Abstract
Bcl-xl and the hepatocyte growth factor (HGF) receptor c-Met are both highly expressed in mesotheliomas, where they protect cells from apoptosis and can confer resistance to conventional therapeutic agents. In our current study, we investigate a model for the transcriptional control of Bcl-xl that involves ETS transcription factors and the HGF/Met axis. In addition, the effects of activated c-Met on the phosphorylation of the ETS family transcriptional factors were examined. The transient expression of ETS-2 and PU.1 cDNAs in mesothelioma cell lines resulted in an increase in the promoter activity of Bcl-xl and consequently in its mRNA and protein expression levels, whereas the transcriptional repressor Tel suppressed Bcl-xl transcription. The activation of the HGF/Met axis led to rapid phosphorylation of ETS family transcription factors in mesothelioma cells through the mitogen-activated protein kinase pathway and via nuclear accumulation of ETS-2 and PU.1. A chromatin immunoprecipitation assay further demonstrated that the activation of c-Met enhanced the binding of ETS transcriptional factors to the Bcl-x promoter. Finally, we determined the Bcl-xl and phosphorylated c-Met expression levels in mesothelioma patient samples; these data suggest a strong correlation between Bcl-xl and phosphorylated c-Met levels. Taken together, these findings support a role for c-Met as an inhibitor of apoptosis and an activator of Bcl-xl.
Malignant mesotheliomas are aggressive tumors commonly associated with asbestos exposure.1 Although there has been some progress in the treatment of these cancers, the overall prognosis remains very poor. Bcl-xl is a key antiapoptotic protein expressed in many tumor types and its overexpression is believed to contribute to chemotherapeutic resistance in mesotheliomas. In previous studies, Bcl-xl expression was found to be influenced by a variety of transcription factors and signal transduction pathways. In addition to nuclear factor-κB (NF-κB) and signal transducers and activators of transcription (STATs), analysis of human Bcl-xl promoter has revealed nine potential ETS-binding sites.2,3,4 Bcl-xl overexpression in various tumors is known to contribute to tumorigenesis and resistance to therapeutic agents. By decreasing Bcl-xl expression through antisense or small interfering (si) RNAs and inhibiting the Bcl-xl protein using BH3 mimetics, an apoptotic response is induced, and the tumor cells are rendered sensitive to chemotherapy.5,6,7,8
The ETS family of transcription factors consists of more than 30 members, which are conserved from sea urchin to human beings. Each ETS family member contains a conserved DNA-binding domain of 85 amino acids, the ETS domain, which binds to a purine-rich GGAA/T core sequence.9 ETS proteins bind to DNA as monomers and can activate transcription alone or in conjunction with other transcription factors. Most ETS proteins are nuclear targets of diverse signaling pathways such as the mitogen-activated protein (MAP) kinase signaling pathway and undergo post-translational modifications including phosphorylation, glycosylation, acetylation, ubiquitination, and sumoylation.10 These modifications have a profound impact on the activity and subcellular localization of the ETS proteins.
It has been reported that several receptor tyrosine kinases (RTKs) are activated in mesothelioma, including epidermal growth factor receptor, platelet-derived growth factor receptor, and hepatocyte growth factor (HGF) receptor (c-Met). Clinical trials of imatinib (a platelet-derived growth factor receptor inhibitor) and gefitinib (epidermal growth factor receptor inhibitor) in mesothelioma tumors have shown limited success. Thus, there is great interest in identifying an alternative receptor tyrosine kinase target in these cancers. c-Met is overexpressed and activated in most cases of mesothelioma in comparison with normal adjacent tissue.11,12 In addition, the circulating serum levels of HGF are twofold greater in patients with mesothelioma compared with the healthy population.13 There are numerous signal transduction cascade mechanisms that are activated on HGF stimulation, c-Met phosphorylation, MAP kinase activation, and phosphatidylinositol 3-kinase kinase activation.12,13 Attenuation of c-Met through siRNA and the small molecular inhibitor SU11274 has been found to inhibit both tumor cell growth and migration.14
The association between Bcl-xl and c-Met expression levels was well established in a number of previous studies.15,16 The elevation of Bcl-xl in both tumor and normal cells on HGF exposure indicates a role for activated c-Met in Bcl-xl transcriptional regulation.17,18,19 The anti-apoptotic survival role of activated c-Met has also been partly explained by its activation of the phosphatidylinositol 3-kinase-AKT kinase pathway12 and its angiogenic properties.20 There has been no report to date, however, that has addressed the mechanism underlying the up-regulation of Bcl-xl after c-Met activation. Given also that Bcl-xl and Akt are independent guardians of the mitochondria, which provide the gateway to the intrinsic apoptosis pathways, it is of some importance to elucidate the mechanism by which HGF up-regulates Bcl-xl expression.
We have examined the role of HGF signaling in controlling apoptosis in a human mesothelioma model. The role of c-Met activation in the regulation of Bcl-xl expression via the ETS family of transcription factors has been further clarified.
Materials and Methods
Cell Lines and Reagents
The human mesothelioma cell lines H28, HAY, I45, MSTO, REN, and ROB were maintained in RPMI 1640 medium (Invitrogen, Grand Island, NY) containing 10% fetal bovine serum (Invitrogen). The mesothelioma cell lines SF.HAT and SF.ORT were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) also containing 10% fetal bovine serum. The human lung cancer cell line, H1299, was maintained in RPMI 1640 medium containing 10% fetal bovine serum. I45 cells are sarcomatous subtypes of mesothelioma. REN and H28 are epithelial subtypes. MSTO is a biphasic subtype. The subtypes of HAY, ROB, SF.HAT, and SF.ORT are unknown. Hepatocyte growth factor was obtained from R&D Systems (Minneapolis, MN). Anti-Bcl-xl antibody and all anti-MAP kinase antibodies were purchased from Cell Signaling Technology (Beverly, MA). All antibodies used to detect ETS family transcriptional factors were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-actin monoclonal antibody was purchased from Sigma-Aldrich (St. Louis, MO). Mitogen-activated protein kinase (MEK) inhibitor (MEK inhibitor I) and c-Jun NH2-terminal kinase (JNK) inhibitor (SB203580) were obtained from Calbiochem (San Diego, CA). P38 kinase inhibitor (SB202190) was purchased from LC Laboratories (Woburn, MA).
