Abstract
AAA+ proteases are ATP-fueled machines that bind protein substrates via a degradation tag, unfold the molecule if necessary, and then translocate the polypeptide into a chamber for proteolysis. Tag recognition is normally viewed as a passive reaction. By contrast, for the AAA+ Lon protease, we show that degron tags are also regulatory elements that determine protease activity levels. Indeed, different tags fused to the same protein change degradation speeds and energetic efficiencies by 10-fold or more. Degron binding to multiple sites in the Lon hexamer appears to differentially stabilize specific enzyme conformations, including one with high protease and low ATPase activity, and results in positively cooperative degradation. These allosteric mechanisms allow Lon to operate in either a fast or slow proteolysis mode, according to specific physiological needs, and may help maximize degradation of misfolded proteins following stress-induced denaturation.
Keywords: AAA+ protease, allosteric control, degradation tags
Intracellular degradation frees the cell of unfolded potentially toxic proteins and regulates processes such as DNA replication, gene expression, and differentiation (1). Because degradation is irreversible, these reactions are carefully regulated and generally executed by ATP-dependent AAA+ proteases. Proteins are safe from these proteases, unless they bear specific recognition elements, known as degrons or degradation tags (2). All AAA+ proteases adopt multisubunit structures with an internal proteolytic chamber, accessible through narrow channels that exclude native proteins (3). This mechanism protects most cytoplasmic proteins from degradation and requires that specific substrates be recognized by the AAA+ protease, unfolded if necessary, and then translocated into the chamber for degradation.
Lon degrades damaged proteins in bacteria and eukaryotic organelles (4–6). For example, Lon degrades approximately 50% of the misfolded proteome in Escherichia coli (6). Functional Lon is a homohexamer. Each subunit consists of an N domain, an AAA+ ATPase domain that provides mechanical power, and a peptidase domain (7–8). Lon's ability to distinguish misfolded variants from correctly folded proteins depends on recognition of degradation tags rich in aromatic and hydrophobic residues, which tend to be hidden in the core of native proteins but become accessible upon denaturation (9). Lon also possesses an unfoldase activity, and some substrates are native proteins with accessible degrons (9–13).
Prior studies have shown that substrates and other ligands interact with Lon allosterically (7, 14–15). However, little is known about the importance of these mechanisms in controlling protein degradation. Here, we show that degradation tags play unanticipated and highly active roles in programming the operation of the Lon machine. Swapping one degradation tag for another can substantially change the speed and energetic efficiency at which Lon degrades otherwise identical proteins. We present evidence that degron binding controls Lon proteolysis by altering the equilibrium distribution of distinct enzyme conformations with very different proteolytic activities. This mechanism provides opportunities for matching Lon proteolysis to specific physiological needs. During normal growth, for example, a slow degradation mode might allow denatured proteins multiple chances to refold before destruction by Lon becomes likely. When stress conditions result in massive protein unfolding, by contrast, Lon degradation could proceed at the fastest speed possible in an attempt to avert catastrophic aggregation.
Results
Tags Determine Maximal Rates of Lon Degradation.
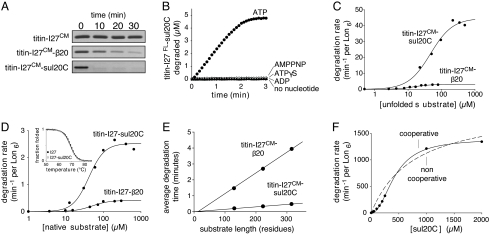
Both a 20-residue β-galactosidase sequence (β20) and a sequence at the C terminus of the SulA protein function as degradation tags for E. coli Lon (9, 16–17). To compare the properties of these tags in directing Lon degradation, we appended either β20 or the C-terminal 20 residues of SulA (sul20C) to the C terminus of the I27 domain of human titin. Native titin-I27 can be unfolded by carboxymethylation of its cysteines (titin-I27CM), simplifying analysis of proteolysis reactions by eliminating the need for unfolding (18). Lon degraded sul20C-tagged titin-I27CM rapidly, degraded b20-tagged titin-I27CM more slowly, and did not detectably degrade titin-I27CM without an added tag (Fig. 1A). As shown previously (9), ATP hydrolysis was required for degradation of unfolded substrates (Fig. 1B).
Fig. 1.
