Abstract
Animal development is remarkably robust; cell fates are specified with spatial and temporal precision despite physiological and environmental contingencies. Favorable conditions cause Caenorhabditis elegans to develop rapidly through four larval stages (L1–L4) to the reproductive adult. In unfavorable conditions, L2 larvae can enter the developmentally quiescent, stress-resistant dauer larva stage, enabling them to survive for prolonged periods before completing development. A specific progression of cell division and differentiation events occurs with fidelity during the larval stages, regardless of whether an animal undergoes continuous or dauer-interrupted development. The temporal patterning of developmental events is controlled by the heterochronic genes, whose products include microRNAs (miRNAs) and regulatory proteins. One of these proteins, the DAF-12 nuclear hormone receptor, modulates the transcription of certain let-7-family miRNAs, and also mediates the choice between the continuous vs. dauer-interrupted life history. Here, we report a complex feedback loop between DAF-12 and the let-7-family miRNAs involving both the repression of DAF-12 by let-7-family miRNAs and the ligand-modulated transcriptional activation and repression of the let-7-Fam miRNAs by DAF-12. We propose that this feedback loop functions to ensure robustness of cell fate decisions and to coordinate cell fate with developmental arrest.
Keywords: gene regulation, microRNA, nuclear hormone receptor
The complexity of animal development requires the coordination of temporal, spatial, and physiological cues. Successful development is necessary to achieve optimal fitness, and natural selection has favored developmental programs that robustly produce a desired outcome in a range of physiological and environmental conditions. Forward genetics has revealed much of the internal programming of several developmental processes in standardized laboratory conditions. A fundamental question emerging from these studies is how these genetic circuits are affected by conditions that are more similar to natural, variable environments. Caenorhabditis elegans has been an excellent system to uncover developmental mechanisms because of its well-defined cell lineage and tractability to genetic analysis. C. elegans development is robust in a wide range of environmental conditions, providing an excellent opportunity to determine how genetic pathways are modulated in these diverse environments (1).
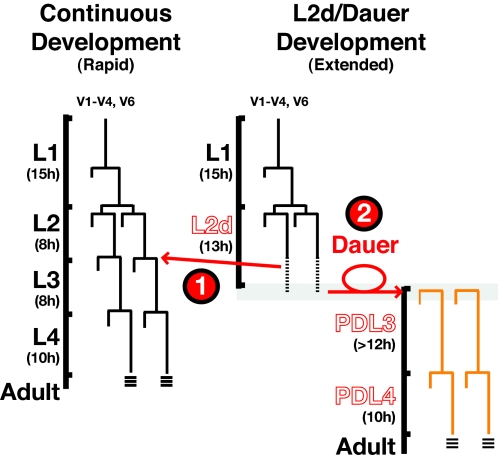
There are two distinct life histories a C. elegans larva may follow: a rapid, continuous life history that occurs in favorable environments and an extended, interrupted life history (2) that occurs in response to increased population density, decreasing food supplies, and elevated temperature (Fig. 1) (3). Within the continuous life history, animals progress through four larval stages (L1–L4) to a reproductively competent adult within ≈40–50 h after hatching (4). By contrast, unfavorable environments sensed during L1 stage cause larvae to enter the predauer L2d stage, which is ≈50% longer than the normal L2 (Fig. 1) (2). L2d stage larvae assess their environment while delaying the expression of L3 cell fates. By the end of the L2d stage, the larva commits to one of two developmental programs: (i) expressing L3 cell fates and resuming continuous development or (ii) entering the developmentally arrested, stress-resistant dauer larva diapause enabling animals to survive for extended periods (5).
Fig. 1.
Temporal development can be modulated by environmental conditions. Wild-type seam cell lineage diagrams for both continuous (Left) and dauer (Right) life histories. In harsh environments, wild-type animals extend the chronological interval between the L1 and L2 molts (L2d). During the L2d, animals enter a critical period in which one of two life histories must be chosen to (1) resume continuous development or (2) arrest as a dauer larva. Once committed to dauer program, wild-type animals can remain in this stress-resistant form for hours to months (2). When food supplies return to favorable conditions, dauer animals will continue development to a reproductively competent adult.
Regardless of whether C. elegans larvae develop continuously or interrupt development with the dauer diapause, they execute the same specific temporal pattern of cell fates, as exemplified by the lateral hypodermal stem cell (seam cell) lineages (Fig. 1) (6). Each seam cell executes a single asymmetric self-renewing division at each larval stage, with one additional, symmetric division inserted specifically in the beginning of the L2 (or L2d) stage (4). The heterochronic genes regulate the temporal patterning of these seam cell division programs. Mutations in heterochronic genes that specify the L2 cell fates cause a precocious phenotype, where the L2-specific symmetric seam cell division is skipped. By contrast, mutations in genes that promote L3 cell fates cause a retarded phenotype, where the L2-specific symmetric division is reiterated at later larval stages (7). Key to the proper specification of L2 and L3 seam cell fates is a progressive developmental down regulation of the transcription factor Hunchback-Like-1 (HBL-1) (7). hbl-1 gene expression is modulated by several regulatory inputs, including the let-7-family miRNAs miR-48, miR-241, and miR-84 (“let-7-Fam miRNAs”). These miRNAs are transcriptionally activated in L2 larvae and are redundantly required to down-regulate hbl-1 via its 3′ UTR (8).
