Abstract
Viral DNA induces potent antiviral immunity by activating dendritic cells; however, the mechanism governing viral DNA-mediated triggering or aggravation of glomerulonephritis is unknown. Glomerular endothelial cells (GEnCs) do not express toll-like receptor (TLR)9, the only DNA-specific TLR. We therefore hypothesized that DNA could activate GEnCs via the recently discovered TLR-independent viral DNA recognition pathway. Indeed, double-stranded non-CpG (B-) DNA activated GEnCs to produce interleukin-6, CCL5/RANTES, CCL2/MCP-1, CXCL10/IP10, interferon (IFN)-α, and IFN-β when cationic lipids facilitated intracellular DNA uptake. This cytokine production was inhibited by chlorpromazine, suggesting that clathrin-dependent endocytosis is required for B-DNA entry. However, chloroquine and MyD88 inhibition did not affect GEnC activation, suggesting TLR-independent DNA recognition. In addition, IFN-β activated cytokine and chemokine mRNA expression, although only CXCL10/IP10 was induced at the protein level, and type I IFN did not activate GEnC in an autocrine-paracrine auto-activation loop. B-DNA complexes induced intercellular adhesion molecule-1 expression at the GEnC surface and increased intercellular adhesion molecule-1-dependent leukocyte adhesion and microvascular extravasation in vivo. Furthermore, B-DNA complexes increased albumin permeability of GEnC monolayers in culture or microvascular dextran leakage in vivo. In addition, B-DNA complexes impaired GEnC proliferation. Thus, complexed B-DNA activates GEnC to produce cytokines, chemokines, and type I IFNs, increases leukocyte adhesion and microvascular permeability, and reduces GEnC proliferation via a MyD88-independent cytosolic DNA recognition pathway. This innate antiviral response program suggests a novel pathomechanism regulating DNA virus-mediated induction or aggravation of glomerulonephritis.
Extrarenal viral infections can trigger de novo immune complex glomerulonephritis, eg, hepatitis C virus-associated glomerulonephritis, but more frequently, acute viral infections trigger disease activity of pre-existing glomerulonephritis, as in IgA nephropathy, lupus nephritis, or renal vasculitis.1 Viral entry activates systemic antiviral immune responses, which may trigger disease flares of glomerulonephritis by enhancing autoantibody production, immune complex formation or by systemic interferon (IFN) release.2 Rapid production of type I IFN is a central element of antiviral immunity, since type I IFNs inhibit viral replication in infected cells and have pleiotrophic immunomodulatory effects on macrophages, T cells, and natural killer cells.3 In the intravascular compartment, plasmacytoid dendritic cells are the main type I IFN-producing cell type on recognition of viral proteins via Toll-like receptors (TLRs) 2 and 4 at the cell surface.2,4,5,6 Viral nucleic acids activate type I IFN production not before being taken up into intracellular endosomes where they activate TLR3 (double-stranded [ds]RNA), TLR7/8 (single-stranded RNA), or TLR9 (CpG-DNA).6,7 DAI/ZBP1 has been proposed as one of potentially more cytosolic sensors of viral DNA but little is yet known about their structure, expression and cell type-specific function.8,9 HIN-200 proteins also contribute to cytosolic DNA signaling, however, predominantly activate the inflammasome caspase-dependent release of interleukin (IL)-1β.10,11,12
Despite 50 years of IFN research and the clinical association of viral infection and glomerulonephritis, little is known about local production of type I IFN in glomerulonephritis.13 We hypothesized that glomerular cells like glomerular endothelial cells (GEnCs) would also produce type I IFN on viral DNA exposure. GEnCs line the intraglomerular capillaries, acting as a barrier, but also contribute to fluid filtration, and therefore interact with circulating viral particles.14,15 Similar to other microvascular endothelia, GEnCs can contribute to local inflammation by secreting pro-inflammatory cytokines and by fostering glomerular leukocyte recruitment via presenting chemokines and adhesion molecules at the luminal GEnC membrane.16 GEnCs express a limited pattern of TLRs, ie, TLR1-6, but lack the expression of the only DNA-specific TLR, ie, TLR9.17 We hypothesized that GEnCs harbor a TLR-independent cytosolic DNA recognition pathway and that viral ds(non-CpG) DNA can trigger antiviral immunity in GEnC.
Materials and Methods
GEnC Culture and Biochemicals
GEnCs, prepared from ts A58 immorto mice,18 were grown in medium RPMI 1640 (Invitrogen, Paisley, UK) with 10% heat-inactivated fetal calf serum. GEnCs were stimulated with endotoxin-free poly dA/dT (B-DNA, Sigma-Aldrich, St. Louis, MO) either pure or complexed with the cationic lipid (CL) lipofectamine 2000 (Invitrogen) for 24 hours in medium containing 2% fetal calf serum. B-DNA and CL were pre-incubated with polymyxin B (Invivogen, San Diego, CA) before use to block any residual lipopolysaccharaide (LPS) contamination. Murine IFN-α was purchased from Serotec (Oxford, UK), IFN-β from PBL (Piscataway, NJ), IFN-γ from PeproTech (Rocky Hill, NJ), chloroquine and methyl-β-cyclodextrin from Sigma-Aldrich (Steinheim, Germany), chlorpromazine from Merck (Darmstadt, Germany), cytochalasin D from BIOMOL (Plymouth Meeting, PA), MyD88 homodimerization inhibitory peptide or control peptide from Imgenex (San Diego, CA),19 and Pam3CSK from Invitrogen.20 Rat monoclonal antibodies against mouse IFN-α and IFN-β (32100-1 and 32400-1, PBL Interferon Source, Piscataway, NJ) were used to neutralize GEnC-derived IFN in vitro. They both were added simultaneously with different concentrations of B-DNA, as indicated. Cytokine levels were measured in cell supernatants using commercial enzyme-linked immunosorbent assay (ELISA) kits for IL-6 (OptEiA, BD, Mannheim Germany), IFN-α (PBL), IFN-β (PBL), IFN-γ (BD), CCL2/MCP-1 (BD), CCL5/RANTES, and CXCL10/IP-10 (R&D Systems, Minneapolis, MN). GEnC proliferation was determined after 72 hours using CellTiter 96 cell proliferation assay (Promega, Madison, WI) reading absorbance at 492 nm. For cellular uptake studies GEnCs were stimulated with 5′-rhodamine-labeled B-DNA. After 2 hours the cells were fixed with 2% paraformaldehyde, 10 mmol/L Pipes, 15% saturated picric acid at pH 6.0. Cells were incubated overnight with fluorescein isothiocyanate (FITC)-phalloidin (1:200, Invitrogen) before scanning with a LSM510 laser microscope (Carl Zeiss, Jena, Germany).