Lon degradation of denatured and native proteins. (A) SDS/PAGE assays of Lon6 (0.15 μM) degradation of untagged and tagged variants of unfolded titin-I27CM (5 μM). (B) Lon6 (0.3 μM) cleavage of sul20C-tagged titin-I27 (5 μM), unfolded by fluorescein modification of its cysteines, was assayed in the presence of no nucleotide or 2 mM ATP, ATPγS, AMPNP, or ADP. (C) Steady-state rates of Lon6 (0.1 μM) degradation of different concentrations of sul20C-tagged or β20-tagged 35S-titin-I27CM. Lines are fits to the Hill equation (rate = Vmax·[S]n/(KMn + [S]n). (D) Steady-state rates of Lon6 (0.3 μM) degradation of native 35S-titin-I27-sul20C or 35S-titin-I27-β20 were fit to the Hill equation. Inset, Titin-I27 (closed symbols) and titin-I27-sul20C (open symbols) had the same stability to thermal unfolding monitored by changes in circular dichroism. (E) Average degradation times normalized to Lon6 (0.3 μM) are plotted as a function of substrate length for β20-tagged (20 μM) or sul20C-tagged (50 μM) substrates containing 1–3 copies of titin-I27CM. Values are averages (n = 3), with standard deviations smaller than the plot symbol. (F) Degradation of sul20C peptide by Lon6 (0.3 μM) shows strong positive cooperativity. The solid and dashed lines are fits to the Hill equation (n = 2.2) and a noncooperative equation, respectively.
To determine steady-state kinetic parameters, we used 35S-labeled variants of unfolded titin-I27CM proteins with the β20 or sul20C tags and determined rates of Lon degradation at different substrate concentrations (Fig. 1C). At substrate saturation, Lon degraded sul20C-tagged titin-I27CM approximately 9-fold faster than the β20-tagged variant (Fig. 1C and Table 1). Because the only difference between these substrates was the tag, its identity must determine the maximal rate at which the attached protein is degraded. This result was surprising, as Vmax for degradation of very similar unfolded proteins might be expected to be a function of the enzyme and not the substrate.
Table 1.
Degradation parameters
| Substrate | Vmax (min−1·Lon6−1) | KM (μM) | Hill(deg.) | ATPasemax (min−1·Lon6−1) | Efficiency (substrate/100 ATP) |
|---|---|---|---|---|---|
| Unfolded (130 residues) | |||||
| Titin-I27CM-sul20C | 48 ± 3 | 53 ± 11 | 1.3 ± 0.3 | 129 ± 15 | 37 ± 5 |
| Titin-I27CM-sul20C YA | 22 ± 0.6 | 50 ± 3 | 2.2 ± 0.2 | 656 ± 26 | 3.4 ± 0.2 |
| Titin-I27CM-sul20C HA | 15 ± 0.6 | 64 ± 7 | 1.7 ± 0.2 | 594 ± 61 | 2.5 ± 0.3 |
| Titin-I27CM-β20 | 5.3 ± 0.3 | 24 ± 2 | 2.1 ± 0.4 | 509 ± 58 | 1.1 ± 0.4 |
| Native (130 residues) | |||||
| Titin-I27-sul20C | 2.5 ± 0.1 | 40 ± 4 | 2.0 ± 0.4 | ND | ND |
| Titin-I27-β20 | 0.4 ± 0.02 | 50 ± 6 | 1.9 ± 0.4 | ND | ND |
| Unfolded (23 residues) | |||||
| F-β20-Q | 10 ± 0.26* | 4.6 ± 0.2* | 1.4 ± 0.1* | ND | ND |
| F-sul20C-Q | 1396 ± 20 | 431 ± 15 | 2.2 ± 0.1 | ND | ND |
*Values taken from ref. 9. ND, not determined.
Lon degraded β20- and sul20C-tagged variants of native titin-I27 with Vmax values that differed by approximately 6-fold (Fig. 1D and Table 1). Importantly, Lon degraded each native variant substantially slower than the corresponding denatured variant, establishing that enzymatic unfolding is rate limiting for degradation of both native substrates. Thermal denaturation monitored by circular dichroism revealed identical melting curves for titin-I27 and titin-I27-sul20C (Fig. 1D Inset), as previously shown for titin-I27 and titin-I27-β20 (9). Thus, tag effects on the stability of these substrates cannot explain the differences in Lon degradation.
