Abstract
The P-cluster of nitrogenase is one of the most complex biological metallocenters known to date. Despite the recent advances in the chemical synthesis of P-cluster topologs, the biosynthetic mechanism of P-cluster has not been well defined. Here, we present a combined biochemical, electron paramagnetic resonance, and X-ray absorption spectroscopy/extended X-ray absorption fine-structure investigation of the maturation process of P-clusters in ΔnifH molybdenum-iron (MoFe) protein. Our data indicate that the previously identified, [Fe4S4]-like cluster pairs in ΔnifH MoFe protein are indeed the precursors to P-clusters, which can be reductively coupled into the mature [Fe8S7] structures in the presence of Fe protein, MgATP, and dithionite. Moreover, our observation of a biphasic maturation pattern of P-clusters in ΔnifH MoFe protein provides dynamic proof for the previously hypothesized, stepwise assembly mechanism of the two P-clusters in the α2β2-tetrameric MoFe protein, i.e., one P-cluster is formed in one αβ dimer before the other in the second αβ dimer.
Keywords: assembly, biosynthesis
Biological nitrogen fixation is a remarkable chemical feat accomplished by a select group of microorganisms. These microorganisms have a complex metalloenzyme, nitrogenase, which is capable of reducing atmospheric dinitrogen (N2) to bioavailable ammonia (NH3) under ambient conditions. The most extensively studied member of this enzyme family is the molybdenum (Mo)-nitrogenase of Azotobacter vinelandii, which consists of two redox-active proteins (1). One, designated iron (Fe) protein (encoded by nifH), is a 60-kDa α2 homodimer containing one [Fe4S4] cluster at the subunit interface and one MgATP binding site in each subunit. The other, termed molybdenum-iron (MoFe) protein (encoded by nifD and nifK), is a 230-kDa α2β2 heterotetramer containing one P-cluster ([Fe8S7]) at each α/β-subunit interface and one iron-molybdenum cofactor (FeMoco) ([MoFe7S9X-homocitrate], where X = C, N, or O) within each α subunit (2). It is believed that, concomitant with ATP hydrolysis, Fe protein undergoes repeated association/dissociation processes with MoFe protein, donating electrons from its [Fe4S4] cluster, through the P-cluster, to the FeMoco of the MoFe protein, where substrate reduction eventually takes place.
The structure of P-cluster can be viewed as a symmetric double cubane in which two [Fe4S4] cubanes share a central μ6-sulfur (S) atom. Such a geometry suggests that the P-cluster is likely assembled by the fusion of two [Fe4S4]-like subclusters (3). This reaction mechanism is well established in synthetic inorganic chemistry and successfully realized by the recent synthesis of P-cluster topologs (4–6). Biological evidence in this regard was supplied by a FeMoco-deficient form of MoFe protein (designated ΔnifH MoFe protein), which was isolated from a nifH-deletion strain of A. vinelandii (7). Extended X-ray absorption fine structure (EXAFS) (8) and magnetic circular dichroism (MCD) (9) analyses show that the ΔnifH MoFe protein contains, instead of the two “standard” [Fe8S7] P-clusters, two pairs of [Fe4S4]-like clusters that can undergo further maturation upon incubation with dithionite, Fe protein (encoded by nifH), and MgATP (10). These observations suggest that the paired [Fe4S4]-like clusters are likely the precursors to P-clusters and the maturation of P-clusters requires the presence of reductant (i.e., dithionite) and ATP-dependent reductase (i.e., Fe protein/MgATP). However, although the further-matured “P-clusters” in ΔnifH MoFe protein are more reduced, they remain as the [Fe4S4]-like fragments despite the fact that the ΔnifH MoFe protein containing these further-matured P-clusters can be activated to a certain degree upon the insertion of isolated FeMoco (10).
Here, we present a combined biochemical, electron paramagnetic resonance (EPR) and X-ray absorption spectroscopy (XAS)/extended X-ray absorption fine structure (EXAFS) investigation of the maturation process of P-clusters in ΔnifH MoFe protein. Our data indicate that the previously identified [Fe4S4]-like cluster pairs in ΔnifH MoFe protein are indeed the precursors to P-clusters, which can be reductively coupled into the mature [Fe8S7] structure in the presence of increased concentrations of dithionite and Fe protein. Moreover, our observation of a biphasic maturation pattern of P-clusters in ΔnifH MoFe protein provides dynamic proof for the previously hypothesized, stepwise assembly mechanism of the two P-clusters in the α2β2-tetrameric MoFe protein, i.e., one P-cluster is formed in one αβ dimer before the other in the second αβ dimer.