Real-Time PCR and Reverse Transcription
Complementary DNA was generated from total RNA using random priming and MMLV reverse transcriptase (Invitrogen). Real-time PCR was performed using SYBRGreen PCR master mix or on Roche Light Cycler 480 (Roche Diagnostics, Mannheim, Germany). All values were normalized to 18s rRNA. The oligonucleotide primer sequences are listed in table 1.
Table 1.
Murine Probes Used for Real-Time RT-PCR
| Gene | Sequence |
|---|---|
| 18S | |
| Forward | 5′-GCAATTATTCCCCATGAACG-3′ |
| Reverse | 5′-AGGGCCTCACTAAACCATCC-3′ |
| ZBP1 | |
| Left | 5′-TATGACGGACAGACGTGGAA-3′ |
| Right | 5′-TGCTGACAAATAATCGCAGG-3′ |
| CCL2 | |
| Left | 5′-CCTGCTGTTCACAGTTGCC-3′ |
| Right | 5′-ATTGGGATCATCTTGCTGGT-3′ |
| CCL5 | |
| Left | 5′-CCACTTCTTCTCTGGGTTGG-3′ |
| Right | 5′-GTGCCCACGTCAAGGAGTAT-3′ |
| CXCL10 | |
| Left | 5′-GGCTGGTCACCTTTCAGAAG-3′ |
| Right | 5′-ATGGATGGACAGCAGAGAGC-3′ |
| MX1 | |
| Left | 5′-TCTGAGGAGAGCCAGACGAT-3′ |
| Right | 5′-CTCAGGGTGTCGATGAGGTC-3′ |
| IFNB1 | |
| Left | 5′-CCCTATGGAGATGACGGAGA-3′ |
| Right | 5′-CCCAGTGCTGGAGAAATTGT-3′ |
Immunoblotting
GEnCs were washed and lysed in lysis buffer (1% Triton, 10 mmol/L Tris, 10 mmol/L NaCl, 10 mmol/L EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mmol/L phenylmethylsulfonylfluoride, pH 7.4). Total protein aliquots of 25 to 50 μg of lysate was separated by 8% SDS-polyacrylamide gel electrophoresis and transferred to an Immobilon-P membrane (Millipore, Eschborn, Germany). The blots were blocked in 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween 20 for 1 hour and then incubated with primary antibodies overnight at 4°C as follows: phosphorylated interferon-related factor (pIRF)-3, 1:2500; β-actin, 1:5000 (all from Cell Signaling, Danvers, MA); pIRF-3, 3 μg/ml (Abcam, Cambridge, MA). Horseradish peroxidase-conjugated anti-rabbit IgG (Cell Signaling) and anti-goat IgG (Jackson ImmunoResearch Laboratories, Suffolk, UK) were diluted to 1:20,000 in blocking solution (5% nonfat dry milk in buffer containing 0.1% Tween 20. Blots were developed with enhanced chemiluminescence (Amersham Pharmacia, Freiburg, Germany).
Flow Cytometry
B-DNA-stimulated GEnCs were washed with PBS and incubated with binding buffer containing either FITC-anti-annexin V (BD, Franklin Lakes, NJ) or propidium iodide (PI, BD) for 15 minutes at room temperature. In other experiments, cells were then washed with PBS and incubated with PBS containing FITC-anti-CD106 (VCAM-1) from BioLegend (San Diego, CA) or FITC-anti-CD54 (ICAM-1) from BioLegend for 45 minutes at room temperature The cells were analyzed by flow cytometry (FACS Calibur, BD) with acquisition of 30,000 events/sample.
In Vitro Fluorescein-Albumin Permeability Assay
GEnCs were grown to confluent monolayers hanging on 1-μm pore size inserts (Millipore, Billerica, MA) in 24-well plates with 0.5 ml media on both sides of the membrane.17,21 FITC-labeled bovine serum albumin (Invitrogen) was added to inserts and the cells were stimulated with 10 μg/ml B-DNA/CL or 6 μg/ml CpG-DNA. The filtrate was sampled at different time points and OD was measured at 485 nm and emission at 535 nm.
RNA Silencing Studies
DAI/ZBP1 small interfering (si)RNA and negative control siRNA (Ambion/Applied Biosystems, Darmstadt, Germany) sequences were as follows: DAI/ZBP1, 5′-ACAGUCCAGACAGUCCACAUCAAAU-3′; control, 5′-GGCAACAAGAUGACCAUCCACCUUA-3′, 5′-GGAAGACACAGGUACAA GCUCUGAA-3′; control, 5′-GAAGAGCACGAGAUAGCAAUU-3′. GEnCs (1 × 105) were plated in 12-well plates in 2% fetal calf serum-RPMI medium. Forty nmol/L siRNA was transfected twice with CL as above; 24 hours later, GEnCs were stimulated with 6 μg of B-DNA/CL. Knock-down efficacy of DAI/ZBP1, as well as CXCL10, IL-6, and IFN-β mRNA expression were determined by real-time reverse transcription-PCR after 6 hours and by Western blot after 24 hours.
