). As anticipated, GATA-4, a cardiac-restricted marker, was observed in Ad-LacZ and Ad-Cre-transduced cells (Fig. 4 O and P). qRT-PCR performed with mRNA harvested 72 h after AV infection revealed marked suppression of genes encoding myofibrillar proteins, including α-MyHC, α-cardiac actin, MLC2v, tropomyosin, α-actinin, and desmin (see Fig. 4Q). By contrast, comparable levels of GAPDH, SRF, and Mrtf-b mRNA were observed in Ad-Cre- and Ad-LacZ-transduced myocytes (see Fig. 4Q).
Fig. 4.
Cell-autonomous loss of myocardin in cardiomyocytes causes disruption of the contractile apparatus and apoptosis. (A–P) Replicate cultures of primary neonatal cardiomyocytes isolated from MyocdF/F mice were infected with Ad-LacZ (A, C, E, G, I, K, M, O) or Ad-Cre (B, D, F, H, J, L, N, P). Seventy-two hours after infection, cells were harvested and immunostained with antibodies that recognize Cre (A, B), myocardin (C, D), α-cardiac actin (E, F), tropomyosin (G, H), caspase 9 (I, J), Bcl-xS/L (K, L), p53 (M, N), and GATA-4 (O, P), respectively. (A–D) High-efficiency gene transduction was demonstrated by Cre-expression (red stain) accompanied by loss of myocardin immunostaining (green stain) in Ad-Cre transduced myocytes. (E–H) Confocal microscopy revealed loss myofibrils in Ad-Cre-transduced cells including loss of cardiac actin- (green stain, in E, F) and tropomyosin- (red stain in G, H). (I–N) Induction of apoptosis was observed in Ad-Cre transduced cells demonstrated by induction of caspase 9- (I, J) and Bcl-xS/L-immunostaining (K, L). p53 was induced in Ad-Cre-transduced myocytes and translocated from the nucleus to the cytoplasm (M, N). GATA-4 is observed in Ad-LacZ and Ad-Cre-transduced myocytes (O, P). (Q) Cardiomyocyte gene expression was quantified by qRT-PCR performed 72 h after transduction of MyocdF/F neonatal cardiomyocytes with Ad-Cre- (n = 3) (dark blue bars) or Ad-LacZ- (n = 3) (light blue bars). The relative levels of GAPDH, myocardin (Myocd), α-MyHC, α-cardiac actin, MLC2v, tropomyosin (tropmyo), α-actinin, desmin, β-actin, Mrtf-b, and SRF gene expression are expressed as mean gene expression (arbitrary units) ± SEM. Original magnification, ×400 (A–P).
Discussion
These data demonstrate that myocardin acts in a cell-autonomous fashion to reinforce and regulate structural organization of the cardiomyocyte and to oppose programmed cell death. These findings reveal a conserved function for myocardin in the cardiac and SMC lineages, where it acts as a transcriptional coactivator integrating and transducing biomechanical and intracellular signals regulating the contractile unit and adaptive response of the cell to stress (4–7). In the heart, myocardin binds directly to SRF, activating a subset of cardiomyocyte-restricted genes encoding myofibrillar, cytoskeletal, and structural proteins (2, 3). These transcriptional targets ultimately establish cardiomyocyte cell structure and organization underlying the unique physiological properties of the cardiomyocyte cell lineage. The profound derangements in sarcomeric organization and intercalated disc structure, accompanied by repression of SRF-dependent genes encoding structural proteins in myocardin-deficient hearts and isolated cardiomyocytes, provides direct evidence that in the adult heart myocardin is required for maintenance of cardiomyocyte structure and contractile function. Similarly, the unique contractile properties of the SMC lineage are directed by SRF/myocardin-regulated genes encoding SMC-restricted contractile proteins (4–7). Ablation of the Myocd gene in vascular SMCs causes markedly attenuated expression of SMC contractile protein isoforms accompanied by a dramatic change in phenotype from a spindle-shaped muscle cell to a cell resembling a fibroblast (6).
The reparative capacity of the adult heart is limited by the regenerative potential of cardiomyocytes, which respond to stress by undergoing hypertrophic growth or apoptosis (16). Hypertrophic adaptation of the heart is accompanied by induction of multiple SRF/myocardin-regulated genes associated with the fetal gene program, including SM22α and α-skeletal actin (17). Not surprisingly, induction of genes associated with the fetal/hypertrophic program was not observed in hearts harvested from 3- or 10-month-old αMyHC-Cre/MyocdF/F-mutant mice (see Fig. 2V). The demonstration that loss of myocardin signaling in the cardiomyocyte triggers apoptosis suggests strongly that maintenance of cardiomyocyte structural organization and contractile function is intimately linked to cardiomyocyte cell survival. In support of this hypothesis, α-cardiac actin-null mutant mice exhibit sarcomeric derangements and increased rates of cardiomyocyte apoptosis during embryonic development (18). Of note, this myocardin-dependent cardiomyocyte survival pathway differs fundamentally from the previously described cardiotrophin 1 (CT-1)/gp130 cardiomyocyte, which is only activated in response to pathophysiological cardiomyocyte cell stress (19, 20).
