Abstract
The polyunsaturated fatty acids (PUFAs) linoleic acid (18:2) and α-linolenic acid (18:3) in triacylglycerols (TAG) are major factors affecting the quality of plant oils for human health, as well as for biofuels and other renewable applications. These PUFAs are essential fatty acids for animals and plants, but also are the source of unhealthy trans fats during the processing of many foodstuffs. PUFAs 18:2 and 18:3 are synthesized in developing seeds by the desaturation of oleic acid (18:1) esterified on the membrane lipid phosphatidylcholine (PC) on the endoplasmic reticulum. The reactions and fluxes involved in this metabolism are incompletely understood, however. Here we show that a previously unrecognized enzyme, phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT), encoded by the Arabidopsis ROD1 gene, is a major reaction for the transfer of 18:1 into PC for desaturation and also for the reverse transfer of 18:2 and 18:3 into the TAG synthesis pathway. The PDCT enzyme catalyzes transfer of the phosphocholine headgroup from PC to diacylglycerol, and mutation of rod1 reduces 18:2 and 18:3 accumulation in seed TAG by 40%. Our discovery of PDCT is important for understanding glycerolipid metabolism in plants and other organisms, and provides tools to modify the fatty acid compositions of plant oils for improved nutrition, biofuel, and other purposes.
Keywords: Arabidopsis, lipid metabolism, oilseeds
Triacylglycerols (TAG) from vegetable oils are a major source of essential fatty acids in the human diet. Linoleic acid (18:2) and α-linolenic acid (18:3) are required for mammalian survival, because these 18- carbon polyunsaturated fatty acids (PUFAs) are precursors in the synthesis of 20- and 22-carbon PUFAs, including arachidonic, eicosapentaenoic, and docosahexaenoic acids, which are important membrane components and substrates for the synthesis of prostaglandins, leukotrienes, and other signaling molecules (1, 2). But trans fats produced during the partial hydrogenation of high-PUFA oils are associated with an increasing prevalence of adult and childhood disorders of lipid metabolism, obesity, and related diseases (3, 4). Studies indicate that increasing the proportion of monounsaturated oleic acid (18:1) in vegetable oils provides significant health benefits, as well as improved oxidative stability, which is important both for food uses and for the production of biodiesel and other renewable resources (5, 6). High-oleic, low-PUFA oils are a workable option nutritionally because the requirement for essential fatty acids in the human diet is met with other foods (7, 8). For these reasons, the enzymology and regulation of TAG synthesis and mobilization in both plants and animals remain very active areas of investigation (9–11). The discovery of new enzymes of lipid metabolism in recent years (10, 12, 13) underscores the need to develop comprehensive and correct models of the pathways involved in both mammals and plants that are the major source of dietary essential fatty acids and TAG.
In oil-accumulating cells of plant seeds, 18:1, which is synthesized from acetyl-CoA in the plastids, is desaturated to 18:2 and 18:3 by 2 desaturase enzymes of the endoplasmic reticulum, FAD2 and FAD3 (14, 15). Before desaturation, 18:1 must be incorporated into phosphatidylcholine (PC), the only substrate recognized by the FAD2 and FAD3 desaturases (16). Interestingly, many modified fatty acids, including those with hydroxyl, epoxy, or acetylene groups or conjugated double bonds, also are synthesized on PC by enzymes that likely evolved from ancestral FAD2 proteins (17–19). In current models of seed lipid metabolism, 18:1 is incorporated into PC by 1 of 2 routes. Direct incorporation from 18:1-CoA exported by the plastids occurs, likely through the action of acyl-CoA:lyso-phosphatidylcholine acyltransferase (LPCAT) (20, 21). Alternatively, 18:1 may be incorporated into diacylglycerol (DAG) by reactions of the Kennedy pathway, after which 18:1-DAG is converted to PC by CDP-choline:diacylglycerol cholinephosphotransferase (CPT) (22, 23). It has been proposed that the CPT reaction is reversible and provides a mechanism for the production of polyunsaturated DAG for the synthesis of TAGs containing 18:2 and 18:3 (22). Here we present genetic and biochemical evidence for a previously unrecognized enzyme, phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT), that interconverts DAG and PC during TAG synthesis in developing seeds of Arabidopsis. PDCT is a gatekeeper enzyme that provides a major route through which 18:1 enters PC for desaturation to 18:2 and 18:3, as well as an important route for the desaturation products, 18:2 and 18:3, to be returned to the DAG pool. Its discovery has important implications for understanding TAG synthesis in seeds and possibly other aspects of lipid metabolism in both plants and animals as well.
Results
Genetic Analysis of the Arabidopsis rod1 Mutant.