In Vivo Microscopy on Cremaster Muscles
In vivo microscopy was performed as described.22,23 In brief, 6-week-old male C57BL/6 mice (Charles River, Sulzfeld, Germany) received an intrascrotal injection of PBS, CL, or 10 μg B-DNA/CL (n = 4 in each group) 6 hours before microscopic analysis. Some mice received 100 μg of an anti-CD54/ICAM-1 antibody (clone YN1/1.7.4, BioLegend, San Diego, CA) or isotype control antibody by intravenous injection, 5 minutes before intrascrotal injection of DNA. Images were obtained with water immersion lenses (×20/numerical aperture 0.5 and ×40/numerical aperture 0.8). For measurement of centerline blood flow velocity, green fluorescent microspheres (2 μm diameter; Molecular Probes, Leiden, The Netherlands) were injected via a arterial catheter, and their passage through the vessels of interest was recorded using the FITC filter cube under appropriate stroboscopic illumination (exposure 1 ms, cycle time 10 ms, λ = 488 nm). From measured vessel diameters and centerline blood flow velocity, apparent wall shear stress was calculated, assuming a parabolic flow velocity profile over the vessel cross section. The quantitative analysis of leukocyte migration was performed using Cap-Image software (Dr. Zeintl, Heidelberg, Germany). Firmly adherent cells were determined as those resting in the associated blood flow for more than 30 seconds and related to the luminal surface per 100 μm vessel length. Transmigrated cells were counted in regions of interest covering 75 μm on both sides of a vessel over 100-μm vessel length. Dextran permeability was determined as described.23
In brief, FITC-dextran (5 mg in 0.1 ml saline, Mr150,000, Sigma-Aldrich) was infused intra-arterially after DNA stimulation. Five postcapillary vessel segments, as well as the surrounding perivascular tissue were excited at 488 nm, and emission >515 nm was recorded by a CCD camera (Sensicam, PCO, Kelheim, Germany) 30 minutes after injection of FITC-dextran using an appropriate emission filter (LP 515). Mean gray values of fluorescence intensity were measured by digital image analysis (TILLvisION 4.0, TILL Photonics) in six randomly selected regions of interest (50 × 50 μm2), localized ~50 μm distant from the postcapillary venule under investigation. The average of mean gray values was calculated.
Statistical Analysis
Data were expressed as mean ± SEM. Groups were compared by two-tailed t-test or one-way analysis of variance. A value of P < 0.05 was considered to be statistically significant.
Results
B-DNA Activates GEnC to Secrete Cytokines, Chemokines, and Type I Interferons
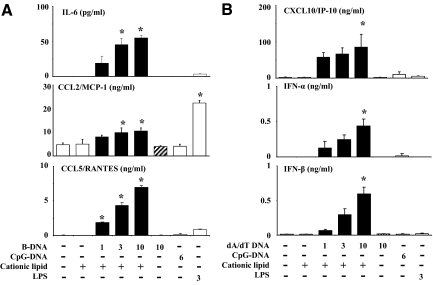
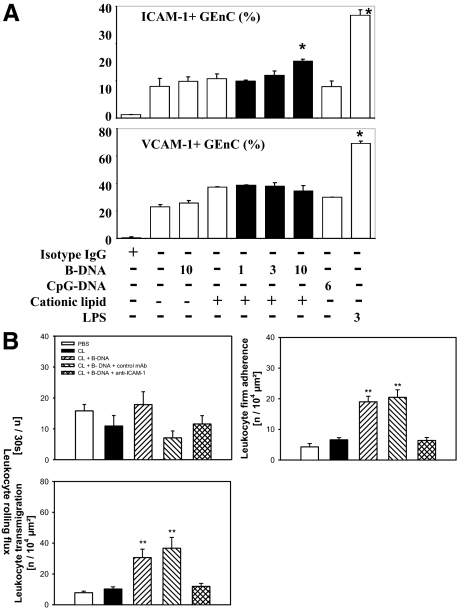
As GEnCs lack the only DNA-specific TLR (TLR9),17 it is questionable whether DNA can at all activate GEnCs. In fact, B-DNA and CpG-DNA both did neither induce IL-6, CCL2/MCP-1, CCL5/RANTES, CXCL10/IP-10, IFN-α, nor IFN-β secretion in cultured GEnCs (Figure 1, A and B). However, when being complexed with CL, low concentrations of B-DNA activated GEnCs to secrete all of these factors in a dose-dependent manner (Figure 1, A and B). CL alone had no stimulatory effect on GEnCs. B-DNA induced higher IL-6, CCL5/RANTES, and CXCL10/IP-10 levels as 3 μg/ml LPS, a potent endothelial cells (EC) activator (Figure 1, A and B). GEnCs did not produce IFN-γ (not shown). Thus, low amounts of B-DNA activate GEnCs to secrete cytokines, chemokines, and type I (but not type II) IFN only when being complexed with CL.
Figure 1.
B-DNA activates GEnCs to produce multiple pro-inflammatory mediators. GEnCs were exposed to increasing doses of B-DNA complexed or not complexed with CL as indicated. Exposure to CL, CpG-DNA, or LPS was used as controls. Supernatants were harvested after 24 hours and the concentrations of various cytokines, chemokines, and interferons were determined by ELISA. Data are means ± SEM from three experiments each analyzed in duplicate. *P < 0.05 vs. medium.