The fact that the specification of L2 and L3 seam cell fates occurs properly regardless of whether the L3 follows rapidly after the L2 (in continuous development) or whether the L3 is delayed indefinitely (by dauer arrest), implies that the regulation of heterochronic genes must be integrated with the genetic programming of dauer larva formation. First, L3 seam cell fates must be postponed during the extended L2d stage. Second, at the end of the L2d stage, seam cells must commit to one of two distinct choices: either expression of L3 fates and the rapid completion of larval development or indefinite postponement of L3 fates in the dauer larva (Fig. 1). This suggests that L2 larvae undergo a complex decision-making process that incorporates an assessment of environmental conditions into the programming of cell fates. Little is currently understood about how genes that regulate dauer formation are integrated with heterochronic gene activity to coordinate this complex decision.
The decision to enter dauer diapause or to develop continuously centers on the activity the DAF-12 nuclear hormone receptor (9). Direct binding of an endogenous steroid ligand, dafachronic acid (DA), modulates the activity of DAF-12 protein (10). Synthesis of DA ligand is coupled to environmental conditions (5), and DA regulates DAF-12 activity by binding and displacing a corepressor DIN-1S (10, 11). In favorable culture conditions, DA ligand is abundant, and liganded DAF-12 activates transcription. In harsh environments, DA production is dramatically reduced, so most DAF-12 is unliganded and hence free to bind the corepressor DIN-1S (10, 11). The DAF-12:DIN-1 repressive complex is required for dauer formation.
Recently an additional role for DAF-12 has been described whereby DAF-12 directly affects the transcription of several let-7-Fam miRNAs, either positively (as liganded DAF-12) or negatively (as unliganded DAF-12:DIN-1) (12). Importantly, this activity of DAF-12 in let-7-Fam transcriptional regulation appears to be modulatory; null mutations of daf-12 (daf-12(0)) do not display overt heterochronic phenotypes (9, 12, 13), so the temporal activation of these miRNAs occurs independently of DAF-12. However, certain daf-12 mutations that affect ligand binding have severe heterochronic phenotypes, indicating that DAF-12 has a strong potential activity in the heterochronic pathway that is normally held in check by the DA ligand (9, 13). These observations suggest a complex role in developmental timing for hormone modulated transcriptional regulation by DAF-12.
In this article, we provide evidence for a gene regulatory circuit linking hypodermal seam cell fate specification and environmental signals controlling dauer larva arrest. The essence of this circuit is a complex feedback relationship between DAF-12 and the let-7-Fam miRNAs, wherein DAF-12 regulates let-7-Fam miRNA expression and the let-7-Fam miRNAs temporally down-regulate DAF-12. We show that the DAF-12 component of the pathway modulates the normal temporal transcriptional up-regulation of the let-7-Fam of miRNAs. Specifically, liganded DAF-12 accelerates let-7-Fam miRNA accumulation when environmental conditions favor developmental progression. In contrast, unliganded DAF-12 inhibits let-7-Fam-triggered cell fate transitions when environmental conditions favor developmental arrest. Moreover, we show that let-7-Fam miRNAs feed back and regulate DAF-12 expression, so that the accumulation of let-7-Fam miRNAs governs the sensitivity of the animal to the environmental cues that control DAF-12-mediated developmental arrest.
Results
Unliganded DAF-12 Exerts din-1-Dependent Inhibition of let-7-Family miRNA Expression.
DAF-12 can directly regulate the transcription of let-7-Fam miRNAs either positively or negatively, depending on the presence of DA ligand (12). However, daf-12(0) mutants develop almost normally (9), suggesting that the role for DAF-12 in developmental timing is modulatory rather than essential. We reasoned that the daf-12(rh61) mutation, encoding a DAF-12 protein lacking the ligand-binding domain (11, 13), would result in DAF-12 acting as a constitutive repressor of let-7-Fam miRNA expression. Indeed daf-12(rh61) animals exhibit developmental timing defects similar to those of let-7-Fam compound mutants (8, 9). To test the hypothesis that the activity of rh61 mutant DAF-12 results in repression of let-7-Fam miRNAs, we assayed the expression of 107 C. elegans miRNAs by using real-time-PCR-based TaqMan (ABI) assays on RNA samples extracted from synchronized populations of wild-type or daf-12(rh61) mutant larvae at the L2-molt/L3 stage. We found that daf-12(rh61) mutant larvae display a dramatic reduction in several let-7-Fam miRNAs (12- to 30-fold reduction in miR-795, miR-48, miR-241, and let-7, P < 0.01) (Fig. S1 and Table 1). miR-84 levels were reduced in only one biological replicate and did not qualify as statistically significant in our analysis (see SI Text). This is consistent with the more complex regulation of mir-84 by DAF-12 previously described (12). Of the remaining two let-7-Fam miRNAs, miR-794 was not detectably expressed in any of our samples, and miR-793 did not appear to be affected by daf-12(rh61) (Fig. S1, Table 1).
