Abstract
Reactive oxygen intermediates generated by neutrophils kill bacteria and are implicated in inflammatory tissue injury, but precise molecular targets are undefined. We demonstrate that neutrophils use myeloperoxidase (MPO) to convert methionine residues of ingested Escherichia coli to methionine sulfoxide in high yield. Neutrophils deficient in individual components of the MPO system (MPO, H2O2, chloride) exhibited impaired bactericidal activity and impaired capacity to oxidize methionine. HOCl, the principal physiologic product of the MPO system, is a highly efficient oxidant for methionine, and its microbicidal effects were found to correspond linearly with oxidation of methionine residues in bacterial cytosolic and inner membrane proteins. In contrast, outer envelope proteins were initially oxidized without associated microbicidal effect. Disruption of bacterial methionine sulfoxide repair systems rendered E. coli more susceptible to killing by HOCl, whereas over-expression of a repair enzyme, methionine sulfoxide reductase A, rendered them resistant, suggesting a direct role for methionine oxidation in bactericidal activity. Prominent among oxidized bacterial proteins were those engaged in synthesis and translocation of peptides to the cell envelope, an essential physiological function. Moreover, HOCl impaired protein translocation early in the course of bacterial killing. Together, our findings indicate that MPO-mediated methionine oxidation contributes to bacterial killing by neutrophils. The findings further suggest that protein translocation to the cell envelope is one important pathway targeted for damage.
Keywords: bactericidal, microbicidal
Neutrophils (polymorphonuclear leukocytes [PMN]), a key component of the innate immune response, engulf and kill invading microorganisms. Microbes ingested by neutrophils are exposed to a variety of antimicrobial systems, including myeloperoxidase (MPO), H2O2, and a halide (1–3). MPO released into the phagosome reacts with H2O2 generated by NADPH oxidase to convert the halide to the corresponding hypohalous acid. One important product is hypochlorous acid (HOCl), the primary product of chloride oxidation (4–6).
HOCl is a powerful oxidant that attacks microorganisms in a variety of ways (7, 8). Initial studies concentrated on halogenation in which the halide is bound in covalent linkage, generally to tyrosine residues of proteins (9–12). Although halogenation of tyrosine is a convenient marker of the action of the MPO–H2O2–halide system, the reaction is inefficient and there is no evidence that halogenation per se is toxic to the ingested microorganism. The sulfur-containing amino acids methionine and cysteine, and protein iron–sulfur clusters are much more reactive with HOCl (13, 14). Of these, methionine oxidation produces a stable product, methionine sulfoxide [Met(O)], that can be detected readily by mass spectrometry, as an increase in 16 atomic mass units. Thus, methionine oxidation might serve as a sensitive and stable marker for proteins oxidized by MPO.
In the current studies, we used liquid chromatography coupled with electron spray ionization and tandem mass spectrometry (LC-ESI-MS/MS) to identify methionine residues susceptible to oxidation in Escherichia coli and to investigate the role of methionine oxidation in the microbicidal actions of PMN, HOCl, and the MPO system. Our observations implicate methionine oxidation as a contributor to bacterial killing by the MPO system and demonstrate that Met(O) formation is a sensitive and specific marker for the activity of MPO in PMN. The findings further suggest a microbicidal mechanism: oxidative inhibition of pathways that translocate proteins from cytoplasm to the cell envelope.
Results
Neutrophils Oxidize Bacterial Methionines by MPO-Dependent Pathways.
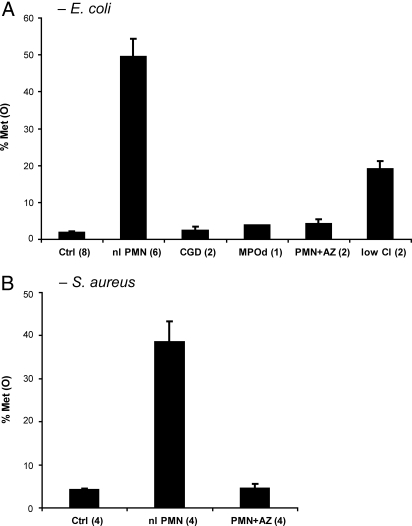
PMN oxidized 50% ± 5% (mean ± SEM, n = 6) of recovered E. coli methionines during a 1-h incubation, under conditions that support efficient phagocytosis (Fig. 1A). Control conditions, omitting serum and divalent cations, were associated with negligible phagocytosis and Met(O) levels of 2% ± 0.4%. Neutrophils lacking in either MPO [4% Met(O)] or the capacity to generate H2O2 [2% ± 1% Met(O)] failed to oxidize methionine above baseline. Similarly, normal PMN incubated with the MPO inhibitor, 0.1 mM sodium azide [4% ± 1% Met(O)] or in chloride-depleted medium [19% ± 2% Met(O)] exhibited a diminished capacity to oxidize bacterial methionine residues. The absence of functional MPO or its cofactors also associated with decreased bacterial killing (Table 1), indicating that MPO contributes to the PMN microbicidal effect under these conditions. Interestingly, the inhibition of PMN microbicidal activity by azide was greater than what was observed in the simple absence of MPO or H2O2, suggesting that, even at these low doses (10−4 M), azide effects go beyond simple inhibition of MPO activity. In similar studies using Staphylococcus aureus (strain 502A), a Gram-positive organism, very similar effects on bacterial methionine oxidation and PMN microbicidal activity were observed (Fig. 1B and Table 1).