Plasmids and DNA Transfection
A 1.2-kb fragment of the human Bcl-xl promoter was cloned into the luciferase reporter plasmid pGL2 (Promega, Madison, WI) to generate pXLPR. All deletion mutant constructs were generated by PCR and fully sequenced for verification. The cytomegalovirus (CMV) promoter-based ETS-1, ETS-2, PU.1, and Tel cDNA expression vectors were purchased from Origene (Rockville, MD). Each of these plasmids was purified using a Qiafilter Maxi kit (Qiagen, Valencia, CA). I45 cells were transfected in 24-well plates using FuGENE 6 (Roche, Indianapolis, IN) with 200 ng of pXLPR, and each of the serial deletion plasmid constructs was supplemented with 20 ng of pCMV-βGal as an internal control for transfection efficiency. Several independent experiments using I45 cells were performed in triplicate. At 48 hours after transfection, cell lysates were prepared in 25 mmol/L Tris (pH 7.5)-10% glycerol-1% Triton X-100–2 mmol/L dithiothreitol and analyzed for luciferase and β-galactosidase activities as described by the manufacturer (Applied Biosystems, Foster City, CA). All luciferase activities were normalized to the β-galactosidase internal control.
Immunoprecipitation and Western Blotting
Western blot analyses were performed using a standard method. In brief, cells were lysed in Laemmli buffer, and equal amounts of total protein were electrophoresed on 4 to 20% polyacrylamide/bisacrylamide gels. The proteins resolved were then transferred to a nitrocellulose membrane and incubated with Bcl-xl, actin, MAP kinase, and ETS antibodies. Signals were visualized using the ECL system (GE Amersham, Little Chalfont, Buckinghamshire, UK). For immunoprecipitation experiments, I45 cells were transfected with Tel, ETS-2, and PU.1 expression plasmids using FuGENE 6 and then cultured for 24 hours. These cells were then either untreated or treated with 100 ng/ml HGF for 30 minutes and harvested in 750 μl of lysis buffer (50 mmol/L Tris-HCl [pH 7.4], 150 mmol/L NaCl, 0.05% SDS, 1% deoxycholic acid, 1% Triton X-100, 500 U/ml ulinastatin, and 2 mmol/L phenylmethylsulfonyl fluoride) per 100-mm diameter culture dish. Immunoprecipitations were performed using Tel, ETS-2, and PU.1 antibodies and the Catch and Release V.20 kit (Upstate, Charlottesville, VA). The signals were detected by electrophoresis and autoradiography.
Immunohistochemistry and Immunofluorescent Microscopy
The expression of Bcl-xl and c-Met was determined by immunohistochemical analysis on formalin-fixed and paraffin-embedded mesothelioma tissues arrays. This study was approved by the Scott & White Memorial Hospital Texas Health Science Center Institutional Review Board. Five-micrometer-thick sections of these mesothelioma tissue arrays were deparaffinized in xylene substitute and rehydrated in PBS. Antigen retrieval was performed with citrate buffer (pH 6.5) for 20 minutes at 99°C, followed by the block of endogenous peroxidase activity (3% hydrogen peroxide for 10 minutes). Sections were incubated with blocking serum in PBS containing 5% bovine serum albumin, followed by incubation with rabbit anti-human Bcl-xl polyclonal antibody (Cell Signaling Technology) or with rabbit anti-phosphorylated human c-Met polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hour, followed by incubation with a biotinylated goat secondary anti-rabbit antibody (1:200 dilution, Vector Laboratories, Burlingame, CA). Immunoreactive signals were detected using a streptavidin-biotin-peroxidase complex from Vector Laboratories, according to the manufacturer’s recommended procedures. All of the slides were counterstained with hematoxylin (Sigma-Aldrich). For the negative control, slides were subjected to the same procedures, including antigen retrieval, except for treatment of samples with control rabbit IgG. This negative control clearly demonstrated the specificity of the immunostaining that we observed.
Subcellular localization of ETS proteins was detected by indirect immunofluorescence. In brief, ETS-2-, PU.1- or Tel-transfected I45 cells were plated on coverslips in RMPI 1640 medium containing 10% fetal bovine serum. The cells were then serum-starved or grown in 10% fetal bovine serum for 24 hours. The serum-starved cells were exposed to 100 ng/ml HGF for 20 minutes, fixed, and then stained with ETS-2, PU.1, or Tel antibodies. Positive immunostaining was detected by incubation with a fluorescein isothiocyanate-conjugated secondary antibody plus a 5 ng/ml concentration of Hoechst dye and visualized using epifluorescence microscopy (Olympus BX70).
Quantitative Measurements of Bcl-xl mRNA
Bcl-xl mRNA levels in both patient samples and cell lines were measured using real-time PCR. Total RNAs were extracted using TRIzol from Sigma-Aldrich, and 1-μg aliquots of total RNA from each sample were reverse-transcribed using a TaqMan reverse transcription kit (Applied Biosystems). Primers and probes to detect Bcl-xl and glyceraldehyde-3-phosphate dehydrogenase were obtained from Applied Biosystems. Human total RNA was used as a related standard and human glyceraldehyde-3-phosphate dehydrogenase was used as the internal PCR control. Real-time PCR was performed using an MX4000 Multiplex quantitative PCR system (Stratagene, La Jolla, CA). All reactions were performed in triplicate.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was performed essentially as described by Saccani et al21 with minor modifications. In brief, I45 cells were treated with 1% formaldehyde for 15 minutes. Cross-linked chromatin was then prepared and sonicated to an average size of 1000 bp before being immunoprecipitated with antibodies against Tel, PU.1, or ETS-2 or with control rabbit IgG at 4°C overnight. After reversal of the cross-linking, the immunoprecipitated chromatin was PCR-amplified with primers specific for the Bcl-xl promoter as follows: forward 5′-GCCTAAGGCGGATTTGAATGTAG-3′); reverse 5′-GAAGGGAGAGAAAGAGATTCAGGAA-3′.
Statistical analysis
Relationships between Bcl-xl and phosphorylated c-Met were analyzed statistically using χ2 analysis.
Results
Bcl-xl Is Highly Elevated in Human Mesotheliomas
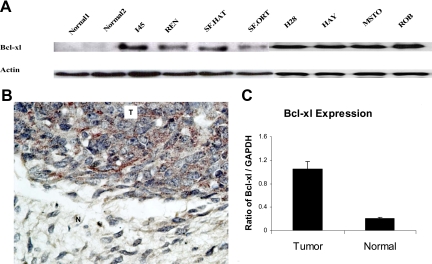
Bcl-xl expression levels in mesothelioma cell lines and in normal lung and pleural tissue (NL1 and NL2) were evaluated by Western blotting with an anti-human Bcl-xl polyclonal antibody. The robust expression of Bcl-xl was evident in all mesothelioma cell lines in contrast with the two normal tissues examined (Figure 1A). Differential Bcl-xl expression in human tumor samples was demonstrated by immunohistochemical analysis in which a strong Bcl-xl signal was detected in the tumor area, whereas the adjacent normal tissue showed no expression of this protein (Figure 1B). The differences in the Bcl-xl RNA levels between the mesotheliomas and normal tissue were further confirmed using real-time PCR analysis of the same human samples used for immunohistochemical staining (Figure 1C).