B-DNA-Driven GEnC Activation Requires Intracellular Uptake via a Clathrin-Dependent Endocytotic Pathway and Is Independent of MyD88 or Endosomal Acidification
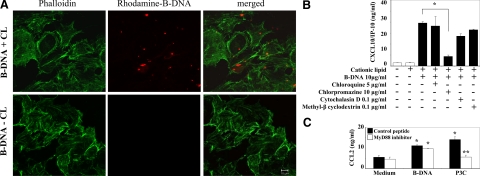
In dendritic cells and embryonic fibroblasts, dsDNA needs to reach the cytosol before it can interact with their putative cytosolic DNA receptors.9,10,11,24 Consistently, rhodamine-labeled dsDNA was not detectable in the intracellular cytosol of GEnCs, unless complexed with CL, as assessed by confocal microscopy (Figure 2A). To identify the possible intracellular uptake mechanism, we stimulated GEnCs with B-DNA/CL in the presence or absence of chlorpromazine, known to specifically block clathrin-dependent endocytosis25,26; methyl-β cyclodextrin, blocking caveolae-dependent endocytosis27; and cytochalasin D, blocking phagocytosis.26 The B-DNA-induced production of CXCL10/IP-10 was inhibited by chlorpromazine, but not by any of the other compounds (Figure 2B), suggesting a role for clathrin in the endocytosis of complexed B-DNA. The endosomal dsRNA receptor TLR3 is the only nucleic acid-specific TLR expressed by GEnCs and should not be involved in innate B-DNA recognition. In fact, B-DNA-induced CXCL10/IP10 secretion was not affected by chloroquine (Figure 2B), a base that inhibits the acidification of endosomes, which is a mandatory requirement for TLR3 signaling. In addition, GEnC activation by B-DNA did not involve MyD88, the signaling adaptor of all other TLR, because GEnC activation was not affected by 100 μmol/L of a MyD88 inhibiting peptide, which entirely prevented GEnC activation by the TLR2/MyD88 pathway agonist Pam3Cys (Figure 2C). Together, complexed B-DNA enters GEnCs via clathrin-dependent endocytosis and activates GEnCs independent of TLR signaling.
Figure 2.
Cationic lipid enhances the uptake of B-DNA in GEnC. A: GENCs were exposed to 5 μg rhodamine-labeled B-DNA in the presence or absence of cationic lipid (CL) for 2 hours. Intracellular uptake was detected by confocal microscopy and appears as red staining inside GEnCs. FITC-labeled phalloidin was used to mark the cytoskeleton of GEnCs and appears as green staining. Note that the intracellular uptake of B-DNA depended on the presence of CL. Images are representative for three independent experiments. Original magnification ×400. B: GEnCs were stimulated with either CL alone or B-DNA/CL in the presence or absence of chlorpromazine, methyl-β cyclodextrin, cytochalasin D, or chloroquine as indicated. CXCL10/IP-10 was determined in supernatants after 24 hours by ELISA. Data are means ± SEM from three experiments each analyzed in duplicates. *P < 0.05. C: GEnC were preincubated with 100 μmol/L MyD88 homodimerization inhibitory peptide or control peptide for 24 hours and then stimulated with either CL alone or B-DNA/CL. CCL2/MCP-1 were determined in supernatants after 24 hours by ELISA. Data are means ± SEM from three experiments analyzed in duplicate. *P < 0.05 vs. medium, **P < 0.05 vs. control peptide.
Interferon-β Induces DAI/ZBP1 Expression and IRF3 Phosphorylation
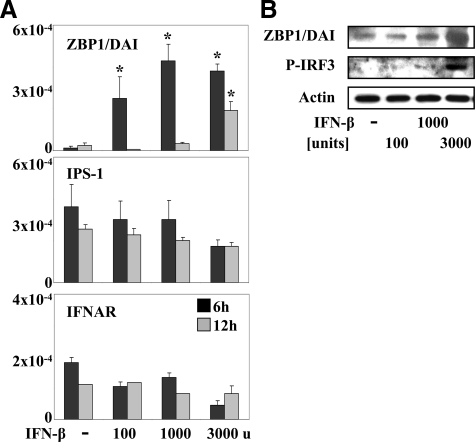
TLR-independent DNA recognition triggering cytokine and type I interferon release may involve DAI/ZBP1 but other, yet unknown, receptors may exist.9 The mRNA expression levels of DAI/ZBP1 were low under basal culture conditions but rapidly increased on IFN-β stimulation (Figure 3A). IFN-β-induced expression of receptor mRNAs was rapid and returned to baseline levels at 12 hours, in the case of IFN-β promoter stimulator-1 and type I interferon receptor (IFNAR) (Figure 3A). DAI/ZBP1 mRNA remained elevated, but only at 3000 U of IFN-β. Immunoblotting of GEnC protein extracts taken after 24 hours of stimulation confirmed that IFN-β induced the expression of DAI/ZBP1 (Figure 3B). IFN-β was also found to trigger the phosphorylation of IRF-3 (Figure 3B), a central transcription factor for type 1 IFN induction and signaling.24,28 By contrast, GEnCs constitutively expressed IFNAR mRNA and interferon-β promoter stimulator-1 mRNA, the latter being a mitochondrial adaptor involved in sensing of viral nucleic acids in the cytosol (Figure 3A). IFN-β stimulation did not affect IFN-β promoter stimulator-1 and IFNAR mRNA levels at 6 and 12 hours with a trend to lower mRNA levels with high doses of IFN-β (Figure 3A). We conclude that IFN-β induces DAI/ZBP1 expression and IRF3 phosphorylation both suggestive of the TLR-independent B-DNA recognition pathway.
Figure 3.
Interferon-β induces innate DNA recognition receptors in GEnC. A: GEnC were cultured in the presence or absence of various concentrations of IFN-β as indicated. Total mRNA was harvested after 6 (black bars) or 12 hours (gray bars) of stimulation and real-time reverse transcription-PCR was performed for ZBP1/DAI and the type I interferon receptor (IFNAR) as indicated. Data are expressed as ratio of the specific mRNA per respective 18S rRNA expression. *P < 0.05 vs. medium. B: In a similar experiment GEnC extracts were harvested after 24 hours for Western blotting with specific antibodies for ZBP1/DAI and phosphorylated (P-) IRF3. Blots shown are representative for each of three comparable experiments.