Results
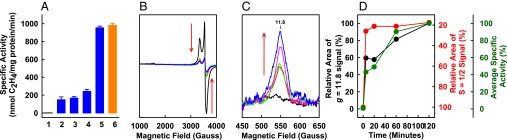
The optimization of P-cluster formation was carried out by incubating ΔnifH MoFe protein with varying concentrations of dithionite and Fe protein and reisolating the protein for reconstitution assays with isolated FeMoco. The reconstituted activity of ΔnifH MoFe protein increases upon maturation with elevated dithionite and Fe protein concentrations and reaches the maximum at a Fe protein:ΔnifH MoFe protein ratio of 2:1 and a dithionite concentration of 20 mM (Fig. 1A, bars 1–5). The maximum activity of the ΔnifH MoFe protein (Fig. 1A, bar 5) is indistinguishable from that of the ΔnifB MoFe protein (Fig. 1A, bar 6), a FeMoco-deficient form of MoFe protein containing intact P-clusters (11)*. This observation signifies a complete transformation of precursors in ΔnifH MoFe protein into mature P-clusters and provides strong evidence that the paired [Fe4S4]-like clusters are indeed the physiologically relevant precursors to P-clusters.
Fig. 1.
Optimization of the maturation conditions of ΔnifH MoFe protein (A) and time-dependent P-cluster formation in ΔnifH MoFe protein (B–D). (A) Reconstitution activities of the as-isolated ΔnifH MoFe protein (bar 1), the ΔnifH MoFe protein matured in the presence of varying concentrations of Fe protein and dithionite (bars 2–5), and the as-isolated ΔnifB MoFe protein (bar 6) are shown. The molar ratios of Fe protein/ΔnifH MoFe protein were 1:4 (bar 2), 1:3 (bar 3), and 2:1 (bars 4 and 5), respectively. A dithionite concentration of 2 mM was used for the maturation of all ΔnifH MoFe protein samples except that represented by bar 5, where a dithionite concentration of 20 mM was applied. (B and C) Perpendicular-mode EPR spectra of dithionite-reduced ΔnifH MoFe protein (B) and parallel-mode EPR spectra of indigo disulfonate (IDS)-oxidized ΔnifH MoFe protein (C) after 0 (black), 5 (red), 20 (green), 60 (pink), and 120 (blue) min of incubation with excess MgATP, Fe protein (molar ratio of Fe protein/ΔnifH MoFe protein = 2:1), and dithionite (20 mM). The concentration of all EPR samples was 10 mg/mL. The decrease of the precursor-associated S = 1/2 EPR signal (B) and the concurrent increase of the P-cluster (P2+)-specific g = 11.8 EPR signal (C) during P-cluster maturation are indicated by the red arrows. (D) Changes in the average specific activity (green) and the relative areas of the S = 1/2 (red) and the g = 11.8 (black) EPR signals of ΔnifH MoFe protein during P-cluster maturation.
To monitor the “real-time” maturation of P-clusters, the ΔnifH MoFe protein was pretreated under optimized conditions for varying lengths of time (i.e., 0, 5, 20, 60, and 120 min) and subsequently reisolated for reconstitution and spectroscopic analyses. Concomitant with an increase in activity with longer incubation, the precursor-specific, S = 1/2 signal of the ΔnifH MoFe protein decreases (Fig. 1B), whereas the P-cluster (P2+)-specific, g = 11.8 signal increases (Fig. 1C) in magnitude, suggesting a gradual conversion of precursors to P-clusters over a certain time period. Interestingly, while the amplitude of the S = 1/2 signal decreases to the near-minimum level within 5 min (Fig. 1D, red line), the intensity of the P2+-spcific signal reaches its maximum much more slowly (Fig. 1D, black line). Further, the increase of the magnitude of the P2+-specific signal (Fig. 1D, black line) aligns well with the increase of activity (Fig. 1D, green line), both following a biphasic pattern with a “lag” period at ≈50% of their respective maximum values (Fig. 1D). Such a lag phase likely marks the formation of approximately half of the P-clusters in the protein.