Characterization of these myocardin-conditional mutant mice provides unanticipated insights into the pathogenesis of dilated cardiomyopathy, a relatively common cause of heart failure associated with a poor prognosis (21). Causal mutations in multiple myocardin-regulated genes encoding cardiac-restricted cytoskeletal and myofibrillar structural proteins, including ACTN2, MYH7, ACTC, TPM1, DES, and DMD, have been reported in patients with dilated cardiomyopathy (22–27). The anatomic and ultrastructural changes in hearts of MyHC-Cre/MyocdF/F- and tamoxifen-treated MerCreMer/MyocdF/F-mutant mice resemble hearts of patients diagnosed with genetic and acquired forms of dilated cardiomyopathy (28). In this regard, it is noteworthy that cleavage of SRF and Rho-associated coiled-coil protein kinase (ROCK-1) by caspase-3 may play a role in the pathogenesis of human heart failure (29, 30). However, we did not observe an increase in cleaved SRF or ROCK-1 protein in protein lysates prepared from tamoxifen-treated MerCreMer/MyocdF/F-mutant hearts, demonstrating that the heart failure observed in myocardin conditional mutant mice is not dependent upon caspase-3-mediated cleavage of SRF or ROCK-1 (Fig. S2). In any case, analyses of myocardin mutant mice suggests strongly that progressive deterioration in cardiac function observed in patients with dilated cardiomyopathy, particularly those cases associated with mutations in myocardin/SRF-activated contractile genes, may result, at least in part, from the cumulative loss of cardiomyocyte mass via programmed cell death. Future studies of mice harboring cardiomyocyte-restricted null mutations of the myocardin gene should provide new insights into the pathogenesis of dilated and arrhythmogenic cardiomyopathy and identify novel therapeutic targets for these debilitating diseases.
Methods
Generation of Mice Containing a Cardiac-Restricted Null Mutation in the Myocd Gene.
Mice containing a conditional null mutation in the Myocd gene (MyocdF/F) were described previously (6). MyHC-Cre mice were provided by M. Schneider (13). MerCreMer mice (cat. #005657) were obtained from Jackson Labs (14). To ablate the Myocd gene in the postnatal heart, 4-month-old MerCreMer/MyocdF/F mice received 65 mg/kg i.p. injection of tamoxifen daily for 4 days. Mice were killed 2, 4, or 6 days following initiation of tamoxifen treatment. Genotyping was performed by Southern blot analysis and PCR as described (6). All animal experimentation was performed under protocols approved by the University of Pennsylvania IACUC and in accordance with National Institutes of Health guidelines.
Cardiac Morphometry and Echocardiography.
Cardiac morphometric and echocardiographic measurements were performed as described previously (31). Echocardiography analyses were performed on a Vevo 770 VisualSonic scanner equipped with a 30-MHz probe, as described previously (31).
Histology, Immunohistochemistry, and Electron Microscopy.
A complete description of the histological methods, including a list of the specific antibodies used, is provided in the SI Text. Histology, immunohistochemistry, and electron microscopy was performed as described previously (6).
Real Time RT-PCR and Western Blot Analysis.
RNA was isolated and quantitative real time RT-PCR was performed using the DNA Engine Opticon 2 Real Time Detection System (Applied Biosystems, Inc.) as described previously (32). Western blot analyses were performed as described previously (6). Antibodies included rabbit polyclonal anti-Cx43 (Invitrogen 71–0700), monoclonal anti-myocardin (R&D Systems, clone 355521), and rabbit polyclonal anti-β-tubulin (Abcam, ab6046). The hybridization signal was quantified using Image Quant 5.0 software, as described by the manufacturer (Molecular Dynamics Inc.).
RdAV Transduction of Neonatal Cardiomyocytes and Quantification of Apoptosis.
Primary cultures of mouse neonatal cardiomyocytes were isolated from 1- or 2-day-old mice harboring an MyocdF/F allele, as described (33). Replicate cultures of cardiomyocytes were grown in low serum medium for 24 h and infected with (10 moi) Ad-LacZ or Ad-Cre (University of Pennsylvania Adenoviral Vector Core). Seventy-two hours after transduction, triplicate cultures were harvested and qRT-PCR analysis or immunostaining was performed as described previously (6). Experiments were repeated three times to ensure reproducibility.
Statistical Considerations.
Comparison of survival rates was performed by Kaplan-Meier analysis with PRISM software (GraphPad). All measurement data are expressed as mean ± SEM. The statistical significance of differences between groups was determined by Student's t test. Differences were considered significant at a P value <0.05.