In a screen for altered seed fatty acid composition in Arabidopsis (24), the rod1 (reduced oleate desaturation1) mutant was identified as having a marked decrease in 18:2 and 18:3 PUFAs and a concomitant increase in 18:1 relative to WT (Table 1). These changes in fatty acid composition were similar to, but smaller than, those observed in the fad2 mutants (24, 25), raising the possibility that rod1 represents a hypomorphic allele of fad2. Whereas mutations at fad2 reduced PUFA synthesis in leaves and roots as well as in seeds, significant changes in fatty acid composition were seen only in seeds of rod1 plants [Table 1; supporting information (SI) Table S1]. Crosses between rod1 and fad2 produced F1 seeds with considerably higher PUFA levels than those of either parent (Table 1), confirming that the rod1 mutation is at a locus distinct from fad2.
Table 1.
Fatty acid composition in seeds of WT, rod1, and fad2 Arabidopsis and the crosses between WT × rod1 and rod1 × fad2
| Mol % of fatty acids in seeds |
||||||
|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:1 | |
| WT | 8.4 ± 0.1 | 3.1 ± 0.2 | 15.1 ± 0.4 | 29.2 ± 0.3 | 19.9 ± 0.3 | 18.6 ± 0.6 |
| rod1 | 8.5 ± 0.2 | 3.3 ± 0.1 | 32.8 ± 0.6 | 13.8 ± 0.2 | 15.6 ± 0.2 | 20.6 ± 0.1 |
| fad2 | 6.0 ± 0.1 | 2.4 ± 0.1 | 65.0 ± 0.5 | 0.2 ± 0.1 | 1.6 ± 0.1 | 24.0 ± 0.5 |
| WT × rod1 | 8.3 ± 0.5 | 3.1 ± 0.2 | 16.9 ± 0.6 | 29.1 ± 0.9 | 20.4 ± 0.7 | 19.9 ± 0.9 |
| rod1 × fad2 | 8.3 ± 0.1 | 2.4 ± 0.1 | 20.1 ± 0.2 | 24.3 ± 0.2 | 21.0 ± 0.4 | 22.4 ± 0.2 |
Data are mean ± SE; n = 6.
To determine the genetic basis of the rod1 mutation, rod1 plants were crossed to Col-0 WT. F1 seeds showed a fatty acid profile similar to that of the WT parent (Table 1). F1 plants were grown and allowed to self-fertilize. Of the 263 F2 plants analyzed, 69 had seed fatty acid profiles similar to that of the original rod1 seeds (>28% 18:1), while the remaining 194 had fatty acid compositions similar to that of WT (<20% 18:1). This pattern of segregation is a good fit to the hypothesized 3:1 ratio (χ2 = 0.21; P > .05), indicating that rod1 is a single, recessive Mendelian mutation.
Growth, development, and seed production were very similar in rod1 plants and WT. In particular, the timing of lipid accumulation in both lines was comparable, a maximum of 7–9 days after pollination. For rod1, both the weight of mature seeds (17.7 ± 0.2 μg/seed; average ± SE) and the oil content (4.9 ± 0.3 μg/seed) were indistinguishable from those of WT (17.9 ± 0.1 μg/seed and 4.6 ± 0.2 μg/seed, respectively).
Reduced Radiolabeling of Phosphatidylcholine in rod1 Seeds.
We analyzed the fatty acid compositions of different classes of glycerolipids extracted from seeds during this stage of maximum TAG synthesis. Compared with WT, the rod1 mutant had substantially lower levels of PUFAs in both TAG and the immediate precursor DAG (Table 2). Surprisingly, however, PC had higher PUFA levels than WT, with the most highly unsaturated fatty acid, 18:3, accounting for 31.7% of total acyl groups, compared to 19.9% in WT (Table 2). The second most-abundant phospholipid in seeds, phosphatidylethanolamine, does not play a major role in TAG synthesis (13); the fatty acid composition of this lipid was similar in the WT and rod1 samples.
Table 2.
Fatty acid compositions of TAG, DAG, PC, and PE isolated from developing seeds of WT and rod1 Arabidopsis
| Fatty acid composition (mol %) |
||||||
|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:1 | |
| TAG | ||||||
| WT | 9.2 | 3.7 | 17.9 | 30.5 | 16.2 | 18.6 |
| rod1 | 9.9 | 3.8 | 39.1 | 14.2 | 12.3 | 17.9 |
| DAG | ||||||
| WT | 13.2 | 4.4 | 14.8 | 36.0 | 17.7 | 6.8 |
| rod1 | 16.1 | 5.3 | 33.8 | 22.2 | 10.9 | 8.6 |
| PC | ||||||
| WT | 17.5 | 2.4 | 7.9 | 45.4 | 19.9 | 3.5 |
| rod1 | 16.1 | 1.3 | 6.6 | 39.8 | 31.7 | 1.1 |
| PE | ||||||
| WT | 30.6 | 3.3 | 7.5 | 35.4 | 18.9 | 1.3 |
| rod1 | 33.2 | 3.2 | 6.4 | 34.9 | 18.3 | 1.6 |
Seeds were harvested 9 days after flowering, when TAG accumulation was proceeding rapidly. A repeat analysis yielded similar results.