Figure 1.
A: Baseline Bcl-xl expression levels are increased in mesothelioma. Eight human mesothelioma cell lines and normal lung and pleura tissues (NL1 and NL2) were evaluated by Western blotting. B: Immunohistochemical staining of a mesothelioma tumor specimen (T) and normal surrounding pleura (N) for Bcl-xl. Note the lack of signal in the normal tissue compared with the tumor. C: Evaluation of Bcl-xl mRNA by real-time PCR in normal pleura and mesothelioma samples.
Functional Analysis of the Bcl-xl Promoter
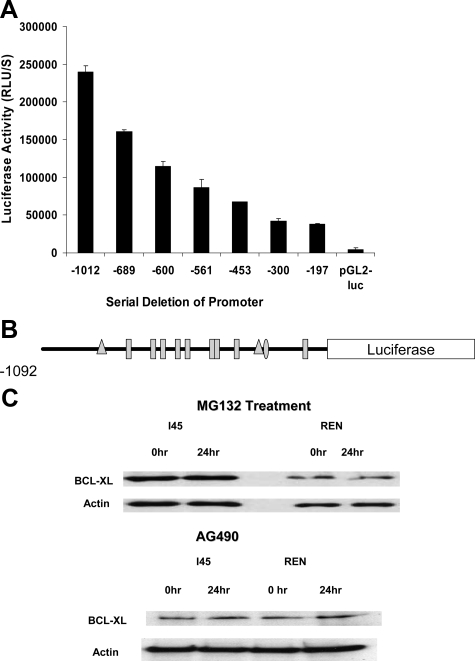
To identify the potential regulatory elements of the human Bcl-xl gene, we performed a transient luciferase assay using a series of 5′ deletions of the Bcl-xl promoter linked to the luciferase reporter gene. pCMV-β-gal cDNA was cotransfected as an internal control (Figure 2A). The data indicate that the Bcl-xl regulatory elements are spread along the entire promoter region. Similar results were obtained in other mesothelioma cell lines. We used the TESS (Transcription Element Search Software) package from the Department of Computational Biology and Informatics Laboratory at the University of Pennsylvania to analyze the putative transcription factor binding sites within the Bcl-xl promoter. Nine ETS-binding sites were identified in the promoter region (Figure 2B) in addition to two NF-κB binding sites and one STAT binding site. Several transcription factors have been reported previously to be involved in the regulation of Bcl-xl expression in a variety of tissues, including ETS-1,2 PU.1, TEL, C-REL, REL A, and STATs.16,22 To evaluate the possible roles of NF-κB and STATs in regulating the Bcl-xl promoter, NF-κB activity was inhibited by the proteasome inhibitor MG132 in the I45 and REN mesothelioma cell lines. Bcl-xl expression was then analyzed by Western blotting but was unaffected at 24 hours after exposure, although the tumor cells had already undergone apoptosis (Figure 2C). The Jak kinase inhibitor, tyrphostin AG490 was used to block the activity of the JAK-STAT pathway in the same mesothelioma cell lines (Figure 2C) but there were no detectable effects on Bcl-xl expression after 24 hours of exposure.
Figure 2.
A: Analysis of Bcl-xl promoter activity by serial deletions of the promoter region. I45 cells were seeded in 96-well plates and transfected with a Bcl-xl promoter reporter, pxl-luc, or a series of 5′ deletions of the Bcl-xl promoter along with a p-CMV-β-galactosidase cDNA control vector using FuGENE 6. A p-GL2 luciferase vector was also transfected as a negative control. At 24 hours after transfection, cells were lysed and luciferase and β-galactosidase activities were measured by luminometry using a dual-light luciferase kit (Applied Biosystems). The luciferase activity was normalized to β-galactosidase activity, and the data shown are the averages of triplicate determinations. This promoter analysis was repeated three times. B: Schematic representation of the location of transcription factor binding sites on the Bcl-xl promoter: triangles, NF-κB; oval, STAT; rectangles, ETS. C: Inhibition of NF-κB or JAK kinase does not suppress Bcl-xl protein expression. The mesothelioma cell lines, I45 and REN, were exposed to proteasome inhibitor MG-132 or JAK kinase inhibitor AG490. These cells were then harvested and Bcl-xl expression was determined by Western blot. RLU, relative light units.
HGF Induces Bcl-xl Expression through the ETS Family of Transcription Factors
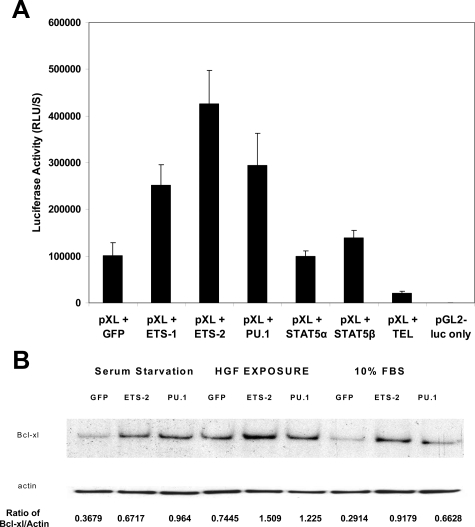
To next determine whether the ETS family of transcription factors regulates Bcl-xl expression, different ETS transcription factor cDNAs or a green fluorescent protein (GFP) cDNA control were cotransfected into I45 cells with the Bcl-xl promoter construct. Cells transfected with the ETS-1, ETS-2, and PU.1 constructs showed much higher luciferase activities than the controls (Figure 3A). We then cotransfected I45 cells with a TEL expression or GFP control vector and the Bcl-xl promoter construct and found from the luciferase activity measurements that the Bcl-xl promoter was much inhibited (Figure 3A).
Figure 3.