Table 1 lists kinetic parameters for Lon degradation of peptides corresponding to the β20 and sul20C sequences. Vmax for the sul20C peptide was much faster than for the β20 peptide, even though both substrates were the same length and degradation of both peptides required ATP hydrolysis. Hence, the ability of different tags to control rates of Lon degradation appears to be an inherent property of the tag sequence itself. KM for Lon degradation of the sul20C peptide was approximately 10-fold higher than for sul20C-tagged unfolded titin or native titin (Table 1). This observation could be explained by the high Vmax for the peptide substrate (KM = KD + Vmax/kassn) or if there were a few additional stabilizing contacts between the unfolded or native titin substrates and Lon.
The experiments above demonstrate that the sul20C tag directs faster Lon degradation of unfolded and folded protein substrates compared to the β20 tag. The tag-dependent differences in Vmax might arise if, after initial binding, Lon's translocation machinery grasped or engaged the sul20C tag sequence in one or a few attempts but required many more attempts to bind the β20 sequence, causing a lag in engagement and degradation. We tested for an initial lag by determining proteolysis rates for β20- or sul20C-tagged substrates containing one to three repeats of titin-I27CM (19). The reciprocals of these rates, which correspond to “average” degradation times, were proportional to substrate length (Fig. 1E), with slopes differing by approximately 9-fold. Importantly, if tag engagement occurred slowly for these substrates, then the y-intercept of the Fig. 1E plots would have a substantial positive value. By contrast, this intercept was negligible for both classes of substrates. Thus, these experiments are inconsistent with models that posit a slow, length-independent step in degradation, such as tag engagement.
The results presented so far, show that different Lon degrons determine the maximum speed at which a substrate can be unfolded, translocated, and eventually degraded. In other words, these degrons appear to program changes in the activity of the Lon machine.
Positive Cooperativity in Lon Degradation.
Substrate interactions with Lon were positively cooperative, with Hill constants (n) ranging from 1.3–2.2 (Table 1). For example, the concentration dependence of sul20C-peptide degradation (Fig. 1F) was fit well by the Hill equation (n = 2.2) but poorly by a noncooperative model. These results suggest that a minimum of two substrates interact with Lon during degradation. It is possible; therefore, that one bound substrate influences the rate at which another substrate is degraded. None of our substrates showed evidence of solution multimerization, making it unlikely that substrate–substrate interactions stabilize the binding of dimers or higher multimers to Lon. Because Lon is a homohexamer, positive cooperativity could occur by an allosteric mechanism in which the binding of one substrate stabilized an enzyme conformation with higher affinity for additional substrates (20).
Evidence for Allosteric Regulation of Lon.
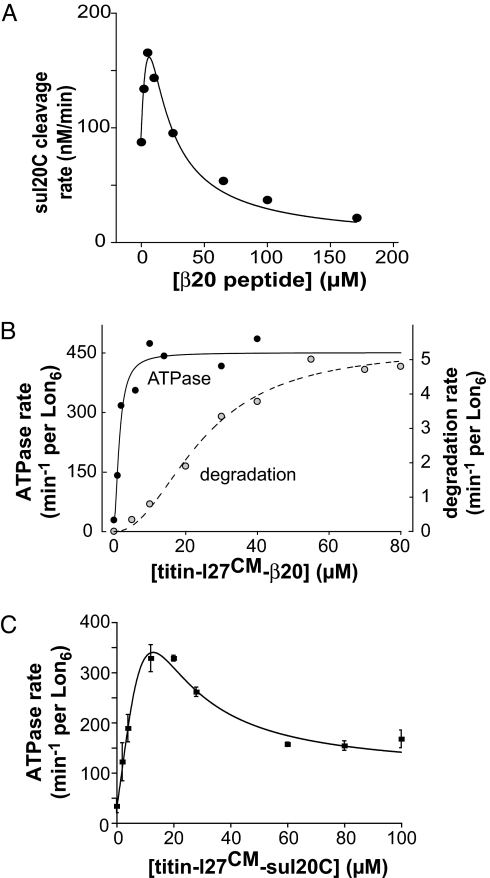
Low concentrations of the β20 peptide stimulated ATP-dependent degradation of the sul20C peptide, whereas higher concentrations inhibited cleavage (Fig. 2A). This result is reminiscent of classical allosteric enzymes, in which low concentrations of “competitive” inhibitors shift an equilibrium toward a higher-affinity conformation with free active sites, allowing more substrate to bind (21–22). At sufficiently high concentrations, such inhibitors fill most sites and prevent substrate binding. Previous studies also showed that protein substrates activate ATP-independent Lon cleavage of very small peptide mimics (14, 7).