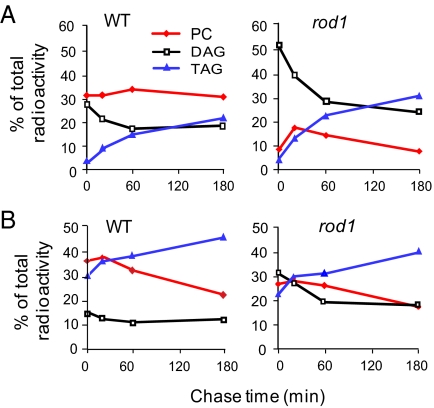
Because PC is the substrate for the FAD2 and FAD3 desaturases that convert 18:1 to 18:2 and 18:3 PUFAs (26), these data indicate the possibility that the rod1 mutation reduces the transfer of 18:1 into PC for desaturation. Current models of TAG synthesis in oil seeds propose that 18:1 can enter the PC pool through the action of either LPCAT or CPT on 18:1-DAG (20, 21, 23). To gain insight into which of these 2 routes might be blocked in rod1, we labeled developing seeds with 14C-glycerol, which predominantly labels the lipid backbone, and 14C-acetate, which labels the acyl groups, in pulse-chase experiments. At the end of the 15-min incubation in 14C-glycerol, WT seeds contained 30% of the total label in PC and 27% of that in DAG, and radioactivity accumulated in TAG during the 3-hour chase (Fig. 1A). In contrast, rod1 seeds contained only 8% of the label in PC but 51% of that in DAG at the end of the pulse; however, radioactivity accumulated in TAG during the chase, as it did in WT. Similarly, the experiment with 14C-acetate revealed reduced incorporation of label into PC at the end of the 15-min labeling pulse in rod1 compared with WT (Fig. 1B). Both sets of data are consistent with rod1 having a defect that reduces the flux of 18:1 into PC, and additional labeling experiments confirmed this conclusion. The experiment with 14C-glycerol suggests a limitation in the de novo synthesis of PC from DAG, because a lesion in LPCAT would not be expected to restrict the flux of glycerol into PC.
Fig. 1.
Lipid synthesis in developing seeds of WT and rod1 mutant. After a 15-min pulse labeling with [14-C]-labeled glycerol (A) or acetate (B), the chase was carried out in unlabelled medium. Radioactivities in PC, DAG, and TAG at 0, 30, 60, and 180 min of chase time were determined.
Identification of the ROD1 Locus.
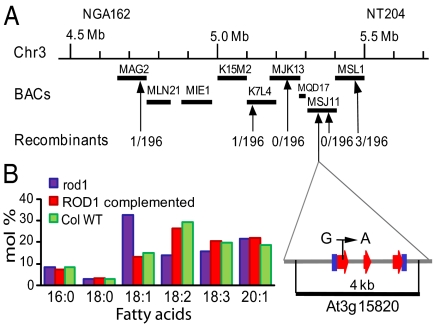
Arabidopsis has 2 genes that encode CPT isozymes, At1g13560 and At3g25585 (27). We determined the sequence of rod1 genomic DNA at both of these loci, but found no changes from WT. We mapped the ROD1 locus, using a population of 800 F2 plants derived from a cross between rod1 and the Landsberg erecta WT (28) (see SI Text), to a region of chromosome 3 covered by BAC clones MJK13, MQD17, and MSJ11 (Fig. 2A). Within this region, 8 genes were annotated as encoding proteins with known or possible functions in lipid metabolism. After considering the literature (29, 30), we amplified, by PCR, rod1 genomic DNA corresponding to 6 of these genes, including At3g15820. We identified a G → A transition in this latter gene that is predicted to change Trp76 to a stop codon. The remaining 5 genes exhibited no changes from WT.
Fig. 2.
The rod1 mutation is in At3g15820. (A) The localization and the structure of the ROD1 gene with the position of the molecular lesion in the mutant. A 4-kb region showing exons (red arrows) and untranslated regions (blue boxes) was used to complement the mutation in rod1. (B) Comparison of seed fatty acid compositions of the At3g15820 transformants (red bars) and WT (green) indicating that At3g15820 fully restored the rod1 mutation (purple), thus confirming the identity of ROD1.