Supplementary Material
Acknowledgments.
We thank Clara Franzini-Armstrong for advice and review of electron micrographs, and Jon Epstein, Mark Kahn, and Ed Morrisey for their advice and helpful comments. This work was supported in part by National Institutes of Health Grants P01-HL075215 and R01-HL094520 and the Commonwealth of Pennsylvania.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910749106/DCSupplemental.
References
- 1.Wang D, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 2.Parmacek MS. Myocardin-related transcription factors: Critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 3.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida T, Kawai-Kowase K, Owens GK. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential embryonic cells. Arterioscler Thromb Vasc Biol. 2004;24:1596–1601. doi: 10.1161/01.ATV.0000137190.63214.c5. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, et al. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J Clin Invest. 2008;118:515–525. doi: 10.1172/JCI33304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long X, Bell RD, Gerthoffer WT, Zlokovic BV, Miano JM. Myocardin is sufficient for a smooth muscle-like contractile phenotype. Arterioscler Thromb Vasc Biol. 2008;28:1505–1510. doi: 10.1161/ATVBAHA.108.166066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci USA. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Small EM, et al. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132:987–997. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- 10.Pipes GC, et al. Stem cells and their derivatives can bypass the requirement of myocardin for smooth muscle gene expression. Dev Biol. 2005;288:502–513. doi: 10.1016/j.ydbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Du KL, et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xing W, et al. Myocardin induces cardiomyocyte hypertrophy. Circ Res. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- 13.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 15.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 16.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 17.Olson EN, Schneider MD. Sizing up the heart: Development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 18.Kumar A, et al. Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc Natl Acad Sci USA. 1997;94:4406–4411. doi: 10.1073/pnas.94.9.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng Z, Pennica D, Wood WI, Chien KR. Cardiotrophin-1 displays early expression in the murine heart tube and promotes cardiac myocyte survival. Development. 1996;122:419–428. doi: 10.1242/dev.122.2.419. [DOI] [PubMed] [Google Scholar]
- 20.Hausenloy DJ, Yellon DM. New directions for protecting the heart against ischaemia-reperfusion injury: Targeting the Reperfusion Injury Salvage Kinase (RISK)-pathway. Cardiovasc Res. 2004;61:448–460. doi: 10.1016/j.cardiores.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 21.Jessup M, Brozena S. Heart failure. N Engl J Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 22.Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farza H, et al. Genomic organisation, alternative splicing and polymorphisms of the human cardiac troponin T gene. J Mol Cell Cardiol. 1998;30:1247–1253. doi: 10.1006/jmcc.1998.0698. [DOI] [PubMed] [Google Scholar]
- 24.Galvagni F, Lestingi M, Cartocci E, Oliviero S. Serum response factor and protein-mediated DNA bending contribute to transcription of the dystrophin muscle-specific promoter. Mol Cell Biol. 1997;17:1731–1743. doi: 10.1128/mcb.17.3.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mericskay M, et al. An overlapping CArG/octamer element is required for regulation of desmin gene transcription in arterial smooth muscle cells. Dev Biol. 2000;226:192–208. doi: 10.1006/dbio.2000.9865. [DOI] [PubMed] [Google Scholar]
- 26.Nelson TJ, Balza R, Jr, Xiao Q, Misra RP. SRF-dependent gene expression in isolated cardiomyocytes: Regulation of genes involved in cardiac hypertrophy. J Mol Cell Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Pasquet S, et al. Transcription enhancer factor-1-dependent expression of the alpha-tropomyosin gene in the three muscle cell types. J Biol Chem. 2006;281:34406–34420. doi: 10.1074/jbc.M602282200. [DOI] [PubMed] [Google Scholar]
- 28.Franz WM, Muller OJ, Katus HA. Cardiomyopathies: From genetics to the prospect of treatment. Lancet. 2001;358:1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI] [PubMed] [Google Scholar]
- 29.Chang J, et al. Inhibitory cardiac transcription factor, SRF-N, is generated by caspase 3 cleavage in human heart failure and attenuated by ventricular unloading. Circulation. 2003;108:407–413. doi: 10.1161/01.CIR.0000084502.02147.83. [DOI] [PubMed] [Google Scholar]
- 30.Chang J, et al. Activation of Rho-associated coiled-coil protein kinase 1 (ROCK-1) by caspase-3 cleavage plays an essential role in cardiac myocyte apoptosis. Proc Natl Acad Sci USA. 2006;103:14495–14500. doi: 10.1073/pnas.0601911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 32.Du KL, et al. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem. 2004;279:17578–17586. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 33.Deng XF, Rokosh DG, Simpson PC. Autonomous and growth factor-induced hypertrophy in cultured neonatal mouse cardiac myocytes. Comparison with rat. Circ Res. 2000;87:781–788. doi: 10.1161/01.res.87.9.781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.