Fig. 2.
Allosteric regulation. (A) Degradation of a fluorescent sul20C peptide (1 μM) by Lon6 (0.15 μM) assayed in the presence of increasing β20 peptide. The line is a fit to the equation rate = C*α*(1 + α + β)/(L + (1 + α + β)2), where C is a scaling factor, α = [sul20C]/KM = 1/430, β = [β20]/K0.5, and L is a conformational equilibrium constant (31). The fitted values were C = 3000 μM/min per Lon6, K0.5 = 2.9 μM, and L = 10.5. (B) Steady-state rates of ATP-hydrolysis and protein degradation by Lon6 (0.15 μM) were measured under identical conditions as a function of increasing titin-I27CM-β20. ATPase data were fit to the equation rate = basal + (Vmax-basal)·[S]/(K0.5 + [S]). Degradation data were fit to the Hill equation. (C) Steady-state rates of ATP hydrolysis by Lon6 (0.15 μM) were measured as a function of increasing titin-I27CM-sul20C. The line is a fit to an equation derived from the Fig. 4A model.
Evidence for two conformations of Lon, whose populations change as a function of substrate binding, was obtained by assaying degradation and ATP hydrolysis under identical conditions as a function of titin-I27CM-β20 concentration (Fig. 2B). Strikingly, substrate binding affected these activities in a noncoincident fashion, with 50% of maximal ATPase activity at 1.5 μM titin-I27CM-β20 and 50% of maximal degradation at 25 μM titin-I27CM-β20. Another Lon substrate, casein, was also shown to activate ATP hydrolysis at lower concentrations than it activated its own degradation (14). These results support the existence of multiple Lon conformations and classes of substrate-binding sites. The degradation curve in Fig. 2B showed strong positive cooperativity (n = 2.1), even though ATPase stimulation was nearly complete before substantial degradation occurred. This observation suggests that substrates must bind to at least two additional sites for full proteolytic activation of Lon.
Taken together, the effects of different degradation tags on maximal rates of Lon proteolysis, the positive cooperativity of degradation, the stimulation of proteolysis of one substrate by a substrate bearing a different degradation tag, and the offset between β20-mediated stimulation of Lon's ATPase and degradation activities provide strong support for a model in which multiple substrates bind simultaneously to a single Lon enzyme, allowing binding to alter the equilibrium distribution of enzyme conformations with different functional properties.
Biphasic Tag Effects on ATP Hydrolysis.
Because protein degradation by Lon requires ATP hydrolysis, it seemed likely that sul20C-saturated Lon, which has a higher protease activity than β20-saturated Lon, would also have a higher ATPase activity. Surprisingly, the opposite result was observed; Lon hydrolyzed ATP substantially more slowly with saturating sul20C substrate (Fig. 2C) than with β20 substrate (Fig. 2B). Moreover, in the sul20C titration, the ATPase activity increased initially and then decreased to a constant value at high substrate concentrations (Fig. 2C). Degradation of the sul20C-tagged substrate appeared to be roughly coincident with the second ATPase phase (cf. Figs. 1C and 2C).
Sul20C-Tag Mutations Alter Catalytic Activities.
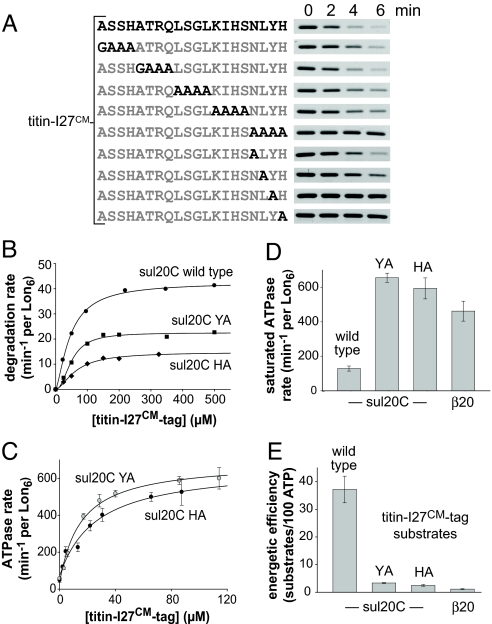
Mutagenesis revealed that alanine substitutions at the C-terminal or penultimate residues of the sul20C tag caused substantial decreases in Lon degradation (Fig. 3A). We constructed, purified, and titrated titin-I27CM-sul20C substrates containing these mutant tags against Lon and assayed steady-state rates of degradation and ATP hydrolysis (Fig. 3 B and C). Both tag mutations reduced Vmax for degradation and increased Vmax for ATP hydrolysis compared to the parental substrate (Table 1). Moreover, in contrast to the wild-type sul20C-tagged substrate, titrations with substrates bearing the mutant sul20C tags resulted in monophasic increases in ATP hydrolysis (Fig. 3 B and C).