We invesigated the possibility that this mutation is the basis of the rod1 lesion and fatty acid phenotype by transforming the mutant with a 4-kb genomic fragment of WT DNA, including the coding region of At3g15820, a total of 2 kb of 5′ and 3′ flanking sequences (Fig. 2A), using a vector containing the DsRed transformation marker (31). The fatty acid composition of the transgenic seeds was nearly identical to that of WT (Fig. 2B), confirming that At3g15820 is indeed the ROD1 locus.
The ROD1 Locus Encodes a PDCT.
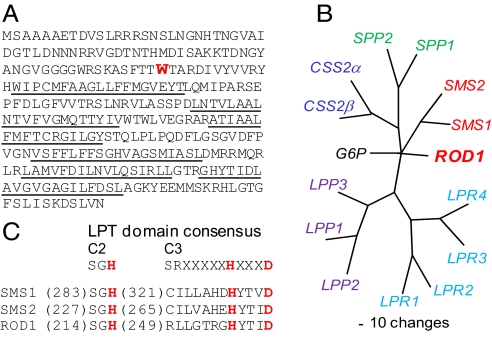
The protein encoded by At3g15820 has been annotated as a phosphatidic acid phosphatase/(PAP2)-related protein containing Pfam profile PF01569—the PAP2 domain (TAIR 9.0, www.arabidopsis.org). But when we used the ROD1 protein sequence (Fig. 3A) to query the Pfam database, the E-value for identification of a PAP2 domain (0.17) was above the recommended cutoff. Only 8 of the 15 most-conserved residues in the Pfam PAP2 profile are present in ROD1. The Arabidopsis genome contains at least 4 genes with clearly identified PAP2 domains (E value <e−40), including Lipid-Phosphate Phosphatase1 (LPP1; At3g02600) and LPP2 (At1g15080), which have both been shown to encode lipid-phosphate phosphatase activities (32). ROD1 contains no substantial sequence similarity to these true PAP2 homologues. In addition, we were unable to detect lipid-phosphate phosphatase activity for the ROD1 protein expressed in yeast (Saccharomyces cerevisiae) cells (Fig. S1).
Fig. 3.
Sequence analysis of the ROD1 protein. (A) Deduced amino acid sequence of At3g15820 with putative transmembrane regions underlined. The site of the W76 stop mutation in rod1 is shown in red. (B) A dendrogram showing the relationship of ROD1 to members of the LPT family in Homo sapiens (34). (C) Sequences of conserved domains in ROD1 and human SMS proteins showing the catalytic triad in red.
When we used the ROD1 amino acid sequence to search the nonredundant protein database using the position-specific iterated BLAST (PSI-BLAST) algorithm (blast.ncbi.nlm.nih.gov), the second iteration identified a mammalian phosphatidylcholine:ceramide cholinephosphotransferase (EC 2.7.8.27). This enzyme, also known as sphingomyelin synthase (SMS), catalyzes the transfer of the phosphocholine headgroup from PC to the alcohol group of ceramide (33). It belongs to the large family of lipid phosphatase/phosphotransferase (LPT) proteins (34). Phylogenetic analysis places ROD1 in close relationship to the SMS1 and SMS2 proteins within the LPT family (Fig. 3B), and topology prediction programs identify ROD1 as an integral-membrane protein with up to 6 putative transmembrane domains (Fig. 3A), similar to predictions for other LPT proteins (34). In addition, 5 highly conserved residues in the C2 and C3 domains of SMS1, SMS2, and other LPT proteins have been identified at comparable positions in the ROD1 protein (Fig. 3 A and C). Plants do not contain sphingomyelin, but the structure of ceramide is analogous in some respects to that of DAG, prompting us to consider the possibility that ROD1 catalyzes the transfer of phosphocholine from PC to DAG in a reaction analogous to that mediated by SMS in animals. Following biochemical convention, we designate this putative enzyme as phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) in the IUPAC subclass EC 2.7.8.