Fig. 1.
Neutrophil-mediated oxidation of bacterial methionines. Bacteria were incubated with neutrophils for 1 h, recovered, and analyzed by LC-ESI-MS/MS. The fraction of bacterial methionine residues in peptides recovered as sulfoxides was monitored as M + 16. Abbreviations: Ctrl, PMN—nonphagocytic conditions (serum and divalent cations omitted [all PMN donors combined]); nl PMN, normal donor PMN—phagocytic conditions (serum and divalent cations present); CGD, neutrophils defective in H2O2 production (isolated from an individual with chronic granulomatous disease, deficient in NADPH oxidase); MPOd, PMN lacking MPO (from an individual with complete hereditary MPO deficiency); PMN+AZ, normal PMN supplemented with, 10−4 M sodium azide, an MPO inhibitor; low Cl, normal PMN, chloride salts in suspension medium replaced with gluconates. Legend parentheses indicate replicates for each condition. (A) Escherichia coli, strain ATCC11775. (B) Staphylococcus aureus, strain 502A.
Table 1.
Polymorphonuclear leukocyte (PMN) microbicidal activity
| PMN | Escherichia coli | Staphylococcus aureus |
|---|---|---|
| Ctrl | 116% ± 12% (8) | 115% ± 15% (4) |
| nl PMN | 15% ± 4% (6) | 16% ± 4% (4) |
| CGD | 31% ± 17% (2) | |
| MPOd | 31% (1) | |
| PMN + AZ | 86% ± 14% (2) | 60% ± 16% (4) |
| Low Cl | 41% ± 11% (2) |
Ctrl, PMN—nonphagocytic conditions (serum and divalent cations omitted [all PMN donors combined]); nl PMN, normal donor PMN—phagocytic conditions (serum and divalent cations present); CGD, neutrophils defective in H2O2 production (isolated from an individual with chronic granulomatous disease, deficient in NADPH oxidase); MPOd, PMN lacking MPO (from an individual with complete hereditary MPO deficiency); PMN+AZ, normal PMN supplemented with, 10−4 M sodium azide, an MPO inhibitor; Low Cl, normal PMN, chloride salts in suspension medium replaced with gluconates. Bacteria were incubated with PMN under conditions described in Fig. 1. Viability was determined by comparing colony counts after 60 min to those immediately after addition of PMN. Results are the mean ± SEM of (n) determinations for n > 2 and mean ± one-half range for n = 2.
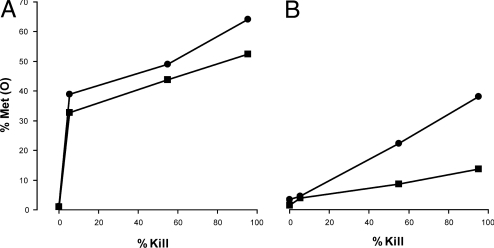
Oxidation of Bacterial Protein Methionine Residues Correlates with Microbicidal Effect of HOCl and MPO.
Fig. 2 compares the oxidation of microbial methionine residues to the loss of viability on exposure of organisms to HOCl and the MPO–H2O2–chloride system. Met(O) increased and viability fell as the HOCl concentration increased from 0 to 200 μM (Fig. 2A). For example, 150 micromolar HOCl produced a 55% loss of viability and oxidized 22% of protein methionine residues. There was no loss of viability and 6% Met(O) in control samples (0 μM HOCl). H2O2 at 50 millimolar produced a 49% loss of viability without any increment in methionine oxidation. Figure 2B compares the loss of viability and the increase in methionine oxidation upon exposure of E. coli to the MPO–H2O2–chloride system. The MPO concentration ranged from 0 to 34 nM, and H2O2 was generated by a glucose plus glucose-oxidase system at an initial rate of 40 μM/min. HOCl formation for the 15-min incubations was measured separately by chlorination of taurine and was in the same range as that shown in Fig. 2A. Exposure of organisms to the 34-nM MPO system (which generated 230 μM HOCl in 15 min) killed 99% of the inoculum and oxidized 15% of the bacterial methionine residues (Fig. 2B). When either MPO or the glucose–glucose-oxidase H2O2 system was omitted or when chloride was replaced by isotonic sodium sulfate, there was no increase in protein methionine oxidation, nor was there loss of viability.