IFN-β Activates GEnCs, but Not in a B-DNA-Induced Autocrine-Paracrine Activation Loop
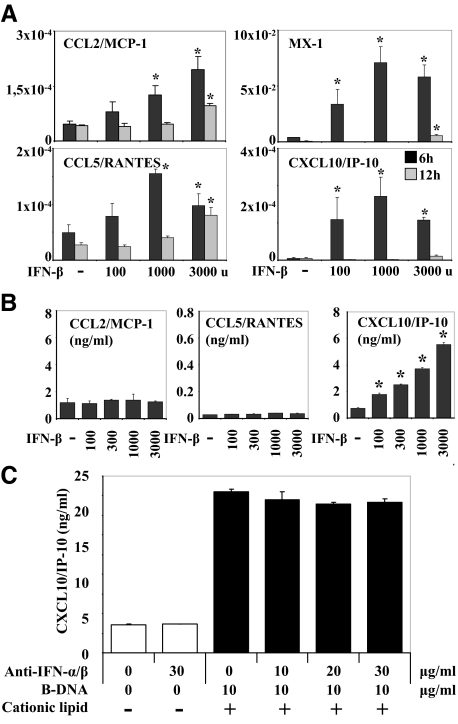
In dendritic cells, the viral nucleic acid–induced release of type I IFN enhances subsequent nucleic acid recognition via autocrine-paracrine type I IFN signaling involving the IFNAR and signal transducer and activator of transcription 1 phosphorylation.29 We have recently shown that viral RNA triggers the same phenomenon in mesangial cells,30 hence, we hypothesized the same for B-DNA in GEnCs. IFN-β stimulation induced the mRNA expression of CCL2/MCP-1, CCL5/RANTES, MX-1, and CXCL10/IP-10 (but not IFN-α and IL-6; not shown) in GEnCs in a dose-dependent manner (Figure 4A). However, at the protein level, IFN-β induced only the secretion of CXCL10/IP-10, while CCL2/MCP-1 and CCL5/RANTES levels remained unaffected by IFN-β stimulation (Figure 4B). The CXCL10/IP-10 levels induced by 3000 U IFN-β were low (<6 ng/ml, Figure 4B) as compared with the levels induced by B-DNA (50 to 100 ng/ml, Figure 1B). To examine whether type I IFN produced by the GEnCs contributes to DNA-induced secretion of CXCL10/IP-10 in an autocrine-paracrine manner, we stimulated GEnCs with B-DNA in the absence or presence of increasing doses of antibodies that neutralize the functions of IFN-α and IFN-β. By blocking both type I IFN the DNA-induced CXCL10/IP-10 production of GEnCs remained unaffected (Figure 4C). These data show that cytosolic DNA signaling activates GEnCs to secrete type I IFN but this does not contribute to GENC activation in autocrine-paracrine activation loop.
Figure 4.
Interferon-β induces cytokines and chemokines in GEnC. A: GEnC were cultured in the presence or absence of various concentrations of IFN-β as indicated. Total mRNA was harvested after 6 (black bars) or 12 hours (gray bars) of stimulation and real-time reverse transcription-PCR was performed for various cytokines and chemokines as indicated. Data are expressed as ratio of the specific mRNA per respective 18S rRNA expression. *P < 0.05 vs. medium. B: GEnC were stimulated as before. Supernatants were harvested after 24 hours for cytokine ELISA. Data represent means ± SEM. *P < 0.05 vs. medium. C: GEnC were exposed to 10 μg/ml B-DNA before increasing doses of neutralizing antibodies against IFN-α and IFN-β were added as indicated. After 24 hours cell culture supernatants were harvested and CXCL10/IP10 concentrations were determined by ELISA. Data represent means ± SEM.
B-DNA Induces ICAM-1 Surface Expression in GEnC and Enhances Leukocyte Adhesion in Vivo
EC activation can induce luminal expression of adhesion molecules, which facilitate the firm adhesion of rolling leukocytes.16 We used flow cytometry for intercellular adhesion molecule (ICAM-1) and vascular cell adhesion molecule (VCAM-1) to test whether B-DNA induces GEnC adhesion molecule expression. The basal GEnC surface expression of VCAM-1 was almost double as high as that of ICAM-1 (Figure 5A). ICAM-1 and VCAM-1 24 hours after B-DNA exposure to GEnCs significantly increased the percentage of GEnCs with ICAM-1 surface expression, but VCAM-1 expression was not induced (Figure 5A). ICAM-1 induction depended on complex formation with CL. However, the B-DNA-related induction of ICAM-1 was lower, as compared with that of LPS (Figure 5A). What is the functional significance of B-DNA-induced ICAM-1 induction in vivo? To answer this question we assessed leukocyte rolling, adhesion, and transendothelial migration using in vivo microscopy. Video analysis was conducted on cremaster muscle postcapillary venules of male C57BL/6 mice 6 hours after intrascrotal injection of PBS, CL, or B-DNA/CL. We found B-DNA/CL, but not CL alone or PBS, significantly induced the numbers of leukocytes adhering to the microvascular endothelium (Figure 5B). The numbers of transmigrating leukocytes were also significantly increased on B-DNA/CL exposure (Figure 5B). This effect was completely blocked by a ICAM-1 neutralizing antibody, suggesting a functional role for B-DNA-induced expression of ICAM in vivo (Figure 5B). By contrast, B-DNA did not affect leukocyte rolling (Figure 5B). Thus, B-DNA has the potential to induce ICAM-1 expression in endothelial cells, which enhances leukocyte adhesion and transendothelial migration both contributing to renal inflammation.
Figure 5.
B-DNA activates GEnC to express adhesion molecules and enhances leukocyte adhesion in vivo. A: GEnC surface expression of ICAM-1 and VCAM-1 was determined by flow cytometry after stimulation with various concentrations B-DNA, cationic lipids, and LPS as indicated. Data represent means ± SEM of four experiments. *P < 0.05. B: In vivo microscopy was performed on cremaster muscle postcapillary venules as described in methods. Four mice in each group were treated with intrascrotal injections of vehicle, cationic lipid (CL), B-DNA/CL, B-DNA/CL+ isotype IgG control, and B-DNA/CL+anti ICAM-1 as indicated in the legend. Leukocyte rolling, firm adhesion, and transendothelial migration were determined 6 hours after injection and are shown as means ± SEM **P < 0.05 vs. PBS.