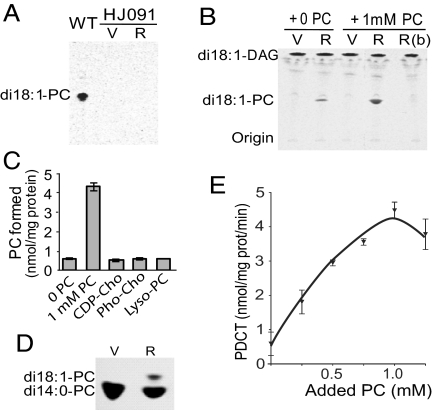
To directly test for the proposed PDCT activity of the ROD1 protein, we placed the coding sequence of At3g15820 under control of a yeast constitutive promoter in vector p424GPD (35) and expressed the protein in strain HJ091 (cpt1::LEU2 ept1−), which lacks CPT activity (36). We first tested microsomal preparations from HJ091 cells expressing ROD1 and from empty-vector controls for the ability to synthesize PC from DAG and CDP-[14C]choline (36). No activity was detected in either control microsomes or those from cells expressing ROD1 (Fig. 4A); however, 14C-labeled PC was produced when ROD1 microsomes were incubated with dioleoyl-[14C]glycerol, and this activity was enhanced in the presence of added PC (Fig. 4B). Control microsomes did not exhibit activity in this assay, and ROD1 microsomes that had been boiled before the assay were inactive as well. Because the [14C] radiolabel was in the glycerol moiety of the [14C]-DAG substrate, these assays indicate that ROD1 synthesizes [14C]-PC through transfer of the phosphocholine headgroup from PC to [14C]-DAG. The activity observed in assays without added PC presumably relied on endogenous PC of the yeast microsomes.
Fig. 4.
ROD1 is a phosphatidylcholine:diacylglycerol cholinephosphotransferase. (A) Radio-TLC image of CPT assays. (B) Radio-TLC image of PDCT assays. Microsomes from DBY746 (WT) or HJ091 S. cerevisiae cells transfected with p424GPD (V) or p424ROD1 (R) were incubated with CDP-[14C]choline and diolein for CPT (A) or with [14C-glycerol]di18:1-DAG and PC (0 or 1 mM) for PDCT (B). R(b) indicates boiled p424ROD1 microsomal proteins. (C) PDCT assays of microsomes from HJ091 transfected with p424ROD1 incubated with [14C-glycerol]di18:1-DAG and the phosphocholine compounds indicated (at 1 mM concentration). (D) Radio-TLC of PDCT assays. Microsomes of HJ091 cells transfected with p424GPD (V) or p424ROD1 (R) were incubated with di14:0-PC [14C-choline] and di18:1-DAG. (E) PDCT activity as a function of exogenous PC. Data represent mean and SD of 3 independent reactions.
Assays with other possible phosphocholine donors indicated that only the phosphocholine headgroup of PC was accessible to the ROD1 enzyme. The addition of 1 mM CDP-choline, phosphocholine, or lyso-PC did not support [14C]-PC synthesis at rates higher than those of ROD1 microsomes without added PC (Fig. 4C). To specifically test for transfer of the PC headgroup, we incubated microsomes with [14C]choline-labeled dimyristoyl-PC and unlabeled dioleoyl-DAG. In this assay, ROD1 microsomes, but not the control, synthesized dioleoyl-[14C]-PC, which separated from the dimyristoyl-[14C]PC substrate on thin layer chromatography (Fig. 4D). PDCT activity was highest at pH 6.5–7, linear with time up to 3 min, and linear with protein concentration up to 20 μg of microsomal protein (Fig. S2). Under optimized assay conditions, PDCT activity was 0.6 nmol/min/mg microsomal protein in the absence of added PC, increasing to 4.5 nmol/min/mg with 1 mM added PC (Fig. 4E). These results clearly indicate that ROD1 is a PDCT enzyme in Arabidopsis.
ROD1-Related Proteins in Arabidopsis and Other Organisms.
Data from the Arabidopsis Gene Expression Atlas (37) indicate that the level of ROD1 transcript (detected by Affymetrix array element 258249_s_at) is highest in seeds that are accumulating TAG, and that ROD1 is expressed in other tissues as well (Fig. S3). A related gene, At3g15830, cross-hybridizes to array element 258249_s_at, but data from the Arabidopsis Massively Parallel Signature Sequence (MPSS) project (http://mpss.udel.edu/at/) indicate that this gene is expressed only in floral tissues. Our experiments confirmed that At3g15830 is not expressed in seeds (Fig. S4). In addition, no PDCT activity was detected in microsomal preparations from HJ091 yeast cells expressing a cDNA for this gene (see SI Text).
Sequences homologous to Arabidopsis ROD1 are identifiable in many higher plants, including oil crops such as canola (Brassica napus) and castor bean (Ricinus communis) (Fig. S5), indicating that PDCT likely is an important enzyme of TAG synthesis in many plants. Although no readily identifiable homologues are present in animals, the human LPT family contains at least 8 genes that encode proteins of unknown function (34); thus, it remains possible that PDCT will be found to play a role in lipid metabolism in animals as well.