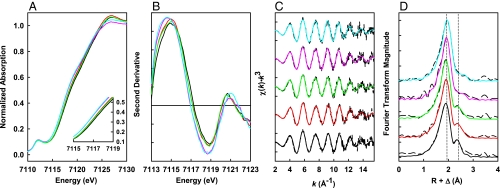
Consistent with the outcome of EPR analysis, the Fe K-edge XAS data show a biphasic pattern of P-cluster formation in ΔnifH MoFe protein. With an increase in the duration of incubation, there is a clear shift in the shoulder feature of the rising edge at 7117 eV (Fig. 2A), which corresponds to a successive reduction of cluster Fe atoms that eventually stabilize at a near all-ferrous oxidation state on average. This transformation is not evenly paced, because the shifts in Fe K-edge energy are more significant between 0–5 and 20–60 min of incubation (Fig. 2B). The Fourier transforms (Fig. 2D) of the EXAFS data (Fig. 2C) show a shift in the first transform peak (at ≈1.9 Å) to a larger distance, which suggests an elongation of the average Fe-S backscattering distance with longer incubation time (8). Additionally, the second Fourier transform peak (at ≈2.4 Å) undergoes a substantial change in shape and intensity (Fig. 2D), which reflects a change in the Fe-Fe backscattering components during the course of incubation. Such a Fourier transform pattern is consistent with the conversion of the precursor in ΔnifH MoFe protein to a cluster species that closely resembles the mature P-cluster in ΔnifB MoFe protein (8).
Fig. 2.
Overlay of the Fe K-edge XAS absorption spectra (A), the second derivatives (B), the Fe K-edge EXAFS (C), and the nonphase shifted Fourier transforms (D) of ΔnifH MoFe protein during P-cluster maturation. The ΔnifH MoFe protein was matured by incubation with excess MgATP, Fe protein (molar ratio of Fe protein/ΔnifH MoFe protein = 2:1) and dithionite (20 mM) for 0 (black), 5 (red), 20 (green), 60 (pink), and 120 (cyan) min. The change in the rising edge at 7,117 ev (A) corresponds to a reduction of cluster Fe atoms, and the most significant shifts in Fe K-edge energy are between 0–5 and 20–60 min (B). The change in the EXAFS spectra (C) is reflected by the change in the Fe-Fe scattering peak at ≈2.4 Å and by the shift in the Fe-S scattering peak at ≈1.8 Å in the Fourier transforms (D), indicating a structural rearrangement upon P-cluster maturation. The Fourier transforms of the ΔnifH MoFe protein matured beyond 60 min (D, pink and cyan) closely resemble that of the ΔnifB MoFe protein (8).
EXAFS fit results over a k range of 2 to 15.7 Å−1 suggest, once again, a significant rearrangement of the P-cluster species of ΔnifH MoFe protein in two distinct steps. While each Fe atom can be modeled as being coordinated to an average of 3.5 S atoms at a steadily increased distance from 2.30 to 2.32 Å throughout the duration of incubation, there is a noticeable increase in the number of short Fe-Fe distances and a concomitant decrease in the number of long Fe-Fe distances (Table 1). The ratio of short to long Fe-Fe distance changes over the course of incubation from 1:2.25 to 1:1.16, which is consistent with a conversion of the immature precursor (with a ratio of short to long Fe-Fe distance of ≈1:2) in ΔnifH MoFe protein to a fully matured P-cluster (with a ratio of short to long Fe-Fe distance of ≈1:1) in ΔnifB MoFe protein (8). Moreover, as early as 60 min into the incubation, the Fe-Fe bond lengths (2.55 Å and 2.77 Å) and the Fe-S bond lengths (2.32 Å) are already in excellent accord with the corresponding distances determined by crystallographic analysis of the mature P-cluster in ΔnifB MoFe protein (11).
Table 1.
EXAFS fitting results for ΔnifH MoFe protein samples matured for varying lengths of time
| Incubation time, min | Fe-S |
Fe-Fe (short) |
Fe- Fe (long) |
ΔE0, eV | Error | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | R, Å | σ2, Å2 | N | R, Å | σ2, Å2 | N | R, Å | σ2, Å2 | |||
| 0 | 3.50 | 2.30 | 0.0060 | 0.50 | 2.49 | 0.0040 | 2.50 | 2.70 | 0.0119 | −12.7 | 0.352 |
| 5 | 3.50 | 2.31 | 0.0054 | 1.00 | 2.52 | 0.0065 | 2.25 | 2.73 | 0.0099 | −12.0 | 0.303 |
| 20 | 3.50 | 2.31 | 0.0054 | 1.00 | 2.52 | 0.0063 | 2.25 | 2.72 | 0.0104 | −12.6 | 0.323 |
| 60 | 3.50 | 2.32 | 0.0048 | 1.50 | 2.55 | 0.0116 | 1.75 | 2.77 | 0.0100 | −10.8 | 0.332 |
| 120 | 3.50 | 2.32 | 0.0051 | 1.50 | 2.55 | 0.0094 | 1.75 | 2.76 | 0.0097 | −10.8 | 0.336 |
The data were fit over the k range 2 to 15.7 Å−1. The variables are coordination number, N; interatomic distance, R (Å); mean-square thermal and static deviation in R, σ2 (Å2); and the shift in the threshold energy, ΔE0 (eV). R, σ2, and ΔE0 were allowed to float during the fitting process in which N was systematically varied; a single ΔE0 was used for each fit. The estimated uncertainties in R and σ2 are ± 0.02 and ± 0.0001 Å2, respectively (30). There is a 10–20% uncertainty in N (31). Error is reported as F/no. of points, where F = [Σk6(χexptl − χcalcd)2/ Σk6χexptl2] (27).