A: ETS family transcription factors up-regulate Bcl-xl promoter activity. I45 cells seeded in 96-well plates were cotransfected with Bcl-xl promoter p-XL and either GFP, ETS-1,−2, PU.1, Tel, or STAT expression vectors along with p-CMV-β-galactosidase. A p-GL2 luciferase vector was transfected as a negative control. Twenty-four hours after transfection, the cells were lysed, and the luciferase and β-galactosidase activities were measured with a luminometer. The luciferase activities were normalized to those of β-galactosidase, and the data shown are the average of triplicate determinations. This experiment was repeated twice. B: HGF stimulates Bcl-xl protein expression after ETS-2 and PU.1 transfection. I45 cells were seeded into six-well plates (106 cells) and were transfected with ETS-2, PU.1 and GFP control expression vectors. The cells were then cultured under normal conditions, serum starvation, or serum starvation plus HGF (100 ng/ml). Bcl-xl expression was then determined by Western blot. Protein expression was quantified using an UN-SCAN-IT automated digitalized system (Silk Scientific Corporation, Orem, UT). RLU, relative light units.
We next investigated whether a connection existed between the HGF receptor, c-Met, and Bcl-xl expression in mesothelioma cells and whether overexpressed ETS transcriptional factors could increase the Bcl-xl expression levels. We expressed ETS-2, PU.1, and GFP control cDNA in I45 cells under normal growth conditions or under serum starvation conditions and then exposed the cells to HGF. Compared with the serum-starved samples, Bcl-xl expression was found to be significantly elevated in the untreated I45 cells expressing ETS-2 and the same cells exposed to HGF, respectively (Figure 3B). These results indicate that ETS transcription factors and exposure to HGF activate Bcl-xl gene expression.
HGF Induces the Phosphorylation of ETS Transcription Factors
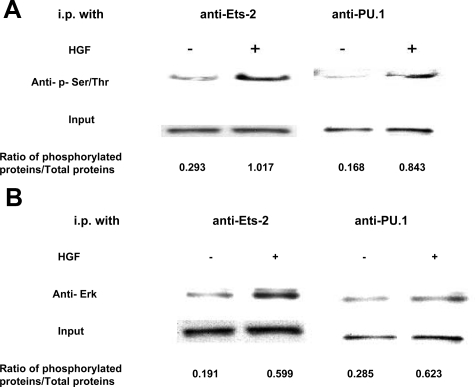
Post-translational modifications are known to influence transcription factor activities. In this regard, the ETS proteins have been reported to be regulated by phosphorylation, glycosylation, acetylation, ubiquitination, and sumoylation.10 To further examine how HGF may affect ETS functions, we analyzed the levels of phosphorylation of the ETS-2 and PU.1 proteins in I45 cells under conditions of serum starvation or HGF stimulation by immunoprecipitation and Western blot analysis. Cell lysates was immunoprecipitated using ETS-2 and PU.1 antibodies, and the phosphor-serine and -threonine levels were detected using phosphor-serine-specific antibodies. Whereas the total ETS levels were observed to be equivalent in the cells, the levels of phosphorylated ETS-2 and PU.1 were clearly elevated (Figure 4A). We next determined whether physical binding occurs between extracellular signal-regulated kinase, ETS-2, and PU.1. ETS-2 and PU.1 proteins were immunoprecipitated from I45 cell lysates that had been treated with PBS or HGF for 30 minutes and subjected to Western blotting. The signals on these blots demonstrated that extracellular signal-regulated kinase is indeed associated with these ETS proteins (Figure 4B).
Figure 4.
HGF induces the phosphorylation of ETS-2 and PU.1 in human mesothelioma cells. The mesothelioma cell line, I45, was grown under conditions of serum starvation or serum starvation plus 20 minutes of HGF (100 ng/ml) stimulation. The endogenous ETS-2 and PU.1 proteins were precipitated from the cell lysates using respective antibodies. The immunoprecipitates were then analyzed by Western blot using phosphor-Ser/Thr antibodies (A) and an ERK antibody (B). The loading was normalized by measuring the ETS-2 and PU.1 expression levels in the cell lysates by Western blotting. Protein expression was quantified using an UN-SCAN-IT automated digitalized system.
HGF Stimulates Bcl-xl Expression by Enhancing Bcl-xl Promoter Transcriptional Activity
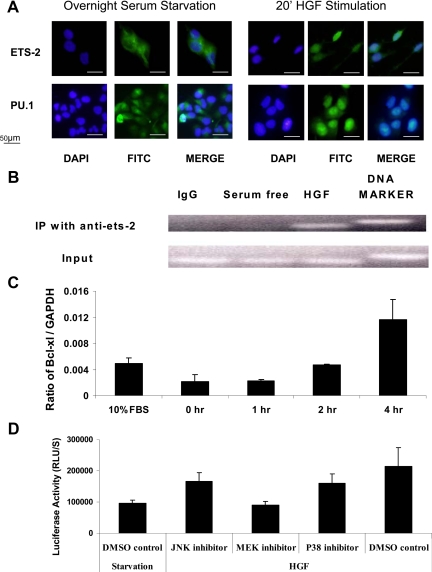
We analyzed the subcellular distribution of ETS-2 and PU.1 using fluorescent microscopy. Twenty minutes after HGF stimulation in serum-starved I45 cells, the ETS-2 and PU.1 proteins showed increased nuclear accumulation (Figure 5A). Furthermore, we analyzed the effects of PU.1 and ETS-2 transcriptional factors on the Bcl-xl promoter in vivo via formaldehyde cross-linking followed by chromatin immunoprecipitation with PU.1 and ETS-2 antibodies. PCR amplification of the immunoprecipitated DNA with primers specific for the Bcl-xl promoter region produced a 200-bp fragment. Compared with the unstimulated samples, HGF stimulation resulted in a significantly increased PCR signal from the chromatin precipitated by ETS-2 antibody (Figure 5B). We did not detect any PCR signal from the chromatin precipitated by PU.1 antibody (not shown). This result suggests that PU.1 plays a limited role in regulating Bcl-xl transcription in mesothelioma. Its regulation of Bcl-xl transcription was only focused in hematopoietic cells.
Figure 5.