Discussion
We report a recently discovered enzyme, PDCT, that is required for the efficient synthesis of PUFAs during TAG accumulation in seeds. In WT Arabidopsis, 49.1% of the fatty acids in seed TAG are polyunsaturated 18:2 and 18:3 (Table 1). These are synthesized from 18:1 on PC of the endoplasmic reticulum by the FAD2 and FAD3 fatty acid desaturases. In the PDCT-defective rod1 mutant, these fatty acids represent only 29.4% of the total, indicating that 40% (100 × [49.1 - 29.4]/49.1) of the 18:1 that is converted to 18:2 and 18:3 enters PC via the PDCT enzyme.
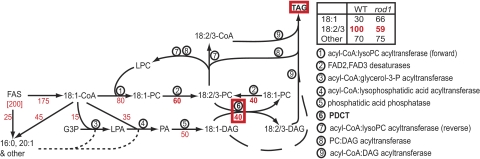
The potentially large numbers of molecular species and subcellular pools of intermediates in the pathways of seed TAG synthesis (9) make it difficult to represent the relationships comprehensively; however, the simplified scheme shown in Fig. 5 illustrates the role of PDCT and the main fluxes of PUFA-TAG synthesis. For convenience, the fluxes shown in Fig. 5 were calculated for 100 U of 18:2 and 18:3 accumulating in TAG of WT (Top Right, box), corresponding to 200 U of total fatty acids from the fatty acid synthase. As shown, 18:1-CoA is the main product exported from the plastid (175 U). Approximately half of this (80 U) is incorporated directly in PC, presumably through the action of LPCAT (21) (reaction 1 in Fig. 5), and much of this (ca. 60 U) is desaturated to 18:2 or 18:3 (reaction 2). In addition, 18:1 is incorporated into DAG by reactions 3–5 (50 U), with most of this transferred into the PC pool by the PDCT enzyme (reaction 6) to be made available for desaturation. The CPT reaction (not shown) is required for de novo PC synthesis, but the fatty acid composition of rod1 seeds (Table 1) and other evidence (9) suggest that the PDCT reaction is responsible for most of the conversion of DAG to PC in developing seeds. Because PDCT is a symmetrical reaction (with 1 DAG molecule generated for each DAG consumed), we assume that 18:2-DAG and 18:3-DAG are produced. This outcome does not necessarily require substrate selectivity of the PDCT enzyme (i.e., 18:2-PC and 18:3-PC over 18:1-PC; 18:1-DAG over 18:2-DAG and 18:3-DAG), because the desaturases will enrich the PC pool with PUFAs. Along with PDCT, 18:2 and 18:3 leave PC through a reverse action of LPCAT (reaction 7) and phosphatidylcholine:DAG acyltransferase (reaction 8), both of which generate lyso-PC. The 18:2-CoA and 18:3-CoA produced by reaction 7 are available to aycl-CoA:DAG acyltransferase (reaction 9). In WT seeds, PDCT provides a substantial proportion of the 18:2-DAG and 18:3-DAG substrate. In the rod1 mutant deficient in PDCT, more 18:1-DAG is incorporated in TAG (dashed line). In both rod1 and WT, 18:2-CoA and 18:3-CoA can be converted to DAG by reactions 3–5; however, this possibility does not prevent the rod1 mutation from reducing the accumulation of 18:2 and 18:3 in TAG by 40%.
Fig. 5.
A simplified scheme of seed TAG synthesis incorporating the PDCT enzyme. The number key lists the enzymatic reactions shown. The fatty acid compositions of WT and rod1 seeds (Top Right and Table 1) were used to calculate the fluxes shown in red for 200 units of fatty acid produced by fatty acid synthase and providing 100 units of PUFA in the TAG.
The high expression of the ROD1 gene in oil-accumulating seed tissue is consistent with our mutant analysis and enzymology showing that interconversion of PC and DAG by PDCT is an important mechanism for PUFA enrichment of TAG. ROD1 also is expressed in vegetative tissues (Fig. S3), and PC is a major substrate for 18:1 desaturation in these tissues as well (25). Although the rod1 mutation does not result in substantial changes in leaf or root fatty acid compositions, PDCT possibly may play a role in lipid homeostasis in vegetative cells of the plant or in remodeling of membrane lipids in response to temperature changes or other environmental perturbations.
PDCT is structurally related to animal SMS and contains the C2 and C3 domains characteristic of the broader LPT family (Fig. 3). These C2 and C3 domains include the histidine and aspartate that are proposed active-site residues involved in phosphate–ester bond cleavage (34). This supports the notion that PDCT uses a catalytic mechanism analogous to that of the SMS enzymes. Further studies are needed to confirm the importance of these residues in PDCT and to investigate whether other aspects of PDCT are analogous to those of the more extensively studied SMS proteins.