B-DNA Increases Albumin Permeability of GEnC Monolayers and Enhances Microvascular Permeability in Vivo
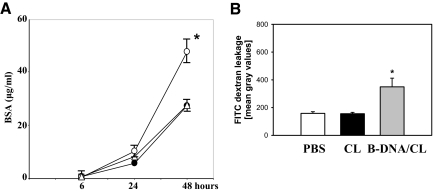
GEnCs contribute to the glomerular filtration barrier and systemic exposure to viral nucleic acids including B-DNA increases proteinuria in nephritic mice.17,31 To test whether B-DNA directly affects GEnC permeability we cultured GEnC monolayers in well inserts and added FITC-labeled bovine serum albumin to the upper well. Bovine serum albumin concentrations in the permeate of the lower well were determined after 6, 24, and 48 hours by reading OD at 535 nm. Complexed B-DNA significantly increased GEnC albumin permeability at 48 hours, as compared with cells cultured in medium alone (Figure 6A). B-DNA also increased vascular permeability in vivo, technically shown by the in vivo microscopy approach mentioned before. We used vascular leakage of FITC dextran as a marker of microvascular permeability. B-DNA/CL significantly increased vascular dextran leakage, which was not observed on injection of PBS or CL only (Figure 6B). Thus, B-DNA-induced activation of EC increases albumin permeability of GEnC monolayers and enhances microvascular permeability in vivo.
Figure 6.
B-DNA increase albumin permeability of GEnC monolayer and enhances microvascular permeability in vivo. A: GEnC monolayer albumin permeability was determined by transwell assay as described in methods. Data show FITC-bovine serum albumin concentrations in the lower well after 6, 24 and 48 hours of stimulation with medium (black circle), 10 μg B-DNA/CL (open circle), or 6 μg CpG-DNA (open triangle). Data are means ± SEM of 4 experiments. *P < 0.05 vs. medium. B: In vivo microscopy was performed on cremaster muscle postcapillary venules as described in methods. Four mice in each group were treated with intrascrotal injections of vehicle, cationic lipid (CL) or B-DNA/CL. Microvascular FITC-dextran leakage was determined 6 hours after injection and are shown as means ± SEM. P < 0.05 vs. PBS and vs. CL.
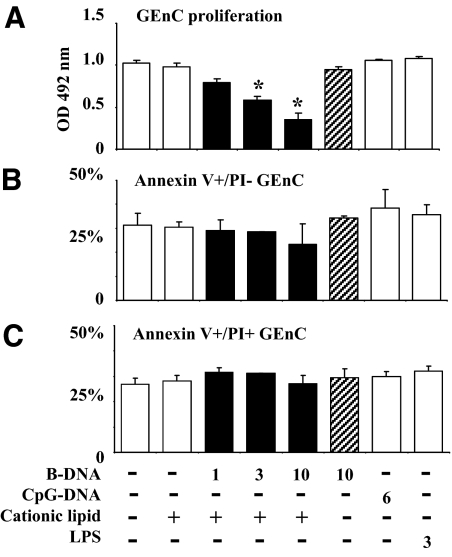
B-DNA Inhibits GEnC Proliferation
Cell proliferation fosters viral replication and spreading. We therefore questioned whether cytosolic recognition of B-DNA may also affect GEnC proliferation and survival. We first determined the GEnC proliferation rate in the presence or absence of B-DNA/CL complexes for a period of 72 hours at subconfluent culture conditions. B-DNA/CL, reduced GEnC proliferation in a dose-dependent manner (Figure 7A). At the same dose of B-DNA, GEnC proliferation was not affected in the absence of CL. CpG-DNA and LPS both did also not alter GEnC proliferation. Is this phenomenon caused by GEnC apoptosis or necrosis? We used flow cytometry to determine the percentages of annexin V–positive (apoptotic) and annexin V/propidium iodide–positive (late apoptotic/necrotic) GEnCs after cell activation. None of the DNA concentrations had a significant effect on the percentages of annexin V–positive (Figure 7B) and annexin V/propidium iodide–positive cells (Figure 7C). Thus, B-DNA inhibits GEnC proliferation without inducing cell death.
Figure 7.
GEnC proliferation and apoptosis. A: GEnC proliferation was determined in a period of 72 hours by bioluminescence assay as described in methods. Data represent mean OD ± SEM of three experiments measured at a wavelength of 492 nm. *P < 0.05 vs. medium. B and C: GEnC apoptosis was determined by flow cytometry. Data represent mean percentages of positive cells ± SEM of three experiments.
Discussion
Renal blood flow and glomerular filtration exposes GEnCs to circulating micro- and macromolecules including viral particles during viral infections. Viral nucleic acids may be protected from nuclease digestion when being complexed to lipids or proteins, eg, in immune complexes.32,33,34 For example, complexed viral nucleic acids often deposit along glomerular capillaries in immune complex disease, which activates GEnCs.35,36 We questioned whether dsDNA can activate GEnCs via the recently discovered TLR-independent cytosolic DNA recognition pathways8,37 that potently activate dendritic cells and embryonic fibroblasts to trigger type I IFN secretion via activating the transcription factor IRF3.9,24,38 Here we first show that ds (B-)DNA activates GEnCs to release type I IFN, as well as other antiviral defense mechanisms that may enhance glomerular inflammation and proteinuria.