Fig. 3.
Mutant sul20C tags alter Lon activity. (A) SDS/PAGE assays of Lon6 (0.15 μM) degradation of titin-I27CM (5 μM) bearing mutant sul20C tags. (B) Steady-state rates of Lon6 (0.3 μM) degradation of different concentrations of 35S-titin-I27CM bearing wild-type, YàA (penultimate residue), or HàA (C-terminal residue) sul20C tags were fit to the Hill equation. (C) ATP hydrolysis by Lon6 (0.15 μM) as a function of [titin-I27CM-sul20C] bearing the penultimate YàA mutation or C-terminal HàA sul20C mutations. Lines are fits to the equation rate = basal + (Vmax-basal)·[S]/(K0.5 + [S]). (D) Rates of Lon ATP hydrolysis at saturating concentrations of titin-I27CM with different degradation tags. Values are averages (n = 3) ± 1 SD. (E) The energetic efficiency of Lon degradation varies dramatically for titin-I27CM variants with different degrons. Error bars are 1 SD.
Tags Cause Dramatic Variations in Energy Efficiency.
Because saturating concentrations of substrates with different tags result in markedly different ATP-hydrolysis and degradation rates (Fig. 3D and Table 1), the energetic efficiency of Lon proteolysis varied markedly. Indeed, when we calculated the average number of substrates that Lon degrades under Vmax conditions, hydrolysis of 100 ATPs hydrolyzed fueled degradation of approximately 35 molecules of titin-I27CM-sul20C but only about one molecule of titin-I27CM-β20 (Fig. 3E and Table 1).
Discussion
It is well established that degradation tags determine the affinity of substrates for bacterial AAA+ proteases, allowing tight binders to be degraded with higher priority than weak binders (2). Here, we have shown that the degrons of Lon substrates also regulate the proteolysis reaction itself. Indeed, different tags attached to the same protein substrate can cause substantial changes in the rates of substrate unfolding, translocation, and degradation, and can dramatically alter the energetic efficiency of the proteolysis reaction. Thus, the degrons of Lon substrates program or determine the operating speed and fuel economy of this AAA+ molecular machine. Our studies support an allosteric model for substrate control of the activities of the Lon proteolytic machine and also illuminate Lon's specialized role in degrading misfolded proteins.
A Working Model for Degron Control of Lon Activity.
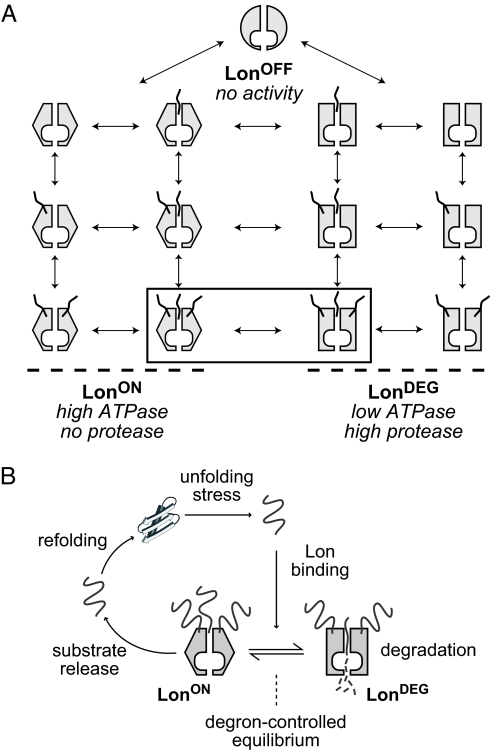
To account for the observed effects of sul20C-tagged substrates and β20-tagged substrates on the ATPase and protease activities of Lon, we propose a model based on MWC allostery (20), that includes three equilibrium enzyme conformations with distinct functional properties (Fig. 4A). In our model, LonOFF has no activity and does not bind substrates (or binds very weakly), LonON has high ATPase but no protease activity, and LonDEG is an active protease with moderately low ATPase activity. By analogy with an automobile, the engine of LonOFF is off, the engine of LonON is revving in neutral, and the engine of LonDEG has engaged the gears of the drive train to power mechanical work.