PDCT also provides a useful tool for using biotechnology to modify the fatty acid composition of plant oils. For example, because PDCT contributes to the control of PUFA synthesis in seeds, suppression of ROD1 expression could reduce the need for hydrogenation of oils, along with the attendant production of unhealthy trans fats (3, 4). This type of genetic engineering also can allow for the production of biofuels with increased oxidative stability (38) and may reduce the incorporation of saturated fatty acids into membrane lipids. Because PC is also the substrate for enzymes that produce hydroxy-, epoxy-, acetylenic and other modified fatty acids (17–19), our discovery of PDCT provides many opportunities to better understand TAG synthesis in different oil seed species and to improve the fatty acid profiles of vegetable oils for both human health and industrial applications.
Materials and Methods
Genetic Analysis of the rod1 Mutant.
Mutant line rod1 in the Arabidopsis thaliana Col-0 background was isolated from an M3 population after mutagenesis with ethyl methanesulfonate (24). Plants were grown on soil in controlled environment chambers at 22 °C under continuous florescent illumination (150 μmol quanta/m2/s). Genetic analysis, map-based cloning of the ROD1 locus, and bioinformatic analyses were performed following conventional approaches (see SI Materials and Methods).
Lipid Analysis and Labeling.
The overall fatty acid compositions of seeds and other tissues were determined as described previously (31). Pulse-chase labeling was carried out in developing seeds harvested from siliques 9 days after flowering, as described previously (22) (see SI Materials and Methods).
ROD1 Enzyme Activity Assays.
Growth of HJ091 yeast cells expressing ROD1, preparation of membrane fractions, and assays of CPT were conducted as described previously (39), using 0.1 μmol diolein and 1 nmol [14C]CDP-choline as substrates. Assays of PDCT activity are detailed in SI Materials and Methods.
Phylogenetic Analyses.
Producing the parsimony bootstrap tree (40) (Fig. 3B) involved 1,000 bootstrap replicate data sets, each of which was analyzed using tree bisection reconnection, steepest decent, and other settings to maximize the detection of global optima or maximization of the parsimony optimality criteria.
Supplementary Material
Acknowledgments.
We thank R. Dewey, C.R. McMaster, and G. Carman for providing strains and advice and M. Schneider, G.-S. Han, M. Lavin, and C. Skidmore for providing technical assistance. This work was supported by grants 2006–35318-17797, 2003–35318-13914, and 2001–345318-10186 from the U.S. Department of Agriculture Cooperative State Research Education and Extension Service, grant DBI-0701919 from the U.S. National Science Foundation, and by the Agricultural Research Center at Washington State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908848106/DCSupplemental.
References
- 1.Funk CD. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 2.Cunnane SC. Problems with essential fatty acids: Time for a new paradigm? Prog Lipid Res. 2003;42:544–568. doi: 10.1016/s0163-7827(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 3.Steinhart H, Rickert R, Winkler K. Trans fatty acids (TFA): Analysis, occurrence, intake and clinical relevance. Eur J Med Res. 2003;8:358–362. [PubMed] [Google Scholar]
- 4.Muoio DM, Newgard CB. Obesity-related derangements in metabolic regulation. Annu Rev Biochem. 2006;75:367–401. doi: 10.1146/annurev.biochem.75.103004.142512. [DOI] [PubMed] [Google Scholar]
- 5.Kinney AJ. Designer oils for better nutrition. Nat Biotechnol. 1996;14:946. doi: 10.1038/nbt0896-946. [DOI] [PubMed] [Google Scholar]
- 6.Knothe G. Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process Technol. 2005;86:1059–1070. [Google Scholar]
- 7.Hunter JE. n-3 fatty acids from vegetable oils. Am J Clin Nutr. 1990;51:809–814. doi: 10.1093/ajcn/51.5.809. [DOI] [PubMed] [Google Scholar]
- 8.Williams CM, Burdge G. Long-chain n-3 PUFA: Plant vs. marine sources. Proc Nutr Soc. 2006;65:42–50. doi: 10.1079/pns2005473. [DOI] [PubMed] [Google Scholar]
- 9.Bates PD, Durrett TP, Ohlrogge JB, Pollard M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 2009;150:55–72. doi: 10.1104/pp.109.137737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann R, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 11.Guo Y, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahlqvist A, et al. Phospholipid diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA–independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cases S, et al. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 14.Arondel V, et al. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science. 1992;258:1353–1355. doi: 10.1126/science.1455229. [DOI] [PubMed] [Google Scholar]