Table 1.
Levels of let-7-Fam miRNAs are specifically reduced in daf-12 mutants or in crowded environments
| miRNA | Fold change wild type versus mutant |
Sparse vs. crowded |
||
|---|---|---|---|---|
| daf-12(rh61) | daf-12(rh61rh411) | din-1; daf-12(rh61) | Wild type | |
| let-7 family | ||||
| mir-48 | −16.0 ± 4.2 | −3.2 ± 0.5 | −3.5 ± 0.8 | −3.2 ± 1.3 |
| mir-241 | −17.8 ± 4.7 | −4.3 ± 0.7 | −4.5 ± 1.1 | −9.4 ± 3.8 |
| mir-84 | −2.7 ± 0.7† | −2.8 ± 0.4† | −1.7 ± 0.6 | −2.6 ± 0.7 |
| let-7 | −12.2 ± 5.5 | −12.3 ± 2.5 | −6.0 ± 1.6 | off* |
| mir-793 | +0.3 ± 0.2 | −0.1 ± 0.3 | +8.5 ± 4.3† | +0.2 ± 0.3 |
| mir-794 | Off* | Off* | Off* | Off* |
| mir-795 | −31.0 ± 17.8 | −5.0 ± 1.2† | −3.8 ± 1.0† | −11.7 ± 5.2 |
| lin-4 family | ||||
| lin-4 | +0.7 ± 0.2 | +0.1 ± 0.1 | −0.03 ± 0.3 | +0.3 ± 0.2 |
| mir-237 | −1.2 ± 0.4 | −0.1 ± 0.2 | −0.75 ± 0.2 | −0.7 ± 0.4 |
| Other miRNAs that were significantly reduced‡ | ||||
| mir-230 | −3.4 ± 1.5 | −1.3 ± 0.2 | −1.1 ± 0.2 | −0.1 ± 0.2 |
| mir-85 | −18.7 ± 5.9 | −2.1 ± 0.6 | −11.0 ± 2.7 | −1.3 ± 0.7 |
| mir-788 | −1.7 ± 0.5 | −1.7 ± 0.3 | −0.3 ± 0.2 | 2.4 ± 0.7 |
Values in boldface indicate miRNA significantly changed, P < 0.01. Negative values indicate miRNA decreased in daf-12 relative to wild type, or in crowded relative to sparse conditions. Values close to 0 indicate miRNA was unchanged. Data are displayed as (mean fold-change) ± (mean estimated noise). For details see Materials and Methods in SI Text.
*Levels were beneath the limit for detection.
†Levels changed in only one biological replicate (see SI Text).
‡No miRNAs were significantly increased in daf-12 mutants relative to wild type. However, crowded wild-type larvae showed a significant increase (2–24 times) of mir-242, mir-243, mir-34, mir-71 and mir-792.
Levels of the lin-4 family and most other miRNAs were unchanged in mutant extracts compared with wild type. Indeed, levels of only two miRNAs that are not part of the let-7-Fam were significantly changed in mutant larvae, suggesting that daf-12(rh61) specifically affects the abundance of certain let-7-Fam miRNAs (Table 1, Fig. S1). We conclude that unliganded DAF-12 represses the expression of a specific subset of C. elegans miRNAs, including four of the seven let-7-Fam miRNAs, plus two additional, non-let-7-Fam miRNAs.
Although the mutant DAF-12(rh61) protein lacks the capacity to bind DA, it retains its ability to bind its corepressor DIN-1S (10), and hence is apparently “locked” in a repressive form. If this constitutively repressive form of DAF-12 is responsible for the strong repression of let-7-Fam miRNAs in daf-12(rh61) larvae, relieving this repression should increase the abundance of let-7-Fam miRNAs. We therefore compared the relative expression levels of miRNAs in daf-12(rh61) animals to those in animals carrying daf-12(rh61) with suppressor mutations that alleviate the severe heterochronic defects of daf-12(rh61) (9, 11). First, removal of the DAF-12 corepressor DIN-1S [din-1(dh181); daf-12(rh61) animals] partially restored expression of most let-7-Fam miRNAs (Table 1). Furthermore, eliminating the expression of daf-12(rh61) via an intragenic null mutation [daf-12(rh411rh61)] also resulted in the derepression of the let-7-Fam miRNAs (Table 1) to an extent very similar to that observed in din-1(dh181); daf-12(rh61) mutants. The observation that levels of certain let-7-Fam miRNAs are moderately decreased in these suppressor strains indicates that DAF-12 can also positively regulate the expression of let-7-Fam miRNAs, consistent with the previously described role for liganded DAF-12 in activating let-7-Fam miRNA transcription (12). These findings demonstrate that unliganded DAF-12 acts together with its corepressor DIN-1 to inhibit the expression of certain let-7-Fam miRNAs. This suggests that in wild-type development under harsh environmental conditions (low DA concentrations), DAF-12 would attenuate the accumulation of let-7-Fam miRNAs and, hence, retard the down-regulation of let-7-Fam targets, such as hbl-1.