Fig. 2.
Bactericidal activity and protein methionine oxidation by HOCl and an MPO, H2O2, chloride system. The E. coli strain was CFT073. For HOCl (A), the reaction was in PBS, pH 7.4. An equal volume of oxidant, at twice the indicated final concentration, was rapidly mixed with 4 × 109 bacterial cells, and after 2–5 s the reaction was quenched with 33 mM methionine. For the MPO system (B), the reaction was in 40 mM sodium phosphate pH 7.0 containing 10 mM glucose and 0.1 M NaCl. MPO, at concentrations of 0–34 nM, was preincubated with bacteria for 3–5 min. The reaction was started by addition of glucose oxidase sufficient to generate H2O2 at an initial rate of 40 μM/min, and the reaction was incubated for 15 min with vigorous agitation, before quenching with 33 mM methionine. HOCl formation by MPO (x axis, B) was estimated as taurine chlorination (μM/15 min). Bacterial viability (diamonds, % viability) and protein methionine and methionine sulfoxide [circles, Met(O)%] were estimated by LC-ESI-MS/MS and spectral counting.
Cell Location Affects Bacterial Protein Susceptibility to Oxidation.
Is methionine oxidation by HOCl and/or by the MPO antimicrobial system stochastic or are some proteins particularly susceptible? Figure 3 categorizes methionine oxidation by subcellular location and compares the extent of Met(O) formation to loss of viability. As a class, methionine residues of the outer membrane and periplasm were oxidized by either HOCl or MPO to a substantial degree (30–40%) before the onset of a major decline in bacterial viability (Fig. 3A). Outer membrane proteins were more susceptible, showing 55%–60% Met(O) at 95% bacterial viability, than were periplasmic proteins, showing 25%–35% Met(O). For outer envelope proteins, methionine oxidation by HOCl and by the MPO enzyme system was similar. In contrast to the outer cell envelope, inner membrane, cytosolic, and unclassified proteins were oxidized linearly proportionate to bacterial killing (Fig. 3B). For these proteins, HOCl-mediated oxidations were more extensive than MPO-mediated oxidations.
Fig. 3.
Protein location affects susceptibility to oxidation by HOCl and an MPO system. Methionine oxidation plotted against extent of bacterial killing for HOCl (circles) and an MPO system (squares). (A) Cell envelope proteins: outer membrane and periplasm. (B) All other proteins (inner membrane, cytoplasm, and location not specified). Data are from experiments described in Fig. 2. Proteins were assigned to cellular locations as described in Materials and Methods.
Methionine Sulfoxide Reductases Modulate Susceptibility to HOCl-Mediated Microbicidal Effects.
Although methionine residues are prominent targets for HOCl, they are not the sole target. We estimate that, on average, 20% (MPO system) to 40% (reagent HOCl) of HOCl is consumed by methionine oxidation (based on 135 nmol methionine per 109 E. coli cells [see SI]), leaving up to 60–80% of HOCl available for other oxidation reactions.
We therefore asked whether methionine oxidation in E. coli serves merely as a marker for the presence and penetration of HOCl into the cell or contributes directly to the microbicidal effect. The approach was to evaluate the HOCl susceptibility of bacteria modified for expression of the Met(O)–reducing repair enzymes msrA and msrB. Figure 4 compares the microbicidal effect of HOCl on wild-type E. coli CFT073 and a double mutant deficient in msrA and msrB. In addition, we evaluated the msrA msrB strain upon complementation with a high-copy msrA-expressing plasmid. HOCl at 125, 150, and 200 μM was toxic to a 109/mL suspension of wild-type E. coli. A significant increase in microbicidal activity was observed with the double mutant (P < 0.05, analysis of variance [ANOVA]). The hypersensitivity of the msrA msrB double mutant was more than reversed by complementation with a high–copy number plasmid expressing msrA at 26 times normal levels (15) (P < 0.05 compared with wild type). That methionine sulfoxide repair capacity is an arbiter of HOCl toxicity implies a direct contribution of methionine oxidation to the microbicidal effects of MPO and HOCI.
Fig. 4.
Increased susceptibility of E. coli deficient in methionine sulfoxide reductases A and B (msrA msrB) to microbicidal effects of HOCl. HOCl in PBS, at twice the concentration indicated in the figure, was rapidly injected into an equal volume of E. coli in PBS, pH 7.4. Residual microbial viability was determined by plating dilutions on Mueller Hinton agar. The E. coli strains were wild type, CFT073 (closed circles), an msrA msrB double mutant (closed squares), and the mutant complemented with an msrA-expressing plasmid (open squares). Results are the geometric mean of six independent experiments. Error bars reflect the SEM of log-transformed data.