A: Increased nuclear localization of ETS transcription factors in mesothelioma cells after HGF treatment. I45 cells were transfected with ETS-2 and PU.1 expression vectors. Twenty-four hours after serum starvation, these transfected cells were exposed to HGF stimulation for 20 minutes. The cells were then washed in PBS and fixed with ice-cold acetone. The fixed cells were next permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate for 2 minutes at 4°C and incubated with ETS-2, PU.1, and Tel antibodies followed by a fluorescein isothiocyanate (FITC)-labeled secondary antibody. Fluorescein signals were visualized under an Olympus IX71 fluorescent microscope. 4,6-Diamidino-2-phenylindole (DAPI) was used as a counterstain for nuclear DNA. B: CHIP (IP) analysis of ETS-2 binding to the Bcl-xl promoter. I45 cells were serum-starved for 24 hours and then subjected to treatment with or without HGF (100 ng/ml) for 20 minutes. The results of the CHIP assay demonstrated increased binding of ETS-2 to the Bcl-xl promoter after HGF treatment. C: Effects of HGF stimulation on the Bcl-xl transcript levels in mesothelioma cells. Bcl-xl mRNA levels were assessed by real-time RT-PCR. Serum-starved cells were stimulated with 100 ng/ml HGF for 0, 2, 4, 8, and 12 hours before harvesting. Normal cultured cells were used as the control group. All values shown are the ratio of Bcl-xl to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and are the averages of triplicate determinations. The results demonstrate that Bcl-xl mRNA levels are significantly increased after HGF stimulation. D: Exposure to a MEK inhibitor abolishes the effects of HGF stimulation on the Bcl-xl promoter as assessed by luciferase reported activity. Serum-starved cells were stimulated with 100 ng/ml HGF after pretreatment with MEK, JNK, and P38 kinase inhibitors. The data are the average of triplicate determinations and demonstrate that Bcl-xl promoter activity is significantly decreased after HGF stimulation and MEK inhibition. DMSO, dimethyl sulfoxide; FBS, fetal bovine serum; RLU, relative light units.
Given that HGF exposure was found to stimulate artificial Bcl-xl promoter activity and enhance ETS transcription factor binding to the endogenous promoter, we assessed whether HGF affected the mRNA levels of endogenous Bcl-xl. Total RNAs were isolated from I45 cells under both normal culture and serum starvation conditions at several different time points after HGF stimulation. The Bcl-xl mRNA levels were found to be significantly increased after 4 hours of HGF exposure, compared with those in untreated serum starved and normal cultured cells (Figure 5C). This up-regulation was also specific to Bcl-xl, as we observed no changes in the levels of other Bcl-2 family members, including Mcl-1, Bak, ad Bax (data not shown).
To further elucidate whether HGF stimulates Bcl-xl expression through the MAP kinase pathway, we analyzed HGF-stimulated Bcl-xl promoter activity in the presence or absence of specific inhibitors of MAP kinases. Pretreatment of cells with an MEK inhibitor (MEK inhibitor I) was found to abrogate HGF-stimulated Bcl-xl promoter activity (Figure 5D). In contrast, pretreatment with JNK inhibitor SB203580 and p38 kinase inhibitor SB202190 had no effect.
HGF Enhances Bcl-xl Expression by Removing Repression by Tel on Bcl-xl Promoter
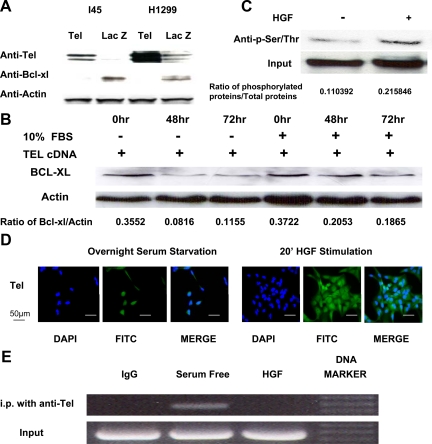
To determine whether Tel would repress bcl-xl expression, Tel and β-galactosidase cDNA expression vectors were transfected into H1299 and I45 cells with high Bcl-xl expression. As shown in Figure 6A, Tel overexpression leads to decreased Bcl-xl expression in both cell lines after 72 hours of transfection. To investigate whether serum starvation could enhance the repressive function of Tel on Bcl-xl expression, we expressed Tel cDNAs in I45 cells under normal growth conditions or under serum starvation conditions for 48 and 72 hours. Bcl-xl expression was found to be significantly decreased in the serum-starved I45 cells in comparison with the I45 cells under normal growth condition (Figure 6B). To examine how HGF may affect Tel functions, we analyzed the levels of phosphorylated Tel protein in I45 cells under conditions of serum starvation or HGF stimulation by immunoprecipitation and Western blot analysis. Tel proteins were immunoprecipitated using Tel antibodies, and phosphorylation levels were detected using phosphor-serine-specific antibodies. Whereas the total Tel levels remained the same in these cells, the levels of phosphorylated Tel were clearly elevated after HGF stimulation (Figure 6C). Next, we analyzed the impact of HGF on subcellular distribution of Tel. As shown in Figure 6D, 20 minutes after HGF stimulation in serum-starved I45 cells, Tel proteins showed increased cytoplasmic accumulation, whereas Tel still remained in nuclear in serum-starved cells. Furthermore, we analyzed the effects of HGF on Tel binding to Bcl-xl promoter using a CHIP assay. Compared with the HGF-stimulated samples, serum starvation resulted in a significantly increased PCR signal of the Bcl-xl promoter from the precipitated chromatin (Figure 6E). Taken together, our results indicate that HGF activates Bcl-xl gene expression through negatively regulating repressive Tel function through phosphorylation.
Figure 6.
A: Overexpressed Tel decreases Bcl-xl protein expression. H1299 and I45 cells were seeded into six-well plates (106 cells) and were transfected with Tel and β-galactosidase expression vectors. Seventy-two hours after transfection, Tel and Bcl-xl expression was determined by Western blot. B: Serum starvation enhanced repression by Tel of Bcl-xl transcription. I45 cells were seeded into six-well plates (106 cells) and were transfected with Tel expression vector. The cells were cultured under normal conditions and serum starvation for 48 and 72 hours. Bcl-xl expression was then determined by Western blot. Protein expression was quantified using an UN-SCAN-IT automated digitalized system. C: HGF induces the phosphorylation of Tel in human mesothelioma cells. The mesothelioma cell line, I45, was grown under conditions of serum starvation or serum starvation plus 20 minutes of HGF (100 ng/ml) stimulation. The endogenous Tel proteins were precipitated from cell lysates using their respective antibodies. The immunoprecipitates were then analyzed by Western blot using phosphorylated-Ser/Thr antibodies. The loading was normalized by measuring the Tel expression levels in the cell lysates by Western blotting. D: Increased cytoplasmic localization of Tel in mesothelioma cells after HGF treatment. Twenty-four hours after serum starvation, I45 cells were exposed to HGF stimulation for 20 minutes. The cells were then washed in PBS and fixed with ice-cold acetone and incubated with Tel antibodies followed by a fluorescein isothiocyanate (FITC)-labeled secondary antibody. Fluorescein signals were visualized under an Olympus IX71 fluorescent microscope. E: CHIP analysis of Tel binding to the Bcl-xl promoter. I45 cells were serum starved for 24 hours and then subjected to treatment with/without HGF (100 ng/ml) for 20 minutes. The results of the CHIP assay demonstrated decreased binding of Tel to the Bcl-xl promoter after HGF treatment. DAPI, 4,6-diamidino-2-phenylindole; FBS, fetal bovine serum.