Fig. 4.
Models for allosteric control of Lon. (A) Model showing equilibrium states of a system with three Lon conformations: LonOFF, LonON, and LonDEG (depicted by different shapes). Degrons are shown as squiggly lines. One degron-binding site in the axial pore of Lon and two flanking allosteric binding sites are depicted. At substrate saturation, the relative affinity of the degron for LonON versus LonDEG determines the equilibrium distribution of the two fully liganded species shown in the rectangle. (B) Model in which an unfolded protein binds Lon and then partitions between a degradation pathway mediated by LonDEG or a refolding pathway mediated by LonON and possibly other cellular chaperones. The substrate degron determines the equilibrium distribution of the two forms of Lon and thus determines which pathway is favored.
Importantly, the relative populations of the three Lon species change in a dynamic fashion that depends both on the protein-substrate concentration and the identity of its degron. LonOFF predominates in the absence of substrate, explaining the low basal rate of ATP-hydrolysis. Because LonOFF binds substrates poorly, saturating concentrations of any substrate will drive most enzymes into a mixture of the fully bound LonON and LonDEG species (shown in the rectangle in Fig. 4A). Under these “Vmax” conditions, the relative affinities of a specific degron for LonON versus LonDEG will determine the equilibrium distribution of these species, and thus will determine the average degradation rate of the attached substrate and the average ATP-hydrolysis rate. For example, our finding that Vmax for β20-tagged unfolded titin is approximately 9-fold slower than for the sul20C-tagged variant is explained if the substrate-saturated population consists of approximately 90% LonON and about 10% LonDEG for the β20 degron, with these proportions reversed for the sul20C degron. Similarly, because the intrinsic ATPase activity is substantially higher for LonON than LonDEG, saturation of the β20 substrate results in faster ATP hydrolysis than saturation of the sul20C substrate. Finally, our finding that the degradation rate remains proportional to substrate length for unfolded proteins with the β20 and sul20C tags follows from the fact that each degron stabilizes a constant but different fraction of enzymes in the proteolytically active LonDEG conformation.
Our model posits two types of substrate-binding sites. One site is located in the Lon translocation pore (Fig. 4A), as binding in this channel is a prerequisite for degradation and certain degrons bind in the axial pores of other AAA+ proteases (23–24). Two additional allosteric sites are shown at positions flanking the pore (Fig. 4A), although the exact number (there might be six in a Lon hexamer) and/or locations of these sites are not crucial aspects of the model. An important feature is that any specific degron (e.g., sul20C or β20) can bind both to the pore site and to the allosteric sites. Without this provision, it would not be possible to account for the positive cooperativity of substrate degradation, to explain noncoincident ATPase and degradation activities, or to account for the biphasic ATPase curve observed for the sul20C-tagged substrate. Although the precise features of our working model remain to be tested, it accounts qualitatively for the experimental data presented here and previously. Moreover, equations derived from the model provide good fits for specific experiments, as shown in Fig. 3C.
There is evidence for substrate-driven changes in Lon conformation (25), and the positive cooperativity of substrate binding arises naturally from our model, accounting for results presented here and previously (9, 13, 26). This model also accounts for the stimulation of Lon degradation of one substrate by another substrate and for previous reports that proteins substrates stimulate Lon peptidase activity (7, 14).
Biological Implications for Regulation of Degradation.
Although the allosteric model explains a wide range of biochemical results, it has one curious feature. Why would some substrates stabilize LonON, which runs at high speed in terms of ATP turnover but apparently performs little or no useful work? The answer is not known, but in view of the robust interaction of Lon with misfolded proteins, we suggest that LonON performs another function, for example serving as a disassembly chaperone to facilitate refolding (Fig. 4B). Indeed, Lon has been proposed to have chaperone activity (27), and recent studies show that Lon binds to the α-crystallin domains of small heat-shock proteins that function as molecular chaperones (S. Bissonnette, T.A. Baker, personal communication).