HOCl Oxidation Impairs Essential Protein Translocation Functions.
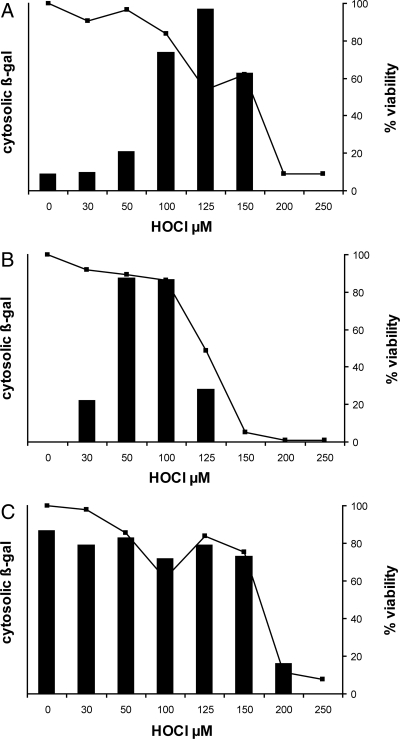
Among the more prominently oxidized cytosolic and inner membrane peptides were several that contribute to the synthesis of cell envelope proteins and export to the cell envelope. The main pathways of export are the general secretory (secA) and signal recognition particle (SRP) pathways. They converge at a highly conserved translocation complex in the inner membrane secYEG. We therefore evaluated the effect of HOCl on the physiology of protein translocation through the secA and SRP pathways as described in Fig. 5. In this system, failure to export an inducible β-galactosidase (lacZ) fusion protein is accompanied by an increase in cytoplasmic β-galactosidase enzyme activity.
Fig. 5.
HOCl effects on protein translocation from cytoplasm to cell envelope. (A) E. coli strain EC626 (MC4100 Φ(lamB-lacZ) Hyb 42–1), which synthesizes a fusion protein that is translocated through the secA pathway, was exposed to HOCl at the indicated concentrations and then induced to synthesize its normally translocated β-gal fusion protein as described in Materials and Methods. Failure to translocate the protein resulted in accumulation of cytoplasmic β-galactosidase activity (bars). Bacterial viability at each HOCl concentration is shown as a line graph (squares, right y axis). (B) Oxidation conditions as for (A) except that the E. coli strain was EC628 (MC4100 Φ(H*lamB‘-’lacZ), H*lamB [diploid]), which synthesizes a fusion protein that is translocated through the signal recognition particle pathway. Induction and β-gal assay conditions were modified to detect the lower overall fusion protein levels in this strain (see Materials and Methods). (C) Oxidation conditions as for (A) except that the E. coli strain was EC627 (MC4100 Φ(lamBΔ60-lacZ) Hyb 42–1), which synthesizes a fusion protein that is predominantly cytosolic (not translocated) under all conditions. This strain serves as an indicator of bacterial capacity, upon HOCl oxidation, to sense an inducing stimulus and respond with new protein synthesis. Each panel reflects a representative experiment from at least three replicates. Maximal ONPG hydrolysis in (A–C) (A420-1.75*A550; see Materials and Methods), was 0.097, 0.210, and 0.083 units, respectively.
Figure 5A describes the HOCl-induced, cytosolic accumulation of a fusion protein that is targeted by its leader sequence to be exported by the secA pathway. At baseline (0 HOCl), bacterial viability is 100% (≈109 cfu/mL), and most of the induced β-gal protein is exported (minimal to no enzyme activity). As the HOCl concentration increases (30–125 μM), viability begins to fall, and the export-targeted protein is confined to the cytoplasm, where ambient conditions cause it to be enzymatically active. At still higher concentrations of HOCl (200–250 μM), oxidation is so extensive that bacteria can no longer respond to the inducing stimulus. Further studies, with a related fusion protein, targeted to be exported by the SRP pathway, gave similar results (Fig. 5B). Thus both the secA and SRP export pathways are inhibited early in the course of HOCl toxicity. Figure 5C illustrates the result with a third fusion construct that has a defective leader sequence so that the protein is largely confined to the cytosol under all conditions. The constant level of β-galactosidase activity in this strain indicates that protein induction and synthesis is constant under these conditions until HOCl oxidation is very extensive and killing exceeds 30%. The secA and SRP export pathways converge on the secYEG translocon, suggesting that this critical site is a likely target for early inactivation by HOCl.