Positive Correlation between the Bcl-xl and c-Met Expression Levels in Primary Mesothelioma Cells
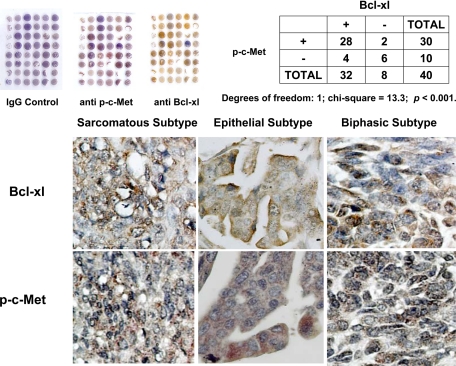
Given the positive association observed between Bcl-xl and c-Met expression in cell culture, we examined whether such a relationship existed in primary human mesothelioma samples. By immunohistochemical staining analysis using mesothelioma tissue arrays, we analyzed the protein expression profile for Bcl-xl and phosphorylated c-Met in 40 patient samples, including 26 epithelial subtypes, 8 sarcomatous subtypes, and 6 biphasic subtypes. χ2 analysis revealed a positive correlation between the levels of Bcl-xl and phosphorylated c-Met (Figure 7).
Figure 7.
Analysis of c-Met and Bcl-xl in human tissue samples. Paraffin-embedded human mesothelioma tissue arrays were stained using primary antibodies against phosphorylated c-Met and Bcl-xl. Rabbit IgG was used as a negative control. A correlation between the Bcl-xl and phosphorylated c-Met expression levels was demonstrated by χ2 analysis (P < 0.01).
Discussion
The c-Met receptor tyrosine kinase has been well studied in malignant mesothelioma and has been shown to be expressed in 82% of human mesothelioma specimens by immunostaining of a tissue array of 66 samples.13 Selective small molecular inhibitors of c-Met kinase have been found to induce apoptosis and suppress cell growth both in vitro and in vivo.23,24 In addition, the activated HGF/Met axis contributes to tumor cell growth and survival,13 and Bcl-xl has been found to be highly expressed in mesothelioma.8 We assessed whether the HGF/Met axis and Bcl-xl were co-expressed in mesothelioma by immunostaining of a mesothelioma tissue array. Our data suggest a strong link between phosphorylated c-MET and Bcl-xl.
Our current data indicate that Bcl-xl is regulated primarily at the transcriptional level in mesothelioma cell lines and patient tumor specimens. Many signal transduction pathways and transcription factors have been reported to be involved in the transcriptional regulation of Bcl-xl. The mechanisms of transcriptional regulation of Bcl-xl vary among different tumor types. NF-κB,2,25 STAT,3 GATA,26 and ETS27 have all been shown to be involved in this process. We aimed to identify the transcription factors and signal transduction pathways involved in Bcl-xl transcription in mesothelioma. Although Bcl-xl is a well known target of NF-κB, NF-κB itself does not play a significant role in Bcl-xl regulation in mesothelioma. Bcl-xl expression did not change when NF-κB activity was reduced by proteasome inhibition, nor was there a change when the activities of STAT transcription factors were blocked by a JAK kinase inhibitor.
In the present study, we have demonstrated that the regulation of Bcl-xl expression is part of the mechanism by which HGF/Met supports tumor survival in mesothelioma, in addition to the many other functions of the HGF/Met axis. ETS transcription factors often function in intracellular regulatory cascades and specific ETS factors have important individual functions in these pathways.28 To identify the ETS transcriptional factors involved in regulating Bcl-xl expression, we functionally examined several family members that regulate Bcl-xl expression. The expression of ETS-2 strongly induced Bcl-xl promoter activity in our experiments. The likelihood that ETS-2 contributes to the induction of Bcl-xl expression in mesothelioma cells was strengthened by our further results showing this ability by exogenous overexpression. In further support of this hypothesis, exogenously expressed Tel was found to repress Bcl-xl promoter activity. MAP kinase-mediated phosphorylation has previously been shown to regulate the transcriptional activation functions of ETS-1 and -2 and also PU.1.29 Our present findings clearly demonstrate that the HGF/Met axis phosphorylates ETS transcriptional factors in mesothelioma cells. Under HGF stimulation, Bcl-xl mRNA and protein levels were elevated, and we observed enhanced binding of ETS-2 to the Bcl-xl promoter.
Our current analyses suggest that post-translational regulation of ETS family proteins regulates Bcl-xl at the transcriptional level. ETS proteins are nuclear proteins although some contain nuclear export signals as well as nuclear localization signals.30,31,32 The phosphorylation of ETS proteins alters their subcellular localization in several cases. We demonstrate that ETS-2 and PU.1 accumulate in the cytoplasm before HGF stimulation. Once HGF has been added to the cell culture, the PU.1 and ETS-2 proteins display nuclear localization. The mechanism underlying this nuclear accumulation is not clear at present. This accumulation could be either the result of increased nuclear import from cytoplasm to nuclei or the result of decreased exportation. The nuclear import of the transcription factor PU.1 occurs via a carrier-independent and energy-dependent process in which PU.1 interacts directly with the nuclear proteins Nup153 and Nup62 through its ETS domain.33,34 The presence of nuclear import signals within the ETS family members also suggests that ETS-2 could be regulated by nuclear import. In addition, PU.1, ETS-1, and ETS-2 could be actively exported from the nucleus to the cytoplasm via a chromosome region maintenance-1/exportin-1-dependent pathway. Chromosome region maintenance/exportin is a nuclear export receptor that exports proteins containing a leucine-rich nuclear export signal to the cytoplasmic compartment.35 The functional nuclear export signal motif was identified within the point domain of the ETS proteins.36 The transcriptional repressors, such as TEL and ERF, are also targets of MAPK.11,37,38 Once phosphorylated, TEL and ERF are removed from the DNA-binding site and their repression of Bcl-xl transcription is abrogated. TEL then interacts with chromosome region maintenance-1 and is exported to the cytoplasm. Other investigators have observed that TEL-induced apoptosis was more dramatic and consistent when cells were cultured in a medium with a lower concentration of serum.39 We propose the following model for how the HGF/Met axis regulates Bcl-xl expression in mesothelioma. High concentrations of HGF constantly activate Met in malignant pleural mesothelioma and in turn activate downstream MAP kinases. These activated MAP kinases can phosphorylate ETS-2 and PU.1, which will stimulate their nuclear import or reduce their nuclear exportation. Phosphorylation of ETS-2 can enhance its function by recruiting the co-activator p300/CBP to the Bcl-xl promoter.40,41 On the other hand, the activated MAP kinases could remove transcriptional repressors from the Bcl-xl promoter by phosphorylating TEL and ERF and facilitating their nuclear export.