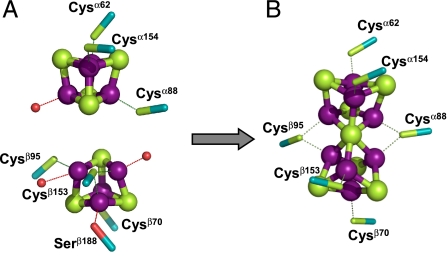
Models for the P-cluster species of ΔnifH MoFe protein at the beginning (i.e., 0 min) and the end (i.e., 120 min) of the maturation process can be derived from these XAS/EXAFS data (Fig. 3). Consistent with the previously reported EXAFS-based models (8), before maturation, the P-cluster species (i.e., precursor) in ΔnifH MoFe protein is composed of a pair of [Fe4S4]-like clusters (Fig. 3A). However, the two [Fe4S4]-like subclusters are likely neither identical nor bridged together, because combined EPR and MCD studies suggest the presence of a [Fe4S4]1+ cluster that is paired with a diamagnetic [Fe4S4]-like cluster in the ΔnifH MoFe protein (7, 9). It is possible, therefore, that one of the subcubanes of the [Fe4S4] cluster pair is a “standard” [Fe4S4]1+ cluster (12); whereas the other subcubane is distorted (e.g., the cage may become asymmetrical if a core sulfide is replaced by a bridging cysteine) and/or coordinated by additional light atoms (e.g., N or O from Asp, His, Ser, or adventitious water) (13, 14), both of which may account for its unusual structural and redox properties (Fig. 3A). Upon maturation, the conformation of the P-cluster species in ΔnifH MoFe protein closely resembles that of the mature P-cluster in ΔnifB MoFe protein (Fig. 3B). However, the P-cluster in ΔnifH MoFe protein is slightly distorted, which is consistent with the fact that the cluster species of the matured ΔnifH MoFe protein contains a somewhat larger population of long Fe-Fe distances than the P-cluster of the ΔnifB MoFe protein (Table 1).
Fig. 3.
Structural models for the cluster species in the ΔnifH MoFe protein before (A) and after (B) P-cluster maturation. The precursor model (A) comprises two individual subclusters, a [Fe4S4] cluster (Upper) and a [Fe4S4]-like cluster (Lower) with a bridging cysteine in place of a core sulfide; whereas the P-cluster model (B) is nearly identical to the mature [Fe8S7] structure in the ΔnifB MoFe protein (Fe, purple; S, green). The P-cluster model was derived by systematically altering the crystallographic coordinates of 1L5H (11) based on the EXAFS fits in Table 1. The possible protein ligands are indicated; additional ligands comprising light atoms (N or O) are represented by red spheres.
Discussion
The complete formation of P-clusters from the previously identified [Fe4S4]-like cluster pairs in ΔnifH MoFe protein (7–9) provides the definitive proof that (i) the paired [Fe4S4]-like clusters are indeed the physiologically relevant precursors to P-clusters; and (ii) unlike FeMoco, which is “preassembled” before being delivered to its targeted site in the MoFe protein, P-cluster is assembled at its destined location within the MoFe protein. Further, the observation that P-cluster is formed at specific concentrations of reductant (i.e., dithionite) and reductase (i.e., Fe protein) (Fig. 1A) suggests a key role of redox chemistry in this process. Interestingly, the optimal solution potential for P-cluster maturation (calculated to be approximately −440 mV for 20 mM dithionite) is the same as that for FeMoco maturation on NifEN (15), both involving the action of Fe protein in an ATP-dependent manner. Thus, the Fe protein likely uses the same electron donor for the in vivo assembly processes of both P-cluster and FeMoco, and the optimal in vitro solution potential for these processes (i.e., approximately −440 mV) could serve as a guideline for future identification of this in vivo electron donor.