DAF-12 Affects Developmental Timing Upstream of hbl-1 Expression.
Three let-7-Fam miRNAs (miR-48, miR-241, and miR-84) redundantly down regulate hbl-1 to allow progression from L2 to L3 cell fates (8). This redundancy suggests that absolute levels of let-7-Fam miRNAs are important for regulation of hbl-1 and is consistent with the hypothesis that quantitative changes in the levels of let-7-Fam miRNAs are responsible for the heterochronic phenotypes of certain daf-12 mutants.
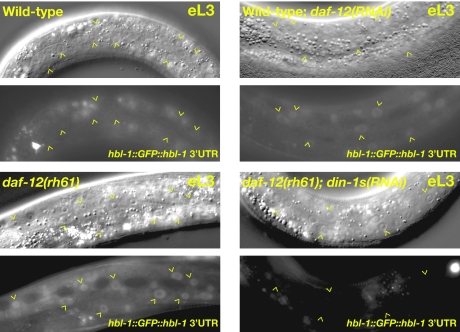
If DAF-12 ultimately affects HBL-1 expression by modulating let-7-Fam miRNA expression, we would expect that ligand-independent DAF-12 activity [daf-12(rh61)] or reduction of DAF-12 [e.g., daf-12(RNAi)] should affect HBL-1 expression in a fashion consistent with the effects on let-7-Fam expression. In particular, because the constitutively repressive form of DAF-12 [daf-12(rh61)] causes a greater reduction in let-7-Fam miRNAs than does daf-12(0) (see above), we predicted that daf-12(rh61) animals should exhibit a greater elevation in HBL-1 expression than daf-12(RNAi) animals. Accordingly, we used strains carrying an hbl-1::GFP::hbl-1 reporter that is normally down-regulated in the hypodermal cells of L3 larvae by let-7-Fam miRNAs (8, 14–16). We find that in daf-12(rh61) mutant larvae the hbl-1 reporter fails to be down regulated in the L3 stage (Fig. 2) (8, 12). We confirmed that this misexpression of hbl-1 in daf-12(rh61) animals depends on DIN-1S, because din-1s(RNAi) suppressed both the heterochronic and hbl-1 misexpression phenotypes of daf-12(rh61) animals (Fig. 2). Importantly, reduction of daf-12 activity by mutation or RNAi does not affect down-regulation of the hbl-1 reporter (Fig. 2). Thus, consistent with both the severe heterochronic phenotypes and the strong and specific repression of let-7-Fam miRNAs in daf-12(rh61) mutant larvae, daf-12(rh61) prevents the normal down-regulation of the hbl-1 reporter more strongly than does a simple reduction of DAF-12 protein. Together, these data suggest that daf-12 is normally not essential for the transcription of the let-7-Fam miRNAs but functions to modulate their expression in response to environmental conditions and impacts stage-specific gene expression by tuning HBL-1 levels.
Fig. 2.
hbl-1 is misexpressed in ligand-binding-defective mutants of daf-12. The hbl-1 3′ UTR reporter (ctIs39) is down-regulated in wild-type and daf-12(RNAi) larvae (eL3), but not in daf-12(rh61) eL3 larvae. Persistence of hbl-1 expression in daf-12(rh61) larvae depends on the DAF-12 corepressor DIN-1S because RNAi of din-1s suppresses the heterochronic phenotypes and corrects the hbl-1 misexpression of daf-12(rh61) animals.
DAF-12 Regulates HBL-1 Expression Independently of lin-28.
HBL-1 expression is tightly controlled by several regulatory pathways in addition to let-7-Fam miRNAs (8). Two well-defined components that regulate HBL-1 in parallel to let-7-Fam miRNAs are lin-28, encoding an RNA-binding protein that promotes hbl-1 translation, and lin-46, encoding a scaffolding protein that negatively regulates hbl-1 (17, 18). Because previous reports have suggested that lin-28 is downstream of daf-12 (9), we asked whether daf-12 regulates stage-specific seam cell fate entirely via modulation of let-7-Fam miRNAs or whether daf-12 might also act via lin-28. We observed LIN-28::GFP expression in wild type, daf-12(lf), and daf-12(rh61) mutant backgrounds. Inconsistent with daf-12 functioning to regulate lin-28 expression, the LIN-28::GFP transgene is down-regulated normally during the second larval stage in daf-12 mutants (Fig. S2) (17). This suggests that daf-12(rh61) does not require lin-28 overexpression to promote L2-specific seam cell fates and is consistent with daf-12 functioning entirely through regulation of let-7-Fam miRNAs. Additional genetic interactions between daf-12 alleles and lin-28, lin-46, and miRISC components are also consistent with daf-12 functioning in the same pathway as the let-7-Fam miRNAs (Table S1, Fig. S3). These data indicate that daf-12 is one of several modulators of HBL-1 expression and that the effect of daf-12 mutations on HBL-1 expression and stage-specific cell fates can be entirely explained by the regulation of let-7-Fam miRNA expression by DAF-12.
let-7-Family miRNAs (miR-48, miR-241, and miR-84) Regulate the Expression of DAF-12.