Discussion
A primary role for PMN is to ingest and kill microbes. Upon phagocytosis, antimicrobial factors concentrate in a membrane-surrounded compartment containing the ingested microbe, antimicrobial proteins, and oxidants generated by an integral membrane NADPH oxidase and by MPO. The exact role of MPO-derived oxidants in microbicidal processes remains unclear, and the cellular targets of oxidants are poorly understood. MPO has even been proposed to protect bacteria by converting peroxide to less toxic oxidants (16, 17). Because methionine is readily oxidized by HOCl, and because oxidation produces predominantly a single stable product, Met(O), we used LC-ESI-MS/MS to quantitatively evaluate the neutrophil-dependent of oxidation of bacterial proteins.
Human PMN incubated with opsonized bacteria for 1 h oxidized 40–50% of all recovered bacterial methionine residues to Met(O) (Fig. 1). Efficient Met(O) formation required phagocytosis, MPO, and a product of the phagocyte NADPH oxidase, likely H2O2. Depletion of chloride from the incubation medium significantly reduced Met(O) formation and low concentrations of sodium azide, an MPO inhibitor, reduced Met(O) to baseline. Taken together the data indicate that, under these conditions, Met(O) formation by PMN is dependent upon MPO. Although other oxidants, such as superoxide anion and H2O2, are present in activated PMN, their reactivity with methionine is extraordinarily low, 3 M−1·s−1 for superoxide (18) and 0.02 M−1·s−1 for H2O2 (19), compared with 3.8 × 107 M−1·s−1 for HOCl (20), providing a kinetic explanation for the nearly exclusive contribution of MPO to Met(O) formation. The data thus indicate that formation of bacterial Met(O) is an excellent and specific measure for the presence and activity of the MPO-mediated antimicrobial system in the human PMN phagosome.
To better understand the effects of the MPO system and its product HOCl on bacteria, we evaluated methionine oxidation in E. coli, comparing the location and extent of Met(O) formation to the microbicidal effect. When sites of Met(O) formation were categorized by cellular compartment, it became evident that both HOCl and the MPO system oxidized methionine residues of the outer membrane and periplasm to a similar extent (Fig. 3A). Also, many methionine residues in the outer membrane and periplasm were oxidized early, with little impact on bacterial viability. Thus, among other functions, these residues consume HOCl and may serve to shield intracellular and inner membrane proteins from oxidative damage.
The detection of Met(O) from PMN and HOCl oxidations does not, by itself, establish methionine oxidation as a cause of viability loss. However, the increased sensitivity to HOCl of an E. coli strain lacking msrA and msrB, and the greater than wild-type resistance when the mutant was complemented with an msrA over-expressing plasmid (15), suggest that methionine oxidation does contribute to bacterial killing (Fig. 4).
MS/MS analysis of oxidized proteins suggested that the general secretion (secA, ATP-dependent) and signal recognition particle (SRP, GTP-dependent) pathways of protein export were important targets for oxidation by MPO and HOCl. The export pathways are essential for translocating outer membrane and periplasmic (secA) or inner membrane (SRP) proteins to their final destinations after synthesis in the cytoplasm. The two export pathways converge at the inner face of the cytoplasmic membrane at a gated membrane channel, secYEG (21).
We monitored HOCl effects on protein translocation using fusion proteins targeted (i) for the secA export pathway, (ii) for the SRP export pathway, or (iii) for no export (22–24) (Fig. 5). The appearance of enzyme activity after inducing fusion protein synthesis reflected failure to translocate, as extracytoplasmic conditions are unfavorable for proper enzyme folding and preclude protein detection by enzyme assay (25). We observed that HOCl induced early impairment of protein export for both secA (Fig. 5A) and SRP (Fig. 5B) pathways, whereas the capacity of bacteria to sense an inducing stimulus and to respond with new protein synthesis remained unimpaired until oxidations were well advanced (Fig. 5C).
These observations prompt the hypothesis that HOCl exerts part of its microbicidal effect by impairing function of the essential secYEG translocon. Impairment of secYEG could occur by depletion of energy sources (26), by oxidation of vulnerable amino acids such as methionine or cysteine (20), or by cross-linking secYEG to peptide chains in process of translocation (27), thereby jamming the channel (25, 28).
In summary, we have shown that HOCl, generated by MPO, targets methionine residues in proteins of phagocytosed bacteria for oxidation, and that this reaction occurs in near quantitative yield. Formation of Met(O) is strongly associated with bacterial killing, and modulation of an enzymatic repair system for Met(O) modulated the microbicidal effects of HOCl, strongly suggesting that methionine oxidation plays a role in bacterial killing. Finally, HOCl mediates a protein translocation defect that may contribute directly to loss of microbial viability.
Materials and Methods
Bacterial Strains.