The Fe protein functions in a similar capacity in the biosynthesis of P-cluster and the turnover of substrates. First, it serves as an ATP-dependent reductase in P-cluster assembly; only in this case, it supplies the electrons for the reduction of Fe atoms of the precursor (Fig. 2A, edge shift) rather than for the reduction of the substrates. Second, it facilitates the structural rearrangement of the MoFe protein during P-cluster assembly; only in this case, it is likely required for positioning the [Fe4S4]-like cluster pairs for the subsequent coupling rather than for satisfying the mechanistic needs of substrate reduction. The interactions between the Fe protein and the MoFe protein during these two processes could also be very much alike, both of which involve the docking of Fe protein on MoFe protein, the conformational rearrangement of both proteins, and the electron transfer between the two proteins. It is interesting to note that the Fe protein-induced conformational changes not only facilitate the formation of P-cluster, but also “open” up the FeMoco site, because FeMoco can be inserted into the MoFe protein only upon the maturation of the P-cluster. Thus, assembly of MoFe protein is a well-coordinated process that requires the precise timing of a series of events.
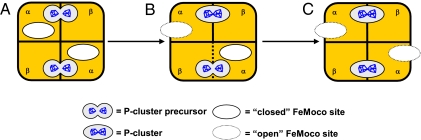
The observation of a biphasic pattern of P-cluster formation in ΔnifH MoFe protein supplies the first dynamic proof for the stepwise assembly mechanism of the two P-clusters in the MoFe protein, i.e., the P-cluster in one αβ dimer is formed before the other in the second αβ dimer (Fig. 4). This “unsynchronized” assembly mechanism was originally proposed based on the “half” P-cluster content (i.e., one P-cluster in one αβ dimer and one precursor in the other) of a ΔnifBΔnifZ MoFe protein, which was obtained upon deletion of nifB (encoding NifB, which is specifically required for FeMoco biosynthesis) and nifZ† (encoding NifZ, which is specifically required for P-cluster biosynthesis) genes (16, 17). Such a “half-assembled” conformation of MoFe protein is once again observed during the maturation of P-cluster in ΔnifH MoFe protein; however, in this case, it is a transient “snapshot” represented by the ΔnifH MoFe protein that is temporarily “stuck” in the “lag” phase (Fig. 1D). Contrary to the maturation of the “second” P-cluster in the ΔnifBΔnifZ MoFe protein, which requires the concerted actions of NifZ and Fe protein (18), formation of both the “first” (before the lag phase) and the “second” (after the lag phase) P-cluster in the ΔnifH MoFe protein only requires the action of Fe protein. This observation is not surprising, because the ΔnifH MoFe protein is expressed in a nifZ-intact background and, therefore, the impact of NifZ on P-cluster formation has already been achieved before the isolation of the ΔnifH MoFe protein. More importantly, the appearance of a distinct lag phase before the formation of the second P-cluster in ΔnifH MoFe protein suggests that the two P-clusters in MoFe protein are indeed formed sequentially. It is likely that, after a fast coupling of the [Fe4S4]-like cluster pair in the first αβ dimer (Fig. 1D, 0–5 min), the MoFe protein undergoes a slow, Fe-protein facilitated conformational rearrangement (Fig. 1D, lag phase, 5–20 min) that positions the [Fe4S4]-like cluster pair in the second αβ dimer in a favorable orientation for the subsequent coupling (Fig. 1D, 20–60 min). Future structural analysis of the various MoFe protein intermediates during P-cluster assembly should provide additional insights into the stepwise assembly mechanism of MoFe protein.
Fig. 4.
Model of the stepwise mechanism of P-cluster formation. Before maturation, the P-cluster species (i.e., precursor) in ΔnifH MoFe protein is composed of a pair of [Fe4S4]-like clusters in each αβ dimer (A). Upon incubation with Fe protein, MgATP and dithionite, the two P-cluster precursors in ΔnifH MoFe protein are matured in a stepwise fashion, i.e., the precursor in one αβ dimer is converted to a fully matured P-cluster (B) before the other in the second αβ dimer (C). Concurrent with the sequential formation of the P-clusters, the FeMoco sites in MoFe protein are converted from a closed conformation to an open one, permitting the subsequent insertion of FeMoco into these sites.
Materials and Methods
Unless noted otherwise, all chemicals and reagents were obtained from Fisher Scientific or Sigma–Aldrich.
Construction of A. vinelandii Variant Strains.
Construction of A. vinelandii nifB- and nifH-deletion strains DJ1143 and DJ1165, which produce His-tagged ΔnifB MoFe protein and ΔnifH MoFe protein, respectively, has been described (7, 19).
Cell Growth and Protein Purification.