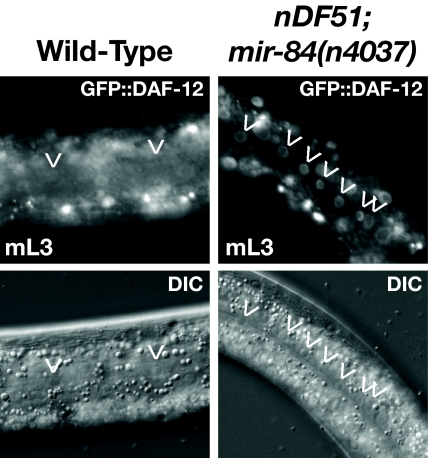
Previous studies have demonstrated that the daf-12 3′ UTR contains let-7 complementary sites that mediate the regulation of daf-12 3′ UTR reporters during late larval development (19). Because DAF-12 and the let-7-Fam miRNAs (miR-48, miR-241, and miR-84) also function during the L2-to-L3 transition, we investigated whether DAF-12 is regulated by the let-7-Fam miRNAs at this stage. Accordingly, we observed the expression of a functional DAF-12::GFP reporter in let-7-Fam mutant larvae. Previous characterization of the DAF-12::GFP reporter in wild-type larvae indicated that DAF-12 is normally down-regulated in hypodermal seam cells (and to a lesser extent in hyp7 cells) after the L2 molt (13). We find that the normal down-regulation of DAF-12::GFP is not observed in L3 stage mir-48(0) mir-241(0); mir-84(0) mutant larvae (Fig. 3). This indicates that these miRNAs contribute to the down-regulation of DAF-12 between the L2 and L3 stages.
Fig. 3.
DAF-12 levels are regulated by let-7-family miRNAs. A translational reporter of DAF-12, containing the daf-12 3′ UTR, is misexpressed in hypodermal seam cells of mir-48(0) mir-241(0); mir-84(0) mutant L3 larvae.
Feedback Between let-7-Family miRNAs and DAF-12 Modulates daf-12-Dependent Developmental Events.
The data reported above indicate a reciprocal regulatory relationship, whereby DAF-12 regulates the abundance of let-7-family miRNAs, and the let-7-family miRNAs regulate the abundance of DAF-12. Because let-7-Fam miRNAs affect DAF-12 levels, we tested whether let-7-Fam miRNAs could also affect daf-12-dependent developmental programs, including gonad migration and dauer formation.
During larval development, the distal tip cells of the somatic gonad guide gonadal growth and morphogenesis. During the L2 and L3 stages, the gonad arms migrate along the ventral surface of the animal. At the L3 molt, the distal tip cells of both gonad arms migrate dorsally and then reflex back toward the middle of the animal (20). The gonad arms of animals expressing mutant DAF-12 proteins that are unable to bind DA fail to reliably execute the L3 molt gonadal turning (9). Similarly, daf-9(0) mutants (which do not make endogenous DA) cultured in the presence of limiting amounts of exogenous DA, exhibit defects in gonadal turning (10). We find that the frequency of gonadal turning defects (the “Mig” phenotype) in daf-9(0) mutants is inversely related to DA concentration consistent with an inhibition of L3 gonadal turning by unliganded DAF-12 (Table 2). This implies that at limiting levels of DA, DAF-12 must be down-regulated by the end of the L3 stage, and/or DA levels must rise, to permit gonadal turning.
Table 2.
Effects on daf-12-dependent activity by reduction of let-7-Fam miRNA levels
| nM DA ligand | daf-9 | mir-241; daf-9 mir-84 | mir-48 mir-241; daf-9 mir-84 |
|---|---|---|---|
| Mig | |||
| 25 | 83 ± 15 (63) | 81 ± 15 (24) | ND |
| 50 | 35 ± 17 (88) | 82 ± 9 (61) | 78 ± 16 (52) |
| 100 | 7 ± 4 (150) | 34 ± 21 (127) | ND |
| dauer | |||
| 0 | 100 ± 0 (46) | 100 ± 0 (33) | ND |
| 25 | 25 ± 13 (138) | 39 ± 4 (95) | ND |
| 50 | 5 ± 2 (108) | 3 ± 1 (79) | 19 ± 8 (95) |
| 100 | 0 ± 0 (150) | 0 ± 0 (127) | ND |
Data are presented as percentage of animals exhibiting the relevant phenotype, plus or minus the standard error of the mean for three independent trials. Total number of animals scored is in parentheses. ND, not determined.