B-DNA dose-dependently activated GEnCs to produce multiple pro-inflammatory cytokines, chemokines, and type I IFN, but only when being complexed with CL, which facilitated the intracellular uptake of the nucleic acids into GEnCs. This process involved clathrin-dependent endocytosis, a mechanism recently shown to mediate the intracellular uptake of dsRNA in Drosophila melanogaster S2 cells25 or of poly I:C RNA into embryonic kidney 293 cells.26 The role of endosomal (Toll-like) receptors, of which GEnCs express only the dsRNA-specific TLR3,17 could be excluded by using the endosomal acidification inhibitor chloroquine. Peptide MyD88 inhibition did also not affect GEnC activation by B-DNA ruling out the contribution of other TLR or of members of the IL-1 receptor family. The nature of the DNA cytosolic recognition receptor that triggers type I interferon signaling still remains unknown. B-DNA can also trigger IL-1β release via an inflammasome-dependent pathway involving HIN-200 proteins.10,11,12
We have recently shown that mesangial cells produce large amounts of type I IFN in response to cytosolic dsRNA.30 Blocking the secreted IFN with neutralizing IFN-α and IFN-β antibodies revealed that type I IFN activates mesangial cell cytokine and chemokine production in an autocrine-paracrine activation loop.30 We hypothesized a similar role of type I IFN release in GEnCs, but we found that identical doses of neutralizing IFN-α and IFN-β antibodies did not affect B-DNA–included CXCL10 release in GEnCs. Consistent with this observation, exogenous IFN-β did not induce CCL2 and CCL5 protein secretion, although both factors were induced at the mRNA level. Local type I IFN release can trigger multiple immunoregulatory functions and antiviral effectors.2,39 For example, GEnCs expressed the nuclear GTPase MX1 that degrades viral nucleocapsids.39 Type I IFN-dependent genes also activate the antiviral endoribonuclease RNase L, a mechanism that initiates the cleavage of viral RNA37 and that can produce small self-RNA cleavage products, which enhance type I IFN signaling in a positive amplification loop.40 B-DNA induced GEnCs to produce various pro-inflammatory factors, of which CXCL10/IP-10 is of particular interest because it is specifically induced by IFN and preferentially recruits cytotoxic T cells via the chemokine receptor CXCR3.41 This mechanism aids to control viral infection by direct killing of infected cells, but may also contribute to glomerular inflammation mimicking virally infected tissue by local type I interferon and CXCL10/IP-10 expression. The specific recruitment of effector leukocytes to the glomerulus may be further supported by the induction of ICAM-1, a surface molecule that mediates the firm adhesion of circulating leukocytes to activated renal endothelia,42 including GEnCs in human postinfectious glomerulonephritis.43 We found a low rate of ICAM-1 expression of the GEnC surface, but ICAM-1 expression was readily induced by DNA exposure. The series of selectin-dependent leukocyte rolling, chemokine-mediated activation of leukocyte integrins, and the subsequent integrin-mediated firm adhesion to the respective adhesion molecules is required to allow the firm leukocyte adhesion to the luminal endothelial cell membrane.44 In fact, B-DNA induced leukocyte adhesion and transendothelial migration in vivo, via the induction of ICAM-1. However, it is likely that B-DNA also activated cytokine release in cells other than ECs, which may indirectly enhance EC activation and adhesion molecule expression.
B-DNA also enhanced albumin permeability of GEnC monolayers or microvascular dextran permeability in vivo, a potential novel mechanism for viral infection-induced proteinuria, which may assist the clearance of viral particles and inflammatory cytokines via urinary excretion. However, systemic exposure of non-CpG-DNA did not enhance proteinuria in nephritic mice.17 Future studies will have to define the molecular mechanisms that link cytosolic B-DNA recognition to GEnC permeability. One simple explanation would have been GEnC apoptosis, which could enhance the permeability of GEnC monolayers simply by GEnC killing. However, B-DNA did not induce GEnC apoptosis (nor necrosis) but these results may be affected by the immortalized GEnC line.
Cell proliferation promotes virion spreading, hence, innate antiviral immunity seeks to limit mitosis or induces apoptotic cell death. We also observed a dose-dependent decrease in GEnC proliferation on exposure to B-DNA/CL complexes. This is in line with the clinical observation that endothelial proliferative glomerulonephritis is more commonly associated with bacterial infection rather than with viral infection. Viral infections such as chronic hepatitis C virus-associated glomerulonephritis are more commonly associated with mesangioproliferative forms of glomerulonephritis. In fact, we recently observed that B-DNA induces mesangial cell proliferation.45 The reasons for this glomerular cell type-specific effect on cell-cycle regulation will be investigated by us in the future.
Together, when entering the intracellular cytosol, B-DNA stimulated GEnCs to produce multiple pro-inflammatory cytokines and type I IFN via a MyD88-independent pathway. GEnC activation also included surface expression of ICAM-1, which enhanced leukocyte adhesion and transmigration, increased microvascular permeability, and reduced GEnC proliferation. All of these mechanisms support a novel pathogenic concept of virally triggered glomerulonephritis: When complexed viral DNA reaches the intracellular cytosol of GEnCs, it will trigger local antiviral defense mechanisms, which altogether promote glomerular inflammation and albuminuria.
Acknowledgments
We thank Dr. Nalan Akis (Division of Microbiology, Uludag University, Bursa, Turkey) for generously providing the GEnCs.
Footnotes
Address reprint requests to Hans-Joachim Anders, M.D., Medizinische Poliklinik, Universität München, Pettenkoferstr. 8a, 80336 München, Germany. E-mail: hjanders@med.uni-muenchen.de.
Supported by the Deutsche Forschungsgemeinschaft: H.J.A, R.A., and R.D.P. via AN372/9-1 and GRK 1202, C.A.R. via RE2885-1/1. Parts of the work were performed by H.H. in a thesis project funded by the Medical Faculty at the University of Munich (FöFoLe program).