Bacterial strains were maintained at −80 °C in 60% Luria Broth (LB), 20% glycerol. E. coli strains were ATCC11775 (EC1) and CFT073 (29, 30). The MsrA MsrB double mutant of E. coli strain CFT073 was constructed by lambda red recombination as described in SI Text. Where indicated, the double mutant was complemented with a high copy plasmid, pAR100, containing msrA from K12 strain MC1061.
Myeloperoxidase.
Myeloperoxidase was prepared as previously described (32) and stored at −80 °C. HOCl formation by MPO was estimated by replacing bacteria with 3 mM taurine and measuring the absorbance of taurine chloramine at 252 nm (ε = 496 M−1·cm−1) (33) after correction for absorbance by glucose oxidase and MPO.
HOCl and H2O2 Oxidations.
Overnight cultures of bacteria in Mueller-Hinton broth were washed and suspended in phosphate-buffered saline (PBS) to 4 × 109 cells per mL. Oxidant (HOCl or H2O2) at twice the desired final concentration in PBS was rapidly mixed with an equal volume of bacterial suspension, followed by quenching with methionine (≈9 mM final). Where indicated in figure legends, bacteria were incubated for 20 min at 37° in PBS/HOCl before quenching. Microbial viability was determined and samples were processed for LC-ESI-MS/MS analysis.
MPO Oxidations.
Components of the reaction mixture (40 mM phosphate, pH 7.0, 10 mM glucose, 100 mM sodium chloride, bacteria and MPO, at concentrations indicated in figure and table legends) were warmed in a 50-ml sterile polypropylene tube at 37°. The oxidation reaction was initiated by addition of glucose oxidase and terminated after 15 min by addition of 1/5 volume of 200 mM methionine.
Neutrophils.
Neutrophils were isolated from venous blood (obtained by an Institutional Review Board–approved protocol) from normal and MPO-deficient individuals and individuals with chronic granulomatous disease as previously described (34). Overnight, trypticase soy broth cultures of EC1 were washed in 0.1 M Na2SO4 and incubated for 30 min at 37 °C, at 1–2 × 109 cfu/mL, in 1.5 mL Hank's balanced salt solution (HBSS) supplemented with 10% fresh human serum, 4 mM CaCl2, and 2 mM MgCl2. For control incubations, serum and divalent cations were omitted. For experiments using diminished levels of chloride, chloride-free HBSS (replacing chloride salts with gluconate) was used to wash and suspend bacteria and PMN. Neutrophils were warmed to 37 °C for 5 min before the addition of an equal volume to the opsonized bacteria. Incubations were tumbled end over end in 4.5-ml snap-cap polypropylene tubes. After 10 min, samples were obtained to assess phagocytosis by cytospin and safranin staining as previously described (34). Omission of serum and divalent cations resulted in negligible phagocytosis of ATCC11775 E. coli.
Samples to determine microbial viability were obtained immediately upon addition of PMN and after 1 h of incubation. The remaining ≈3 mL were diluted 10-fold into 0.1% sodium dodecyl sulfate (SDS), 2 mM methionine, pH 11, to lyse PMN (35). The lysate was sheared by three passes through an 18-G needle and centrifuged over a layer of 24% sucrose, 2 mM methionine. The pellet was washed twice and frozen in 2 mM methionine for subsequent extraction of bacterial proteins.
LC-ESI-MS/MS.
Bacteria were disrupted and fractionated as described in SI Text. Samples were then digested with trypsin and analyzed by LC-ESI-MS/MS, using spectral counting to quantitate results as described in SI Text. Met(O) was detected as Met + 16 in the MS/MS spectra.
Induction of LamB-lacZ Fusion Proteins After HOCl Oxidation.
Strains (28) were grown overnight in 30 mL LB at 37° with end-over-end tumbling, washed once, and resuspended to one-fourth of the original volume in PBS. Bacteria were rapidly mixed with an equal volume of HOCl, at twice the desired final concentration in PBS and promptly quenched with one-twentieth volume of 0.2 M methionine. After oxidation, strains were induced to synthesize fusion protein, as described in SI Text, and then permeabilized and assayed for β-galactosidase (β-gal).
β-Galactosidase Assay.
A 40-μL quantity of induced bacteria was permeabilized with hexadecyl trimethyl ammonium bromide (CTAB) permeabilization buffer and transferred to o-nitrophenyl galactoside (ONPG) assay buffer for 90 min as described in SI Text. The reaction was stopped with 60 μl 1 M Na2CO3, and color development was recorded as A420-1.75*A550 in a microtiter plate reader (Molecular Devices, SpectraMax Plus).
Statistical Analysis.