Because we see that let-7-Fam miRNAs contribute to a down-regulation of DAF-12 between the L2 and L3 stages (see above), we predicted that at constant, limiting concentrations of DA, loss of let-7-Fam miRNAs should result in an increase in the fraction of unliganded DAF-12 and an increase in the Mig phenotype. To eliminate effects on daf-12 activity attributable to variation in endogenous DA levels, we used a daf-9(0) mutation to eliminate endogenous DA, and supplied various levels of synthetic DA exogenously in the culture medium. We find that on plates containing either 50 nM or 100 nM DA, daf-9(0) strains that are also reduced in let-7-Fam miRNAs (because of loss of mir-241 and mir-84) indeed display a more penetrant Mig phenotype than daf-9(0) controls (Table 2). daf-9(0) animals lacking all three let-7-Fam miRNAs [mir-48(0) mir-241(0); mir-84(0)] also display an increased Mig phenotype (Table 2). This observation is consistent with the hypothesis that let-7-Fam miRNAs down-regulate DAF-12 as part of a feedback loop.
Another possible consequence of a feedback loop between daf-12 and the let-7-Fam is that let-7-Fam miRNAs would contribute to the regulation of dauer formation (via the regulation of DAF-12 levels). The ratio of unliganded DAF-12 to liganded DAF-12 during the L2 stage is a critical factor in the decision between dauer vs. reproductive development (for review, see ref. 5) (10). daf-9(0) larvae, which lack DA, constitutively form dauer larvae in a daf-12- and din-1-dependent manner (10, 21). This dauer-constitutive phenotype of daf-9(0) animals can be rescued by addition of synthetic DA in a dosage-dependent manner (Table 2) (10). We reasoned that at a constant, limiting concentration of DA loss of let-7-Fam miRNAs should result in an increase in the fraction of unliganded DAF-12 and hence an increase in the frequency of dauer formation. We found that at a DA concentration of 50 nM, 5% of daf-9(0) mutants formed dauer larvae, whereas 19% of mir-48(0) mir-241(0); daf-9(0) mir-84(0) mutant larvae formed dauer larvae (Table 2). These observations suggest that let-7-family miRNAs affect the dauer formation decision by modulating DAF-12 protein levels and imply that, during wild-type development, the temporal up-regulation of let-7-Fam miRNA levels during the L2 stage could affect the timing and/or frequency of dauer larva arrest.
Note that animals triply mutant for let-7-Fam miRNAs [mir-48(0) mir-241(0); daf-9(0) mir-84(0)] display a stronger dauer formation phenotype than do doubly mutant, mir-48(0) mir-241(0); daf-9(0) animals (Table 2). This indicates a functional redundancy among let-7-family members with respect to regulation of daf-12 and dauer formation, analogous to the redundancy among let-7-Fam miRNAs for the regulation of hbl-1 (8).
Dauer-Promoting Environmental Conditions Affect let-7-Fam Expression in Wild-Type Animals.
One prediction of the proposed feedback loop wherein DAF-12 modulates let-7-Fam miRNA gene expression is that environmental conditions that lead to changes in levels of DA ligand could either accelerate or impede the intrinsic developmental up-regulation of let-7-Fam miRNAs during the L1–L3 stages. If the feedback loop were important for integrating stage-specific cell fate decisions with life-history choice, the critical period for this integration would be the second larval stage. We therefore assessed miRNA levels during the second larval stage of wild-type larvae in two environmental extremes: (i) favorable culture conditions (plentiful food supply and sparse population density) and (ii) unfavorable dauer-inducing conditions (simulated high population density induced by addition of dauer pheromone to the growth medium). Exogenous pheromone in these conditions modulates downstream DAF-12 activity via multiple signaling pathways that ultimately regulate daf-9 expression (21). Larvae on the pheromone-containing plates entered the extended, predauer L2d stage, whereas larvae on the plates without exogenous pheromone developed through the rapid L2 stage. We found that the expression of four of the let-7-family members (miR-48, miR-241, miR-84, and miR-795) was significantly reduced in high-pheromone conditions (≈3- to 12-fold; P < 0.01, Table 1). As was the case for the effect of daf-12 mutations on miRNA expression, the effect of environment was relatively specific for the let-7-family miRNAs; in this case, only one additional miRNA showed moderate but statistically significant (P < 0.01) down-regulation, and five miRNAs showed significant (P < 0.01) up-regulation in crowded conditions. Levels of let-7 and miR-794 were below the limit of detection in some assays, whereas miR-793 was not affected by environmental conditions, consistent with the lack of effect of daf-12 mutations on miR-793 expression (Table 1). These results indicate that environmental conditions, likely mediated by DAF-12, can significantly impact the expression of let-7-Fam miRNAs.