Our finding that degrons control both Lon affinity and maximal degradation rates suggests different strategies of proteolytic regulation depending upon the cellular abundance of a given substrate and the extent to which Lon is saturated with substrates. At substrate concentrations substantially below KM, the degradation rate is determined by the second-order rate constant (Vmax/KM) and changes in either KM and/or Vmax could be used to tune proteolysis to the desired rate. By contrast, when substrates are highly abundant, for example during heat shock, Lon is likely to be saturated. Under these conditions, changes in KM would have little effect and only changes in Vmax would allow alterations in the degradation rate. Evolution has presumably crafted Lon degrons to regulate both affinity and maximal degradation rates, depending on substrate abundance and the need for rapid degradation during normal growth, as well as under stress conditions that result in protein unfolding. For example, SulA is a regulatory protein that needs to be degraded to allow resumption of cell growth after the SOS response to DNA damage (1), and it makes sense that its degron leads to highly efficient Lon degradation.
Lon is the major protease responsible for degradation of misfolded proteins in bacteria and eukaryotic organelles (4–6, 28). Under nonstress conditions, most proteins are natively folded and unfolded or misfolded proteins are present at very low concentrations. These non-native proteins need to be given a chance to fold, either spontaneously or by chaperone-mediated reactions, and then to be degraded if folding fails. The β20 sequence in unfolded β-galactosidase acts in concert with other sequences to provide a very high affinity (<100 nM) for Lon (9). However, our results show clearly that even saturating concentrations of the β20 degron keep Lon in a relatively inactive conformation vis-à-vis degradation. In the cell, this unusual property might limit the possibility of proteolysis and increase the chances of successful β-galactosidase refolding.
Heat shock and other types of environmental stress can cause catastrophic protein unfolding. Under these circumstances, unfolded or misfolded substrates need to be degraded as rapidly as possible to avoid the formation of toxic degradation-resistant aggregates. In this regard, Lon degradation that increases with the second-power of the substrate concentration makes complete biological sense. It is also possible that small-molecule or macromolecular allosteric effectors, which stabilize LonDEG to facilitate very fast protein degradation, are synthesized during heat shock or other global stress responses.
The important result of our studies is that substrates are not passive passengers, but play active roles in controlling the operation of the Lon machine. It will be important to determine if allosteric programming by substrates is a characteristic of other AAA+ proteases. Irrespective of the answer, this method of substrate control greatly expands potential strategies for regulated degradation by AAA+ proteolytic machines.
Materials and Methods
Protein and Peptide Purification and Modification.
Purification of E. coli Lon, purification of His6-tagged variants of human titin-I27, purification of 35S-labeled titin-I27 substrates, and cysteine-modification reactions were performed as described (9). Synthetic peptides were HPLC purified. Concentrations were determined by A280 for proteins (ε280 calculated from sequence), by A381 for peptides containing para-aminobenzoic acid (PABA; ε381 = 2200 M−1cm−1), and by a combination of A280 and A490 for fluorescein-labeled proteins.
Assays.
Lon degradation was performed at 37 °C in a buffer containing T25 (25 mM Tris·HCl, pH 8), 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 2 mM ATP, 80 mM phosphoenolpyruvate, and 10 U/mL pyruvate kinase. Steady-state degradation rates were determined as described (26). Following proteolysis of 35S-labeled substrates, radioactivity of degradation products soluble in 10% cold TCA was used to monitor degradation (29). Some degradation reactions were monitored by changes in fluorescence (excitation 320 nm; emission 420 nm). For ATPase measurements, NADH (1 mM) and lactate dehydrogenease (10 U/mL) were added to the degradation buffer and assays were performed as described (30).
A sul20C peptide variant was synthesized with an N-terminal PABA fluorophore and a nitrotyrosine quencher at the penultimate C-terminal position followed by alanine (F-sul20C-Q). ATP-dependent Lon cleavage of this peptide resulted in a fluorescence increase. A 2-fold dilution of F-sul20C-Q with equimolar sul20C over a range of concentrations resulted in a 2-fold decrease in the degradation rate, suggesting that Lon degrades both peptides at the same rate. The Fig. 2A experiment was performed using a 300:1 mixture of unlabeled sul20C to F-sul20C-Q.
Thermal denaturation [4 μM protein in 10 mM potassium-phosphate (pH 7.6), 100 mM KCl] was performed in a 10-mm path-length cuvette at 1 °C increments using a heating rate of 1 °C/min. After equilibration, the circular-dichroism ellipticity at 228 nM was averaged for 10 s.
Acknowledgments.