References
- Lai AS, Lai KN. Viral nephropathy. Nat Clin Pract Nephrol. 2006;2:254–262. doi: 10.1038/ncpneph0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat Rev Immunol. 2002;2:151–161. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009 doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009 doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current, and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz W. Fenestrated glomerular capillaries are unique. J Am Soc Nephrol. 2008;19:1439–1440. doi: 10.1681/ASN.2008060583. [DOI] [PubMed] [Google Scholar]
- Haraldsson B, Nystrom J, Deen WM. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- Allam R, Pawar RD, Kulkarni OP, Hornung V, Hartmann G, Segerer S, Akira S, Endres S, Anders HJ. Viral 5′-triphosphate RNA and non-CpG DNA aggravate autoimmunity and lupus nephritis via distinct TLR-independent immune responses. Eur J Immunol. 2008;38:3487–3498. doi: 10.1002/eji.200838604. [DOI] [PubMed] [Google Scholar]
- Akis N, Madaio MP. Isolation, culture, and characterization of endothelial cells from mouse glomeruli. Kidney Int. 2004;65:2223–2227. doi: 10.1111/j.1523-1755.2004.00634.x. [DOI] [PubMed] [Google Scholar]
- Loiarro M, Sette C, Gallo G, Ciacci A, Fanto N, Mastroianni D, Carminati P, Ruggiero V. Peptide-mediated interference of TIR domain dimerization in MyD88 inhibits interleukin-1-dependent activation of NF-{kappa}B. J Biol Chem. 2005;280:15809–15814. doi: 10.1074/jbc.C400613200. [DOI] [PubMed] [Google Scholar]
- Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- Satchell SC, Buchatska O, Khan SB, Bhangal G, Tasman CH, Saleem MA, Baker DP, Lobb RR, Smith J, Cook HT, Mathieson PW, Pusey CD. Interferon-beta reduces proteinuria in experimental glomerulonephritis. J Am Soc Nephrol. 2007;18:2875–2884. doi: 10.1681/ASN.2006101104. [DOI] [PubMed] [Google Scholar]
- Reichel CA, Khandoga A, Anders HJ, Schlondorff D, Luckow B, Krombach F. Chemokine receptors Ccr1, Ccr2, and Ccr5 mediate neutrophil migration to postischemic tissue. J Leukoc Biol. 2006;79:114–122. doi: 10.1189/jlb.0605337. [DOI] [PubMed] [Google Scholar]
- Reichel CA, Rehberg M, Bihari P, Moser CM, Linder S, Khandoga A, Krombach F. Gelatinases mediate neutrophil recruitment in vivo: evidence for stimulus specificity and a critical role in collagen IV remodeling. J Leukoc Biol. 2008;83:864–874. doi: 10.1189/jlb.1007666. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Watanabe A, Funami K, Seya T, Matsumoto M. The clathrin-mediated endocytic pathway participates in dsRNA-induced IFN-beta production. J Immunol. 2008;181:5522–5529. doi: 10.4049/jimmunol.181.8.5522. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Ninomiya H, Miwa S, Masaki T. Cholesterol oxidation switches the internalization pathway of endothelin receptor type A from caveolae to clathrin-coated pits in Chinese hamster ovary cells. J Biol Chem. 2000;275:6439–6446. doi: 10.1074/jbc.275.9.6439. [DOI] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Gautier G, Humbert M, Deauvieau F, Scuiller M, Hiscott J, Bates EE, Trinchieri G, Caux C, Garrone P. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med. 2005;201:1435–1446. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flür K, Zecher D, Kulkarni O, Allam R, Lichtnekert J, Schwarz M, Beutler B, Anders HJ. Viral RNA sensors induce type I interferon-dependent cytokine release and cell death in glomerular mesangial cells. Implications for viral infection-induced glomerulonephritis. Am J Pathol. 2009;175:2008–2016. doi: 10.2353/ajpath.2009.080585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patole PS, Grone HJ, Segerer S, Ciubar R, Belemezova E, Henger A, Kretzler M, Schlondorff D, Anders HJ. Viral double-stranded RNA aggravates lupus nephritis through Toll-like receptor 3 on glomerular mesangial cells and antigen-presenting cells. J Am Soc Nephrol. 2005;16:1326–1338. doi: 10.1681/ASN.2004100820. [DOI] [PubMed] [Google Scholar]
- Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen SR, Shlomchik MJ, Viglianti GA, Rifkin IR, Marshak-Rothstein A. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–1177. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Richez C, Maciaszek JW, Agrawal N, Akira S, Marshak-Rothstein A, Rifkin IR. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol. 2007;178:6876–6885. doi: 10.4049/jimmunol.178.11.6876. [DOI] [PubMed] [Google Scholar]
- Savarese E, Chae OW, Trowitzsch S, Weber G, Kastner B, Akira S, Wagner H, Schmid RM, Bauer S, Krug A. U1 small nuclear ribonucleoprotein immune complexes induce type I interferon in plasmacytoid dendritic cells through TLR7. Blood. 2006;107:3229–3234. doi: 10.1182/blood-2005-07-2650. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Lauletta G, Montrone M, Grandaliano G, Schena FP, Dammacco F. Hepatitis C virus RNA and core protein in kidney glomerular and tubular structures isolated with laser capture microdissection. Clin Exp Immunol. 2005;140:498–506. doi: 10.1111/j.1365-2249.2005.02778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai FM, To KF, Wang AY, Choi PC, Szeto CC, Li PK, Leung CB, Lai KN. Hepatitis B virus-related nephropathy and lupus nephritis: morphologic similarities of two clinical entities. Mod Pathol. 2000;13:166–172. doi: 10.1038/modpathol.3880031. [DOI] [PubMed] [Google Scholar]
- Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13:543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MF, Lai SL, Chen JP, Sung JM, Lin YL, Wu-Hsieh BA, Gerard C, Luster A, Liao F. Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J Immunol. 2006;177:1855–1863. doi: 10.4049/jimmunol.177.3.1855. [DOI] [PubMed] [Google Scholar]
- Wuthrich RP. Intercellular adhesion molecules and vascular cell adhesion molecule-1 and the kidney. J Am Soc Nephrol. 1992;3:1201–1211. doi: 10.1681/ASN.V361201. [DOI] [PubMed] [Google Scholar]
- Rastaldi MP, Ferrario F, Yang L, Tunesi S, Indaco A, Zou H, D'Amico G. Adhesion molecules expression in noncrescentic acute post-streptococcal glomerulonephritis. J Am Soc Nephrol. 1996;7:2419–2427. doi: 10.1681/ASN.V7112419. [DOI] [PubMed] [Google Scholar]
- Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat Immunol. 2008;9:970–980. doi: 10.1038/ni.f.213. [DOI] [PubMed] [Google Scholar]
- Allam R, Lichtnekert J, Moll AG, Taubitz A, Vielhauer V, Anders H-J. Viral RNA and DNA sense common antiviral responses including type 1 interferons in mesangial cells. J Am Soc Nephrol. 2009;20:1986–1996. doi: 10.1681/ASN.2008101067. [DOI] [PMC free article] [PubMed] [Google Scholar]