Discussion
The significance of the role for DAF-12 in regulating a choice between continuous vs. dauer-interrupted life history choice in response to environmental conditions has become increasingly clear in recent years, particularly with the identification of DA as a DAF-12 ligand (10). However, the precise role for DAF-12 in influencing developmental timing has remained mysterious. For example, the daf-12(0) allele has a profound defect in dauer formation, but a very minor impact on stage-specific cell fates (9). Yet, because certain nonnull alleles of daf-12 exhibit dramatic developmental timing defects, it was clear that daf-12 could affect stage-specific cell fate decisions in some fashion (9). This work, together with another recent paper (12), establishes that DAF-12 affects developmental timing by modulating the expression of let-7-Fam miRNAs. The mechanism for the modulation of let-7-Fam miRNAs by DAF-12 is similar to that for regulation of dauer formation. Specifically, in both contexts, liganded DAF-12 functions as a gene activator, and unliganded DAF-12 functions as part of a repressor complex.
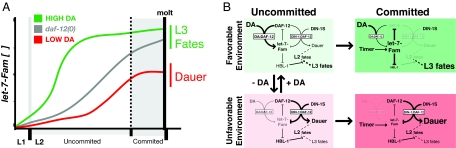
Although unliganded DAF-12 is absolutely required for dauer formation (9), we demonstrate that the role for DAF-12 in regulating let-7-Fam miRNAs is modulatory rather than essential (Table 1 and Table S1). Specifically, we show that levels of let-7-Fam miRNAs are only mildly reduced at the end of L2 in daf-12(0) larvae (Table 1). Furthermore, this mild reduction is not sufficient to affect HBL-1 down-regulation or stage-specific cell fate decisions (Table S1 and Fig. 2). However, although DAF-12 exerts a weak positive modulation of let-7-Fam expression, molecular and genetic analysis of daf-12 mutants that cannot respond to environmental conditions because they do not bind DA indicate that DAF-12 possesses a strong negative regulatory potential that is normally held in check by DA. In this scenario, DAF-12 is locked in its repressive form and can override the developmental timer by dramatically inhibiting the temporal up-regulation of let-7-Fam miRNAs (Fig. 4). In wild-type animals subjected to unfavorable environmental conditions, DA levels are low, and the repressive DAF-12-DIN-1 activity would dampen the accumulation of let-7-Fam miRNAs (Fig. 4A), resulting in a persistence of DAF-12 (and hence promotion of dauer arrest) and HBL-1 (and hence inhibition of L3 cell fates) (Fig. 4B).
Fig. 4.
Model for a feedback circuit that integrates environmental and temporal cues for robust cell fate decisions in different environments. (A) let-7-Fam miRNAs are transcriptionally up-regulated as development progresses independently of daf-12. However, DAF-12 activity modulates the rate at which the let-7-Fam miRNAs are expressed. Liganded DAF-12 (DA:DAF-12) accelerates their up-regulation, whereas unliganded DAF-12 (DAF-12:DIN-1) impedes let-7-Fam up-regulation. (B) The feedback circuit between DAF-12 and the let-7-Fam miRNAs (see Discussion).
A key finding in this work is a reciprocal regulatory relationship wherein DAF-12 and the let-7-Fam miRNAs modulate each other's expression. We demonstrate that the expression of let-7-Fam miRNAs is regulated by DAF-12 and also that let-7-Fam miRNAs down-regulate DAF-12 during larval development (Table 1, Fig. 3). We find that loss of let-7-Fam miRNAs can affect DAF-12-dependent events, including stage-specific gonad turning and dauer larva formation (Table 2). We propose that DAF-12 and let-7-Fam miRNAs comprise a complex feedback loop that is important both to respond to environmental changes during the uncommitted L2 stage and to reinforce the commitment to one choice that incorporates both life history and stage-specific gene expression. A model for this feedback loop is shown in Fig. 4 and discussed below.
In favorable environments where DA concentrations are high, second-stage larvae progress through the rapid L2 stage that leads to L3 and continuous development (2). Levels of let-7-Fam miRNAs rise during this stage in response to temporal cues (Fig. 4A) (8). We interpret the function of the feedback loop in this situation to be to accelerate the up-regulation of let-7-Fam miRNAs. Higher levels of these miRNAs will in turn accelerate the down-regulation of HBL-1, allowing progression to L3 seam cell fates (Fig. 4). Once committed to continuous development and L3 fate choice, the let-7-Fam levels will be high because of the combined effect of temporal cues and DA-DAF-12. Therefore, the levels both HBL-1 and DAF-12 will be reduced, precluding both L2 cell fates and dauer formation, irrespective of new changes in environmental conditions.
By contrast, in poor or marginal environments where DA concentration is low, second-stage larvae progress through an extended L2 stage (L2d) (2). For a period during the L2d, larvae remain uncommitted to either the dauer or continuous life histories while they monitor changes in environmental conditions. This uncommitted status of L2d animals is also accompanied by a postponement of L3 cell fates, compared with the rapid L2 stage. We propose that the feedback circuit contributes to both of these processes—the postponement of L3 fates and the eventual selection of reproductive vs. dauer development.