Our present findings lend further support to the concept of “context-dependent resistance.” Receptors can mediate the activity of several signaling pathways that are part of molecular circuitries shared with other receptors and that are negatively and positively controlled at multiple levels. In this scenario, the inhibition of Met might have no effect if downstream effectors are constitutively activated or if parallel pathways (such as those driven by epidermal growth factor receptor family membranes) are switched on.42 It has been shown that the Met gene is amplified in lung tumors displaying acquired resistance to epidermal growth factor receptor inhibition and the constitutive Met activation leads to the HER3-dependent activation of the phosphatidylinositol 3-kinase kinase-AKT pathway. It has also been demonstrated that the concomitant inhibition of both receptors results in the severe impairment of cell growth and viability.42 Targeting the common downstream proteins of these receptors or common signal transduction molecules such as Bcl-xl, Akt and their associated transcription factors may be a viable alternative to receptor inhibition approaches.
In summary, we show from our current data that the HGF/Met axis regulates the expression of Bcl-xl through the MAP kinase pathway. Altering the balance between the transcriptional activators and transcriptional repressors that target the Bcl-xl promoter controls the transcriptional regulation of Bcl-xl. Further understanding of the relationship between the HGF/Met axis and the ETS family of transcription factors will probably assist with the development of new targeted therapeutic approaches to the treatment of human mesotheliomas.
Footnotes
Address reprint requests to W. Roy Smythe, M.D., Department of Surgery, Scott & White Memorial Hospital, 2401 South 31st St.; Temple, TX 76508. E-mail: rsmythe@swmail.sw.org.
Supported by the National Institutes of Health (grant R01-CA098545-01A1 to W.R.S.) and Mesothelioma Applied Research Foundation (X.C.).
References
- Greillier L, Astoul P. Mesothelioma and asbestos-related pleural diseases. Respiration. 2008;76:1–15. doi: 10.1159/000127577. [DOI] [PubMed] [Google Scholar]
- Chen C, Edelstein LC, Gelinas C. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol Cell Biol. 2000;20:2687–2695. doi: 10.1128/mcb.20.8.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirito K, Watanabe T, Sawada Ki, Endo H, Ozawa K, Komatsu N. Thrombopoietin regulates Bcl-xL gene expression through Stat5 and phosphatidylinositol 3-kinase activation pathways. J Biol Chem. 2002;277:8329–8337. doi: 10.1074/jbc.M109824200. [DOI] [PubMed] [Google Scholar]
- Sevilla L, Aperlo C, Dulic V, Chambard JC, Boutonnet C, Pasquier O, Pognonec P, Boulukos KE. The Ets2 transcription factor inhibits apoptosis induced by colony-stimulating factor 1 deprivation of macrophages through a Bcl-xL-dependent mechanism. Mol Cell Biol. 1999;19:2624–2634. doi: 10.1128/mcb.19.4.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Rodarte C, Zhang L, Morgan CD, Littlejohn J, Smythe WR. Bcl2/bcl-xL inhibitor engenders apoptosis and increases chemosensitivity in mesothelioma. Cancer Biol Ther. 2007;6:246–252. doi: 10.4161/cbt.6.2.3626. [DOI] [PubMed] [Google Scholar]
- Littlejohn JE, Cao X, Miller SD, Ozvaran MK, Jupiter D, Zhang L, Rodarte C, Smythe WR. Bcl-xL antisense oligonucleotide and cisplatin combination therapy extends survival in SCID mice with established mesothelioma xenografts. Int J Cancer. 2008;123:202–208. doi: 10.1002/ijc.23452. [DOI] [PubMed] [Google Scholar]
- Ozvaran MK, Cao XX, Miller SD, Monia BA, Hong WK, Smythe WR. Antisense oligonucleotides directed at the bcl-xl gene product augment chemotherapy response in mesothelioma. Mol Cancer Ther. 2004;3:545–550. [PubMed] [Google Scholar]
- Smythe WR, Mohuiddin I, Ozveran M, Cao XX. Antisense therapy for malignant mesothelioma with oligonucleotides targeting the bcl-xl gene product. J Thorac Cardiovasc Surg. 2002;123:1191–1198. doi: 10.1067/mtc.2002.121684. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays. 2005;27:285–298. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- Arai H, Maki K, Waga K, Sasaki K, Nakamura Y, Imai Y, Kurokawa M, Hirai H, Mitani K. Functional regulation of TEL by p38-induced phosphorylation. Biochem Biophys Res Commun. 2002;299:116–125. doi: 10.1016/s0006-291x(02)02588-3. [DOI] [PubMed] [Google Scholar]
- Ramos-Nino ME, Blumen SR, Sabo-Attwood T, Pass H, Carbone M, Testa JR, Altomare DA, Mossman BT. HGF mediates cell proliferation of human mesothelioma cells through a PI3K/MEK5/Fra-1 pathway. Am J Respir Cell Mol Biol. 2008;38:209–217. doi: 10.1165/rcmb.2007-0206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran R, Ma PC, Seiwert TY, Jagadeeswaran S, Zumba O, Nallasura V, Ahmed S, Filiberti R, Paganuzzi M, Puntoni R, Kratzke RA, Gordon GJ, Sugarbaker DJ, Bueno R, Janamanchi V, Bindokas VP, Kindler HL, Salgia R. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res. 2006;66:352–361. doi: 10.1158/0008-5472.CAN-04-4567. [DOI] [PubMed] [Google Scholar]
- Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, Hansen M, Schaefer E, Naoki K, Lader A, Richards W, Sugarbaker D, Husain AN, Christensen JG, Salgia R. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- Fornoni A, Li H, Foschi A, Striker GE, Striker LJ. Hepatocyte growth factor, but not insulin-like growth factor I protects podocytes against cyclosporin A-induced apoptosis. Am J Pathol. 2001;158:275–280. doi: 10.1016/S0002-9440(10)63966-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad JM, Zeng XR, Boise LH. Regulation of Bcl-xL: a little bit of this and a little bit of STAT. Curr Opin Oncol. 2000;12:543–549. doi: 10.1097/00001622-200011000-00006. [DOI] [PubMed] [Google Scholar]