ANOVA, t tests, and multiple comparisons were performed with SigmaStat. Microbial viability data were log transformed to better fit normality assumptions. For duplicate determinations, error bars reflect one-half of the range; otherwise they reflect the standard error of the mean. Statistical significance was set at α < 0.05.
Supplementary Material
Acknowledgments.
The authors thank Professor Thomas J. Silhavy and his colleagues at Princeton University for the gift of E. coli lamB-lacZ fusion strains and helpful advice. We acknowledge the expert technical contributions of Patrick Lewis and Angela Irwin and gratefully acknowledge support from National Institutes of Health (NIH) grants AI25606 (to H.R.), HL075381 NIH (to X.F.), P30 DK017047, HL030086 (to J.W.H.), and HL086798 (to J.W.H.); and from the University of Washington and DERC Mass Spectrometry Core.
Note Added in Proof.
Another potential mechanism for HOCl-mediated impairment of secYEG translocation would be induced degradation of secY by ftsH protease (38).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909464106/DCSupplemental.
References
- 1.Nauseef WM. How human neutrophils kill and degrade microbes: An integrated view. Immunol Rev. 2007;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 2.Klebanoff SJ. Myeloperoxidase: Friend and foe. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 3.Klebanoff SJ, Clark RA. The Neutrophil. Amsterdam: North-Holland Publishing; 1978. [Google Scholar]
- 4.Harrison JE, Schultz J. Studies on the chlorinating activity of myeloperoxidase. J Biol Chem. 1976;251:1371–1374. [PubMed] [Google Scholar]
- 5.Chapman AL, Hampton MB, Senthilmohan R, Winterbourn CC, Kettle AJ. Chlorination of bacterial and neutrophil proteins during phagocytosis and killing of Staphylococcus aureus. J Biol Chem. 2002;277:9757–9762. doi: 10.1074/jbc.M106134200. [DOI] [PubMed] [Google Scholar]
- 6.Rosen H, Crowley JR, Heinecke JW. Human neutrophils use the myeloperoxidase-hydrogen peroxide-chloride system to chlorinate but not nitrate bacterial proteins during phagocytosis. J Biol Chem. 2002;277:30463–30468. doi: 10.1074/jbc.M202331200. [DOI] [PubMed] [Google Scholar]
- 7.Camper AK, McFeters GA. Chlorine injury and the enumeration of waterborne coliform bacteria. Appl Environ Microbiol. 1979;37:633–641. doi: 10.1128/aem.37.3.633-641.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurst JK, Barrette WC., Jr Leukocytic oxygen activation and microbicidal oxidative toxins. CRC Crit Rev Biochem Mol Biol. 1989;24:271–328. doi: 10.3109/10409238909082555. [DOI] [PubMed] [Google Scholar]
- 9.Klebanoff SJ. Iodination of bacteria: A bactericidal mechanism. J Exp Med. 1967;126:1063–1078. doi: 10.1084/jem.126.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klebanoff SJ, White LR. Iodination defect in the leukocytes of a patient with chronic granulomatous disease of childhood. N Engl J Med. 1969;280:460–466. doi: 10.1056/NEJM196902272800902. [DOI] [PubMed] [Google Scholar]
- 11.Domigan NM, Charlton TS, Duncan MW, Winterbourn CC, Kettle AJ. Chlorination of tyrosyl residues in peptides by myeloperoxidase and human neutrophils. J Biol Chem. 1995;270:16542–16548. doi: 10.1074/jbc.270.28.16542. [DOI] [PubMed] [Google Scholar]
- 12.Hazen SL, Hsu FF, Mueller DM, Crowley JR, Heinecke JW. Human neutrophils employ chlorine gas as an oxidant during phagocytosis. J Clin Invest. 1996;98:1283–1289. doi: 10.1172/JCI118914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Pattison DI, Davies MJ. Reactions of myeloperoxidase-derived oxidants with biological substrates: Gaining chemical insight into human inflammatory diseases. Curr Med Chem. 2006;13:3271–3290. doi: 10.2174/092986706778773095. [DOI] [PubMed] [Google Scholar]
- 15.Rahman MA, Nelson H, Weissbach H, Brot N. Cloning, sequencing, and expression of the Escherichia coli peptide methionine sulfoxide reductase gene. J Biol Chem. 1992;267:15549–15551. [PubMed] [Google Scholar]
- 16.Reeves EP, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 17.Reeves EP, Nagl M, Godovac-Zimmermann J, Segal AW. Reassessment of the microbicidal activity of reactive oxygen species and hypochlorous acid with reference to the phagocytic vacuole of the neutrophil granulocyte. J Med Microbiol. 2003;52:643–651. doi: 10.1099/jmm.0.05181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bielski BH, Shiue GG. Reaction rates of superoxide radicals with the essential amino acids. Ciba Found Symp. 1978;65:43–56. doi: 10.1002/9780470715413.ch4. [DOI] [PubMed] [Google Scholar]