We thank T. Baker, S. Bissonnette, B. Cezairliyan, P. Chien, J. Davis, O. Kandror, M. Laub, A. Martin, J. Sohn, S. Sundar, B. Tidor, and A. Varshavsky for helpful discussions. This work was supported by National Institute of Health Grants AI-15706 and AI-16892.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 2.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: Recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer RT, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 5.NgoJK, Davies KJ. Importance of the Lon protease in mitochondrial maintenance and the significance of declining Lon in aging. Ann NY Acad Sci. 2007;1119:78–87. doi: 10.1196/annals.1404.015. [DOI] [PubMed] [Google Scholar]
- 6.Chung CH, Goldberg AL. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci USA. 1981;78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roudiak SG, Shrader TE. Functional role of the N-terminal region of the Lon protease from. Mycobacterium smegmatis Biochemistry. 1998;37:11255–11263. doi: 10.1021/bi980945h. [DOI] [PubMed] [Google Scholar]
- 8.Rotanova TV, et al. Slicing a protease: Structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 2006;15:1815–1828. doi: 10.1110/ps.052069306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gur E, Sauer RT. Recognition of misfolded proteins by Lon a AAA+ protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez M, Frank EG, Levine AS, Woodgate R. Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: In vitro degradation and identification of residues required for proteolysis. Genes Dev. 1998;12:3889–3899. doi: 10.1101/gad.12.24.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah IM, Wolf RE., Jr Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: Identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J Mol Biol. 2006;357:718–731. doi: 10.1016/j.jmb.2005.12.088. [DOI] [PubMed] [Google Scholar]
- 12.Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gur E, Sauer RT. Evolution of the ssrA degradation tag in Mycoplasma: Specificity switch to a different protease. Proc Natl Acad Sci USA. 2008;105:16113–16118. doi: 10.1073/pnas.0808802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waxman L, Goldberg AL. Selectivity of intracellular proteolysis: Protein substrates activate the ATP-dependent protease (La) Science. 1986;232:500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- 15.Hilliard JJ, Simon LD, Van Melderen L, Maurizi MR. PinA inhibits ATP hydrolysis and energy-dependent protein degradation by Lon protease. J Biol Chem. 1998;273:524–527. doi: 10.1074/jbc.273.1.524. [DOI] [PubMed] [Google Scholar]
- 16.Ishii Y, et al. Regulatory role of C-terminal residues of SulA in its degradation by Lon protease in. Escherichia coli J Biochem. 2000;127:837–844. doi: 10.1093/oxfordjournals.jbchem.a022677. [DOI] [PubMed] [Google Scholar]
- 17.Ishii Y, Amano F. Regulation of SulA cleavage by Lon protease by the C-terminal amino acid of SulA histidine. Biochem J. 2001;358:473–480. doi: 10.1042/0264-6021:3580473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 19.Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 21.Gerhart JC, Pardee AB. The effect of the feedback inhibitor CTP on subunit interactions in aspartate transcarbamylase. Cold Spring Harbor Symp Quant Biol. 1963;28:491–496. [Google Scholar]
- 22.Wittenberger CL, Fulco JG. Purification and allosteric properties of a nicotinamide adenine dinucleotide-linked D(−)-specific lactate dehydrogenase from Butyribacterium rettgeri. J Biol Chem. 1967;242:2917–2924. [PubMed] [Google Scholar]
- 23.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding unfolding and translocation. Cell. 2005;121:1029–10241. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson-Ward J, Huang J, Lee I. Detection and characterization of two ATP-dependent conformational changes in proteolytically inactive Escherichia coli Lon mutants by stopped flow kinetic techniques. Biochemistry. 2007;46:13593–13605. doi: 10.1021/bi701649b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas-Wohlever J, Lee I. Kinetic characterization of the peptidase activity of Escherichia coli Lon reveals the mechanistic similarities in ATP-dependent hydrolysis of peptide and protein substrates. Biochemistry. 2002;41:9418–9425. doi: 10.1021/bi0255470. [DOI] [PubMed] [Google Scholar]
- 27.Van Melderen L, Gottesman S. Substrate sequestration by a proteolytically inactive Lon mutant. Proc Natl Acad Sci USA. 1999;96:6064–6071. doi: 10.1073/pnas.96.11.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg AL. Degradation of abnormal proteins in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norby JG. Coupled assay of Na+K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- 31.Segel IH. New York: John Wiley & Sons, Inc.; 1975. Enzyme kinetics: Behavior and analysis of rapid equilibrium and steady-state enzyme systems. [Google Scholar]