- Fan S, Ma YX, Wang JA, Yuan RQ, Meng Q, Cao Y, Laterra JJ, Goldberg ID, Rosen EM. The cytokine hepatocyte growth factor/scatter factor inhibits apoptosis and enhances DNA repair by a common mechanism involving signaling through phosphatidyl inositol 3′ kinase. Oncogene. 2000;19:2212–2223. doi: 10.1038/sj.onc.1203566. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Mallet VO, Guidotti JE, Goulenok C, Kahn A, Gilgenkrantz H. Liver repopulation by Bcl-xL transgenic hepatocytes. Am J Pathol. 2002;160:31–35. doi: 10.1016/S0002-9440(10)64345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang J, Liu Y. Role of Bcl-xL induction in HGF-mediated renal epithelial cell survival after oxidant stress. Int J Clin Exp Pathol. 2008;1:242–253. [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Karakiewicz PI, Roehrborn CG, Lotan Y, Zlotta AR, Shariat SF. Predictive value of plasma hepatocyte growth factor/scatter factor levels in patients with clinically localized prostate cancer. Clin Cancer Res. 2008;14:7385–7390. doi: 10.1158/1078-0432.CCR-07-5110. [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med. 2001;193:1351–1360. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla L, Zaldumbide A, Pognonec P, Boulukos KE. Transcriptional regulation of the bcl-x gene encoding the anti-apoptotic Bcl-xL protein by Ets, Rel/NFκB, STAT and AP1 transcription factor families. Histol Histopathol. 2001;16:595–601. doi: 10.14670/HH-16.595. [DOI] [PubMed] [Google Scholar]
- Accornero P, Lattanzio G, Mangano T, Chiarle R, Taulli R, Bersani F, Forni PE, Miretti S, Scuoppo C, Dastru W, Christensen JG, Crepaldi T, Ponzetto C. An in vivo model of Met-driven lymphoma as a tool to explore the therapeutic potential of Met inhibitors. Clin Cancer Res. 2008;14:2220–2226. doi: 10.1158/1078-0432.CCR-07-2064. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P, Warn A, Dobbin S, Arakaki N, Daikuhara Y, Jaurand MC, Warn RM. Expression of HGF/SF in mesothelioma cell lines and its effects on cell motility, proliferation and morphology. Br J Cancer. 1998;77:1052–1059. doi: 10.1038/bjc.1998.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YL, Chiang YJ, Chen YC, Papetti M, Juo CG, Skoultchi AI, Yen JJY. MAPK-mediated phosphorylation of GATA-1 promotes Bcl-XL expression and cell survival. J Biol Chem. 2005;280:29533–29542. doi: 10.1074/jbc.M506514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla L, Zaldumbide A, Carlotti F, Dayem MA, Pognonec P, Boulukos KE. Bcl-xL expression correlates with primary macrophage differentiation: Activation of functional competence, and survival and results from synergistic transcriptional activation by Ets2 and PU 1. J Biol Chem. 2001;276:17800–17807. doi: 10.1074/jbc.M008270200. [DOI] [PubMed] [Google Scholar]
- Oettgen P. Regulation of vascular inflammation and remodeling by ETS factors. Circ Res. 2006;99:1159–1166. doi: 10.1161/01.RES.0000251056.85990.db. [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD, Whitmarsh AJ. Transcriptional regulation by the MAP kinase signaling cascades. Gene. 2003;320:3–21. doi: 10.1016/s0378-1119(03)00816-3. [DOI] [PubMed] [Google Scholar]
- Do HJ, Song H, Yang HM, Kim DK, Kim NH, Kim JH, Cha KY, Chung HM, Kim JH. Identification of multiple nuclear localization signals in murine Elf3, an ETS transcription factor. FEBS Lett. 2006;580:1865–1871. doi: 10.1016/j.febslet.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Le Gallic L, Virgilio L, Cohen P, Biteau B, Mavrothalassitis G. ERF nuclear shuttling, a continuous monitor of Erk activity that links it to cell cycle progression. Mol Cell Biol. 2004;24:1206–1218. doi: 10.1128/MCB.24.3.1206-1218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Seo Y, Kim JI, Kim WJ, Choe SY. Identification of the nuclear localization motif in the ETV6 (TEL) protein. Cancer Genet Cytogenet. 2006;167:117–121. doi: 10.1016/j.cancergencyto.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Kwok JC, Perdomo J, Chong BH. Identification of a monopartite sequence in PU.1 essential for nuclear import, DNA-binding and transcription of myeloid-specific genes. J Cell Biochem. 2007;101:1456–1474. doi: 10.1002/jcb.21264. [DOI] [PubMed] [Google Scholar]
- Zhong H, Takeda A, Nazari R, Shio H, Blobel G, Yaseen NR. Carrier-independent nuclear import of the transcription factor PU.1 via RanGTP-stimulated binding to Nup153. J Biol Chem. 2005;280:10675–10682. doi: 10.1074/jbc.M412878200. [DOI] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulli V, Romania P, Morsilli O, Gabbianelli M, Pagliuca A, Mazzeo S, Testa U, Peschle C, Marziali G. Overexpression of Ets-1 in human hematopoietic progenitor cells blocks erythroid and promotes megakaryocytic differentiation. Cell Death Differ. 2006;13:1064–1074. doi: 10.1038/sj.cdd.4401811. [DOI] [PubMed] [Google Scholar]
- Maki K, Arai H, Waga K, Sasaki K, Nakamura F, Imai Y, Kurokawa M, Hirai H, Mitani K. Leukemia-related transcription factor TEL is negatively regulated through extracellular signal-regulated kinase-induced phosphorylation. Mol Cell Biol. 2004;24:3227–3237. doi: 10.1128/MCB.24.8.3227-3237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouras DN, Athanasiou MA, Beal GJ, Jr, Fisher RJ, Blair DG, Mavrothalassitis GJ. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin BJ, Wood LD, Wang L, Fenrick R, Sansam C, Packham G, Kinch M, Yang E, Hiebert SW. TEL, a putative tumor suppressor induces apoptosis and represses transcription of Bcl-XL. J Biol Chem. 2003;278:46378–46386. doi: 10.1074/jbc.M305189200. [DOI] [PubMed] [Google Scholar]
- Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore C, Giordano S. Molecular cancer therapy: can our expectation be MET? Eur J Cancer. 2008;44:641–651. doi: 10.1016/j.ejca.2008.01.022. [DOI] [PubMed] [Google Scholar]