- 15.Okuley J, et al. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stymne S, Appelqvist LA. The biosynthesis of linoleate from oleoyl-CoA via oleoyl-phosphatidylcholine in microsomes of developing safflower seeds. Eur J Biochem. 1978;90:223–229. doi: 10.1111/j.1432-1033.1978.tb12594.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee M, et al. Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science. 1998;280:915–918. doi: 10.1126/science.280.5365.915. [DOI] [PubMed] [Google Scholar]
- 18.Broun P, Shanklin J, Whittle E, Somerville C. Catalytic plasticity of fatty acid modification enzymes underlying chemical diversity of plant lipids. Science. 1998;282:1315–1317. doi: 10.1126/science.282.5392.1315. [DOI] [PubMed] [Google Scholar]
- 19.Cahoon EB, et al. Biosynthetic origin of conjugated double bonds: Production of fatty acid components of high-value drying oils in transgenic soybean embryos. Proc Natl Acad Sci USA. 1999;96:12935–12940. doi: 10.1073/pnas.96.22.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl U, Stalberg K, Stymne S, Ronne H. A family of eukaryotic lysophospholipid acyltransferases with broad specificity. FEBS Lett. 2008;582:305–309. doi: 10.1016/j.febslet.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Stymne S, Stobart AK. Evidence for the reversibility of the acyl-CoA:lysophosphatidylcholine acyltransferase in microsomal preparations from developing safflower (Carthamus tinctorius L) cotyledons and rat liver. Biochem J. 1984;223:305–314. doi: 10.1042/bj2230305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slack CR, Campbell LC, Browse JA, Roughan PG. Some evidence for the reversibility of cholinephosphotransferase-catalyzed reaction in developing linseed cotyledons in vivo. Biochim Biophys Acta. 1983;754:10–20. [Google Scholar]
- 23.Vogel G, Browse J. Cholinephosphotransferase and diacylglycerol acyltransferase: Substrate specificities at a key branch point in seed lipid metabolism. Plant Physiol. 1996;110:923–931. doi: 10.1104/pp.110.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemieux B, Miquel M, Somerville C, Browse J. Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet. 1990;80:234–240. doi: 10.1007/BF00224392. [DOI] [PubMed] [Google Scholar]
- 25.Miquel M, Browse J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis: Biochemical and genetic characterization of a plant oleoyl- phosphatidylcholine desaturase. J Biol Chem. 1992;267:1502–1509. [PubMed] [Google Scholar]
- 26.Wallis JG, Browse J. Mutants of Arabidopsis reveal many roles for membrane lipids. Prog Lipid Res. 2002;41:254–278. doi: 10.1016/s0163-7827(01)00027-3. [DOI] [PubMed] [Google Scholar]
- 27.Goode JH, Dewey RE. Characterization of aminoalcoholphosphotransferases from Arabidopsis thaliana and soybean. Plant Physiol Biochem. 1999;37:445–457. [Google Scholar]
- 28.Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in Arabidopsis: Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan L, Zheng S, Wang X. Antisense suppression of phospholipase D alpha retards abscisic acid– and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell. 1997;9:2183–2196. doi: 10.1105/tpc.9.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heilmann I, Mekhedov S, King B, Browse J, Shanklin J. Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 2004;136:4237–4245. doi: 10.1104/pp.104.052951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C, Fulda M, Wallis JG, Browse J. A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 2006;45:847–856. doi: 10.1111/j.1365-313X.2005.02636.x. [DOI] [PubMed] [Google Scholar]
- 32.Pierrugues O, et al. Lipid phosphate phosphatases in Arabidopsis: Regulation of the AtLPP1 gene in response to stress. J Biol Chem. 2001;276:20300–20308. doi: 10.1074/jbc.M009726200. [DOI] [PubMed] [Google Scholar]
- 33.Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigal YJ, McDermott MI, Morris AJ. Integral membrane lipid phosphatases/phosphotransferases: Common structure and diverse functions. Biochem J. 2005;387:281–293. doi: 10.1042/BJ20041771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 36.Morash SC, McMaster CR, Hjelmstad RH, Bell RM. Studies employing Saccharomyces cerevisiae cpt1 and ept1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J Biol Chem. 1994;269:28769–28776. [PubMed] [Google Scholar]
- 37.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 38.Kinney AJ, Clemente TE. Modifying soybean oil for enhanced performance in biodiesel blends. Fuel Process Technol. 2005;86:1137–1147. [Google Scholar]
- 39.Hjelmstad R, Bell R. sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases in Saccharomyces cerevisiae: Mixed micellar analysis of the CPT1 and EPT1 gene products. J Biol Chem. 1991;266:4357–4365. [PubMed] [Google Scholar]
- 40.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2003. Version 4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.