All A. vinelandii strains were grown in 180-L batches in a 200-L New Brunswick fermentor (New Brunswick Scientific) in Burke's minimal medium. The medium was also supplemented with 2 mM ammonium acetate. The growth rate was measured by cell density at 436 nm by using a Spectronic 20 Genesys spectrophotometer. Cells were grown to an OD436 of 1.0 and harvested immediately afterwards by using a flow-through centrifugal harvester (Cepa). The cell paste was washed with 50 mM Tris·HCl (pH 8.0). Published methods were used for the purification of His-tagged ΔnifB MoFe protein, His-tagged ΔnifH MoFe protein (7, 20), and wild-type Fe protein (21).
P-cluster Maturation in ΔnifH MoFe Protein.
Maturation of the P-clusters in ΔnifH MoFe protein was modified from a previously published method (18). Incubation mixtures designed to convert the ΔnifH MoFe protein-bound P-cluster precursors to mature P-clusters contained, in a total volume of 100 mL, 25 mM Tris·HCl (pH 8.0), 100 mg of purified ΔnifH MoFe protein, 50 mg of Fe protein, 20 mM Na2S2O4, 0.8 mM ATP, 1.6 mM MgCl2, 10 mM creatine phosphate, and 8 units of creatine phosphokinase. These reaction mixtures were incubated at room temperature for 0, 5, 20, 60, and 120 min, respectively, before the ΔnifH MoFe protein samples were reisolated and analyzed for P-cluster maturation by reconstitution assays, EPR, and XAS/EXAFS spectroscopy (see below).
Reconstitution Assays of MoFe Protein.
Assays designed to reconstitute the preincubated ΔnifH MoFe protein samples contained, in a total volume of 0.8 mL, 25 mM Tris·HCl (pH 8.0), 20 mM Na2S2O4, 0.6 mg of reisolated ΔnifH MoFe protein (see above), and 5 μL of isolated FeMoco (equivalent to 10 nmol FeMoco). These reaction mixtures were incubated at 30 °C for 30 min and determined for enzymatic activities as described (22–24).
EPR Analysis.
All samples for analysis by EPR spectroscopy were prepared in a Vacuum Atmospheres dry box with an oxygen level of <4 ppm. The dithionite-reduced ΔnifH MoFe protein samples were in 25 mM Tris·HCl (pH 8.0), 10% glycerol, and 2 mM Na2S2O4. The indigo disulfonate (IDS)-oxidized ΔnifH MoFe protein samples were prepared by incubation with IDS for 5 min followed by removal of excess IDS by a single passage over an anion exchange column (25). All EPR spectra were recorded with a Bruker ESP 300 Ez spectrophotometer interfaced with an Oxford Instruments ESR-9002 liquid helium continuous-flow cryostat using a microwave power of 50 mW, a gain of 5 × 104, a modulation frequency of 100 kHz, and a modulation amplitude of 5 G. The microwave frequencies of 9.62 and 9.39 GHz were used for the perpendicular-mode (5 scans at 10 K) and parallel-mode (10 scans at 4 K) measurements, respectively. Spin quantitation of EPR signals was performed as described (16).
XAS Data Acquisition.
XAS data for all samples were collected at the 20-pole wiggler biological XAS station BL7–3 at the Stanford Synchrotron Radiation Lightsource under 3 GeV and 80- to 100-mA storage ring operating conditions. Rejection of higher harmonics and vertical collimation was provided by a premonochromator flat bent Rh-coated mirror, while a Si (220) double-crystal monochromator was used for energy selection. Protein samples were loaded into Kapton sheathed Lucite cells, flash-frozen, and preserved under liquid nitrogen (LN2). During XAS measurement, an Oxford Instruments CF1208 liquid-helium continuous-flow cryostat held samples at a constant temperature of 10 K. Fe Kα fluorescence data to k of 16 Å−1 were recorded with a Canberra 30-element solid-state Ge detector array with Soller slits and a Mn filter secured between the sample cryostat and the detector window to attenuate signal intensity from inelastic scattering and Fe Kβ fluorescence. Internal energy calibration was affected by concurrent measurement of the absorption of a Fe foil placed between two ionization chambers located after the sample. The first inflection point of the foil XAS edge was assigned to 7,111.2 eV. No signs of photoreduction of the metal sites, as observed by shifts in edge energy with time, were apparent. A total of 37, 43, 40, 35, and 37 scans were measured for samples preincubated for 0, 5, 20, 60, and 120 min, respectively. Concentrations of samples preincubated for 0, 5, 20, 60, and 120 min are 111, 97, 76, 62, and 84 mg/mL, respectively.
XAS Data Analysis.