An important component of our model (Fig. 4) is that in dauer-promoting conditions, the DAF-12-DIN-1-repressive complex would dampened the up-regulation of let-7-Fam miRNAs, delaying the down-regulation of HBL-1 and DAF-12 and hence postponing the expression of L3 seam cell fates (Fig. 4B). In this model, a delayed down-regulation of DAF-12 serves to retain the developmental options of reproductive vs. dauer development during the extended L2d. If the environment remains poor, the larva will eventually commit to dauer arrest, and in our model, the feedback circuit forms part of the commitment mechanism, whereby DAF-12-DIN-1 complex positively autoregulates by repressing let-7-Fam miRNA expression.
If, on the other hand, an uncommitted L2d larva suddenly experiences favorable environmental conditions (and hence a rise in DA level), the direction of the DAF-12-dependent loop can shift, such that reduced DAF-12-DIN-1 repressor complex and increased DA-DAF-12 activator complex would permit accelerated up-regulation of let-7-Fam miRNAs. This environmentally triggered rise in let-7-Fam levels would promote commitment to reproductive development (by repression of DAF-12) and L3 fates (by repression of HBL-1). In either case, once a life history has been chosen, we propose that the feedback loop reinforces the decision and helps to make the two options mutually exclusive.
A reciprocal relationship between miRNAs and transcription factors that regulate the expression of those miRNAs may be a common theme elaborated in a variety of developmental contexts. Previously described miRNA-transcription factor feedback loops have been proposed to “tune” the expression of the transcription factors to optimal levels (22–25), consistent with a role for miRNA–transcription factor feedback contributing to the robustness of developmental decisions. Where the transcription factor is a steroid receptor, as in the DAF-12-let-7-Fam circuit described here, the characteristics of the circuit can be radically affected by ligand, permitting complex responses to environmental and physiological signals. Depending on the abundance of DA, DAF-12 can either accelerate or retard the up-regulation of let-7-Fam miRNAs, enabling ligand-dependent shifts between positive and negative autoregulation. Moreover, DA is a diffusible ligand, enabling the output of the DAF-12 circuit to be coordinated at the organismal level, further ensuring that the distinction between continuous and dauer development is robustly drawn throughout the animal.
Materials and Methods
Nematode Methods.
C. elegans strains were grown under standard conditions (26). Transformation of animals and RNAi were performed as previously described (8, 27).
Microscopy and Phenotypic Analysis.
Lineage analysis and scoring of adult alae phenotypes were performed by picking staged animals of the indicated genotypes and monitoring seam cells derived from the V lineage as described by Sulston and Horvitz (4).
DA Assays.
DA acid experiments were performed as in Motola et al. (10). Genotypes of animals assayed in this manuscript and further experimental descriptions can be found in SI Text.
TaqMan Data Analysis.
Analysis for the TaqMan real-time PCR data were performed similarly to the identification of invariant probes method (28). A detailed description of experimental procedures, calculations and statistical analysis can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank A. Zinovyeva and M. Hammell for critically reading the manuscript and M. Hammell for statistical advice. We also thank Caifu Chen of Applied Biosystems Inc. for development and use of the C. elegans-specific miR-TaqMan assays. Several strains used in this work were kindly provided by A. Antebi and the Caenorhabditis Genetics Center (which is supported by the National Institutes of Health National Center for Research Resources). This work was supported by National Institutes of Health Grants GM30428 (to V.A.), F32 GM69186 (to C.M.H.), and F32 GM73307 (to X.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908131106/DCSupplemental.
References
- 1.Braendle C, Felix MA. Plasticity and errors of a robust developmental system in different environments. Dev Cell. 2008;15:714–724. doi: 10.1016/j.devcel.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- 3.Hu PJ. Dauer (August 8) [Accessed October 1, 2009];WormBook: The C. elegans Research Community. 2007 Available at http://wormbook.org/chapters/www_dauer/dauer.html. [Google Scholar]
- 4.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 5.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Ambros V. Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature. 1991;350:162–165. doi: 10.1038/350162a0. [DOI] [PubMed] [Google Scholar]
- 7.Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- 8.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- 10.Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Ludewig AH, et al. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 14.Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SY, et al. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable MicroRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 16.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 17.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 18.Pepper AS, et al. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development. 2004;131:2049–2059. doi: 10.1242/dev.01098. [DOI] [PubMed] [Google Scholar]
- 19.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 21.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 22.Martinez NJ, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner MJ, Slack FJ. Transcriptional control of microRNA expression in C. elegans: Promoting better understanding. RNA Biol. 2009;6:49–53. doi: 10.4161/rna.6.1.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pradervand S, et al. Impact of normalization on miRNA microarray expression profiling. RNA. 2009;15:493–501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.