- 19.Pan B, et al. Comparative oxidation studies of methionine residues reflect a structural effect on chemical kinetics in rhG-CSF. Biochemistry. 2006;45:15430–15443. doi: 10.1021/bi061855c. [DOI] [PubMed] [Google Scholar]
- 20.Pattison DI, Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 21.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 22.Bieker KL, Silhavy TJ. PrlA (SecY) and PrlG (SecE) interact directly and function sequentially during protein translocation in E. coli. Cell. 1990;61:833–842. doi: 10.1016/0092-8674(90)90193-i. [DOI] [PubMed] [Google Scholar]
- 23.Emr SD, Silhavy TJ. Mutations affecting localization of an Escherichia coli outer membrane protein, the bacteriophage lambda receptor. J Mol Biol. 1980;141:63–90. doi: 10.1016/s0022-2836(80)80029-5. [DOI] [PubMed] [Google Scholar]
- 24.Silhavy TJ, Shuman HA, Beckwith J, Schwartz M. Use of gene fusions to study outer membrane protein localization in Escherichia coli. Proc Natl Acad Sci USA. 1977;74:5411–5415. doi: 10.1073/pnas.74.12.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder WB, Silhavy TJ. Beta-galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J Bacteriol. 1995;177:953–963. doi: 10.1128/jb.177.4.953-963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrette WC, Jr, Hannum DM, Wheeler WD, Hurst JK. General mechanism for the bacterial toxicity of hypochlorous acid: Abolition of ATP production. Biochemistry. 1989;28:9172–9178. doi: 10.1021/bi00449a032. [DOI] [PubMed] [Google Scholar]
- 27.Fu X, Mueller DM, Heinecke JW. Generation of intramolecular and intermolecular sulfenamides, sulfinamides, and sulfonamides by hypochlorous acid: A potential pathway for oxidative cross-linking of low-density lipoprotein by myeloperoxidase. Biochemistry. 2002;41:1293–1301. doi: 10.1021/bi015777z. [DOI] [PubMed] [Google Scholar]
- 28.Bowers CW, Lau F, Silhavy TJ. Secretion of LamB-LacZ by the signal recognition particle pathway of Escherichia coli. J Bacteriol. 2003;185:5697–5705. doi: 10.1128/JB.185.19.5697-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JR, Clabots C, Rosen H. Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1:K1:H7 in a mouse model of ascending urinary tract infection. Infect Immun. 2006;74:461–468. doi: 10.1128/IAI.74.1.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickstein DD, et al. Isolation and characterization of the receptor on human neutrophils that mediates cellular adherence. J Biol Chem. 1987;262:5576–5580. [PubMed] [Google Scholar]
- 32.Rakita RM, Michel BR, Rosen H. Differential inactivation of Escherichia coli membrane dehydrogenases by a myeloperoxidase-mediated antimicrobial system. Biochemistry. 1990;29:1075–1080. doi: 10.1021/bi00456a033. [DOI] [PubMed] [Google Scholar]
- 33.Kooter IM, et al. The sulfonium ion linkage in myeloperoxidase. Direct spectroscopic detection by isotopic labeling and effect of mutation. J Biol Chem. 1999;274:26794–26802. doi: 10.1074/jbc.274.38.26794. [DOI] [PubMed] [Google Scholar]
- 34.Staudinger BJ, Oberdoerster MA, Lewis PJ, Rosen H. mRNA expression profiles for Escherichia coli ingested by normal and phagocyte oxidase-deficient human neutrophils. J Clin Invest. 2002;110:1151–1163. doi: 10.1172/JCI15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decleva E, Menegazzi R, Busetto S, Patriarca P, Dri P. Common methodology is inadequate for studies on the microbicidal activity of neutrophils. J Leukoc Biol. 2006;79:87–94. doi: 10.1189/jlb.0605338. [DOI] [PubMed] [Google Scholar]
- 36.Ingraham JL, Maaloe O, Neidhardt FC. Growth of the Bacterial Cell. Sunderland, MA: Sinauer Associates; 1983. pp. 1–48. [Google Scholar]
- 37.Cox RA. Quantitative relationships for specific growth rates and macromolecular compositions of Mycobacterium tuberculosis, Streptomyces coelicolor A3(2) and Escherichia coli B/r: An integrative theoretical approach. Microbiology. 2004;150:1413–1426. doi: 10.1099/mic.0.26560-0. [DOI] [PubMed] [Google Scholar]
- 38.van Stelten J, Silva F, Belin D, Silhavy TJ. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science. 2009;325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.