After inspection of raw data to remove specious detector channels and scans, average files of all scans for each sample were generated. The average data files for each sample were normalized by using the program PYSPLINE (26) to select control points and fit a first- or second-order polynomial to the preedge region before subtracting this function from the entire data range. The data were normalized to an edge jump of 1.0 at 7,130 eV. A four-region spline function of orders two, three, and three over the postedge region was used to yield EXAFS spectra. EXAFS data over the k range of 2 to 15.7 Å−1 were fit by means of the least-squares fitting program OPT, a component of the EXAFSPAK suite of software, using initial ab initio theoretical phase and amplitude functions calculated from FEFF 7.0 using a starting model based on the 1L5H crystal structure (11, 27, 28). Atomic coordinates from the crystal structure were modulated as fitting progressed to generate more chemically plausible models. During fit optimization, the interatomic distance between the absorbing and backscattering atom (R) and the mean-square thermal and static deviation in R (σ2) were varied for all components. The threshold energy (ΔE0) was allowed to vary for each fit but constrained to the same value for all components. The amplitude reduction factor (S02) was maintained at a value of 1.0 throughout fitting. Coordination numbers (N) were systematically adjusted from crystallographic values to provide the best chemically viable agreement to the EXAFS data and their Fourier transforms. Inclusion or exclusion of various scattering paths was methodically tested to fully explore the average atomic environment at iron (29).
Acknowledgments.
This work was supported by National Institutes of Health Grants GM 67626 (to M.W.R.) and RR 001209 (to K.O.H.) from the National Institute of General Medical Sciences and the National Center for Research Resources. Stanford Synchrotron Radiation Lightsource operations are funded by the Department of Energy Basic Energy Sciences, and the Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the National Institutes of Health National Center for Research Resources Biotechnology Training Program and the Department of Energy Basic Energy Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
nifB encodes for a product that is essential for the biosynthesis of FeMoco but not required for the P-cluster assembly. Thus, deletion of nifB results in the production of a FeMoco-deficient, yet P-cluster intact form of MoFe protein (11).
nifZ encodes for a product that is required for the assembly of the second P-cluster in the α2β2-tetrameric MoFe protein, because deletion of nifZ results in the formation of a half-assembled form of MoFe protein that contains a P-cluster and a FeMoco in one αβ dimer and a P-cluster precursor and no FeMoco in the second αβ dimer (16, 17).
References
- 1.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Einsle O, et al. Nitrogenase MoFe-protein at 1.16-Å resolution: A central ligand in the FeMo cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 3.Hu Y, Fay AW, Lee CC, Yoshizawa J, Ribbe MW. Assembly of nitrogenase MoFe protein. Biochemistry. 2008;47:3973–3981. doi: 10.1021/bi7025003. [DOI] [PubMed] [Google Scholar]
- 4.Zhou HC, Holm RH. Synthesis and reactions of cubane-type iron-sulfur-phosphine clusters, including soluble clusters of nuclearities 8 and 16. Inorg Chem. 2003;42:11–21. doi: 10.1021/ic020464t. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Holm RH. Structural conversions of molybdenum-iron-sulfur edge-bridged double cubanes and PN-type clusters topologically related to the nitrogenase P-cluster. Inorg Chem. 2004;43:674–682. doi: 10.1021/ic030259t. [DOI] [PubMed] [Google Scholar]
- 6.Hlavinka ML, Miyaji T, Staples RJ, Holm RH. Hydroxide-promoted core conversions of molybdenum-iron-sulfur edge-bridged double cubanes: Oxygen-ligated topological PN clusters. Inorg Chem. 2007;46:9192–9200. doi: 10.1021/ic701070w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribbe MW, Hu Y, Guo M, Schmid B, Burgess BK. The FeMoco-deficient MoFe protein produced by a nifH deletion strain of Azotobacter vinelandii shows unusual P-cluster features. J Biol Chem. 2002;277:23469–23476. doi: 10.1074/jbc.M202061200. [DOI] [PubMed] [Google Scholar]
- 8.Corbett MC, et al. Comparison of iron-molybdenum cofactor-deficient nitrogenase MoFe proteins by X-ray absorption spectroscopy: Implications for P-cluster biosynthesis. J Biol Chem. 2004;279:28276–28282. doi: 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
- 9.Broach RB, et al. Variable-temperature, variable-field magnetic circular dichroism spectroscopic study of the metal clusters in the ΔnifB and ΔnifH MoFe proteins of nitrogenase from Azotobacter vinelandii. Biochemistry. 2006;45:15039–15048. doi: 10.1021/bi061697p. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, et al. Nitrogenase reactivity with P-cluster variants. Proc Natl Acad Sci USA. 2005;102:13825–13830. doi: 10.1073/pnas.0506967102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid B, et al. Structure of a cofactor-deficient nitrogenase MoFe protein. Science. 2002;296:352–356. doi: 10.1126/science.1070010. [DOI] [PubMed] [Google Scholar]
- 12.Musgrave KB, Angove HC, Burgess BK, Hedman B, Hodgson KO. All-ferrous titanium(III) citrate reduced Fe protein of nitrogenase: An XAS study of electronic and metrical structure. J Am Chem Soc. 1998;120:5325–5326. [Google Scholar]
- 13.Mansy SS, et al. Crystal structure and stability studies of C77S HiPIP: A serine ligated [4Fe-4S] cluster. Biochemistry. 2002;41:1195–1201. doi: 10.1021/bi011811y. [DOI] [PubMed] [Google Scholar]
- 14.Brereton PS, Verhagen MFJM, Zhou ZH, Adams MWW. Effect of iron-sulfur cluster environment in modulating the thermodynamic properties and biological function of ferredoxin from Pyrococcus furiosus. Biochemistry. 1998;37:7351–7362. doi: 10.1021/bi972864b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshizawa JM, et al. Optimization of FeMoco maturation on NifEN. J Am Chem Soc. 2009;131:9321–9325. doi: 10.1021/ja9035225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Fay AW, Dos Santos PC, Naderi F, Ribbe MW. Characterization of Azotobacter vinelandii nifZ deletion strains: Indication of stepwise MoFe protein assembly. J Biol Chem. 2004;279:54963–54971. doi: 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- 17.Cotton MS, et al. VTVH-MCD study of the ΔnifBΔnifZ MoFe protein from Azotobacter vinelandii. J Am Chem Soc. 2009;131:4558–4559. doi: 10.1021/ja807525m. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Fay AW, Lee CC, Ribbe MW. P-cluster maturation on nitrogenase MoFe protein. Proc Natl Acad Sci USA. 2007;104:10424–10429. doi: 10.1073/pnas.0704297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen J, Goodwin PJ, Lanzilotta WN, Seefeldt LC, Dean DR. Catalytic and biophysical properties of a nitrogenase apo-MoFe protein produced by a nifB-deletion mutant of Azotobacter vinelandii. Biochemistry. 1998;37:12611–12623. doi: 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
- 20.Hu Y, Fay AW, Ribbe MW. Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc Natl Acad Sci USA. 2005;102:3236–3241. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bursey EH, Burgess BK. Characterization of a variant iron protein of nitrogenase that is impaired is its ability to adopt the MgATP-induced conformational change. J Biol Chem. 1998;273:16927–16934. doi: 10.1074/jbc.273.27.16927. [DOI] [PubMed] [Google Scholar]
- 22.Ribbe MW, Burgess BK. The chaperone GroEL is required for the final assembly of the molybdenum-iron protein of nitrogenase. Proc Natl Acad Sci USA. 2001;98:5521–5525. doi: 10.1073/pnas.101119498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess BK, Jacobs DB, Stiefel EI. Large-scale purification of high-activity Azotobacter vinelandii nitrogenase. Biochim Biophys Acta. 1980;614:196–209. doi: 10.1016/0005-2744(80)90180-1. [DOI] [PubMed] [Google Scholar]
- 24.Gavini N, Burgess BK. FeMo cofactor synthesis by a nifH mutant with altered MgATP reactivity. J Biol Chem. 1992;267:21179–21186. [PubMed] [Google Scholar]
- 25.Bursey EH, Burgess BK. The role of methionine 156 in cross-subunit nucleotide interactions in the iron protein of nitrogenase. J Biol Chem. 1998;273:29678–29685. doi: 10.1074/jbc.273.45.29678. [DOI] [PubMed] [Google Scholar]
- 26.Tenderholt A. PYSPLINE. Stanford, CA: Stanford Synchrotron Radiation Laboratory; 2006. [Google Scholar]
- 27.George GN. EXAFSPAK. Stanford, CA: Stanford Synchrotron Radiation Laboratory; 1990. [Google Scholar]
- 28.Rehr JJ. FEFF. Seattle: University of Washington Department of Physics; 2004. [Google Scholar]
- 29.Pickering IJ, et al. X-ray absorption spectroscopy of cuprous-thiolate clusters in proteins and model systems. J Am Chem Soc. 1993;115:9498–9505. [Google Scholar]
- 30.Scott RA. Measurement of metal-ligand distances by EXAFS. Methods Enzymol. 1985;117:414–459. [Google Scholar]
- 31.Zhang HH, Hedman B, Hodgson KO. In: Inorganic Electronic Structure and Spectroscopy, Volume I: Methodology. Solomon EI, Lever ABP, editors. New York: Wiley; 1999. pp. 513–554. [Google Scholar]