Abstract
Gpnmb is a glycosylated transmembrane protein implicated in development of glaucoma in mice and melanoma in humans. It shares significant amino acid sequence homology with the melanosome protein Pmel-17. Its extracellular domain contains a RGD motif for binding to integrin and its intracellular domain has a putative endosomal and/or melanosomal-sorting motif. These features led us to posit that Gpnmb is associated with melanosomes and involved in cell adhesion. We showed that human Gpnmb is expressed constitutively by melanoma cell lines, primary-cultured melanocytes, and epidermal melanocytes in situ, with most of it found intracellularly within melanosomes and to a lesser degree in lysosomes. Our newly developed monoclonal antibody revealed surface expression of Gpnmb on these pigment cells, albeit to a lesser degree than the intracellular fraction. Gpnmb expression was upregulated by UVA (but not UVB) irradiation and by α-MSH (but not β-MSH); its cell surface expression on melanocytes (but not on melanoma cells) was increased markedly by IFN-γ and TNF-α. PAM212 keratinocytes adhered to immobilized Gpnmb in a RGD-dependent manner. These results indicate that Gpnmb is a melanosome-associated glycoprotein that contributes to adhesion of melanocytes with keratinocytes.
Keywords: Adhesion, Gpnmb, keratinocytes, melanocytes, melanoma
Introduction
Subtractive cDNA cloning led us to discover DC-HIL, a type I transmembrane glycoprotein, expressed constitutively by antigen presenting cells (APC), especially dendritic cells (DC), but also macrophages (1). DC-HIL is also referred to as Gpnmb (2,3), osteoactivin (4) and hematopoietic growth factor-inducible neurokinin-1 type (HGFIN) (5). Because DC-HIL first appeared in the melanocyte literature as Gpnmb, we will use this term instead of DC-HIL.
Gpnmb shares highest amino acid sequence homology with: QNR-71 (responsible for melanin production in quail neuroretina cells) (48%); a melanosome protein Pmel-17/gp100 (25%); and lysosome-associated membrane protein-1 (LAMP-1) (13%) (1). Extracellularly, Gpnmb contains: a RGD motif that binds to integrins (6); several N-glycosylation sites; a proline-rich region and an Ig-like polycystic kidney disease (PKD) domain shared by PKD-susceptible gene products (7,8). The intracellular domain contains an immunoreceptor tyrosine-based activation motif (ITAM)-like motif (YxxI) (9). It also contains a di-leucine-based signal (ExxPLL), which is found in the cytoplasmic domain of QNR-71 that is required for its sorting to the endosomal/premelanosomal compartments (10,11). By contrast, the same motif in Pmel-17 acts just as an internalization motif (12).
Mouse and human DC-HIL share 71% amino acid sequence homology and both contain the aforementioned motifs. However, they differ in the number of amino acids (574 for mouse and 572 for human) and of possible N-glycosylation sites (11 in mouse and 12 in human); the latter may account for differences in size of the human and mouse Gpnmb protein. Moreover, human Gpnmb gene produces a truncated form (560 amino acids) with deletion of 12 amino acids in the proline-rich region corresponding to the first 36 nucleotides of exon 7. A corresponding isoform has not been identified in mice.
We showed that Gpnmb binds to heparin/heparan sulfate and adheres to endothelial cells via its RGD motif (1). We also discovered that Gpnmb expressed by APC serves as an inhibitor of T cell activation by binding syndecan-4 on activated T cells (13,14). In melanocyte biology, a premature stop codon mutation in the Gpnmb (GpnmbR150X) gene was shown to cause iris pigment dispersion in mouse pigmentary glaucoma (3). Gpnmb gene expression also correlated inversely with the metastatic capacity of human melanoma (2). Thus, Gpnmb may play important roles in melanocytes biology.
To better characterize Gpnmb expression in melanocytes and melanoma, we developed a monoclonal antibody (mAb) against human or mouse Gpnmb. We showed Gpnmb expression on the surface and intracellularly, with preferential localization within melanosomes. Gpnmb expression was upregulated by UVA (but not UVB) irradiation and by α-melanocyte-stimulating hormone (α-MSH, but not β-MSH), and by IFN-γ and TNF-α. Finally, Gpnmb induced adhesion to keratinocytes in a RGD-dependent manner. Our results suggest that Gpnmb promotes adhesion of melanocytes to keratinocytes.
Methods
Cell culture
B16F10 melanoma, EL-4 T lymphoma, P815 mastocytoma, PAM212 keratinocyte (all mouse origin), SK-MEL-28 melanoma and 293T embryonal kidney cells (both human origin), and COS-1 cells were purchased from ATCC, and all maintained in Dulbecco’s MEM supplemented with 10% FCS. Human primary melanocytes were purchased from Cambrex Bio Science Walkersville Inc. (Walkersville, MD), and maintained in Melanocyte Cell Basal Medium-4 and MGM-4 Single Quots (both from Cambrex). These cells were used experimentally after 5 to 8 passages.
Expression vectors and production of Fc-fused proteins
A plasmid vector encoding mouse or human Gpnmb (pmGpnmb or phGpnmb) was constructed by inserting the full-length cDNA into pcDNA3.1 (Invitrogen, Carlsbad, CA) using HindIII and XbaI restriction enzyme sites. A plasmid vector (pSTB-hDC-HIL-hFc) encoding the extracellular domain of Gpnmb fused to the Fc portion of human IgG was constructed by replacing the extracellular domain of mouse Gpnmb in pSTB-mDC-HIL-hFc (encoding also human IgG-Fc) (13) with that of the human homolog, using HindIII and XbaI sites. We also constructed pSTB-hDC-HIL-mFc, in which the extracellular domain was fused with mouse IgG-Fc, by replacing the Fc portion of pSTB-hDC-HIL-hFc with the corresponding portion of mouse IgG, using XbaI and ApaI. A plasmid encoding Fc-fused RAA mutant (RGD mutated to RAA) was constructed previously (1).
Fc-fusion proteins were produced in COS-1 cells and purified as described previously (13). Purity of the final preparation was very high, judged by a single band on SDS-PAGE and Coomassie Blue staining and in immunoblotting using anti-human (or mouse) IgG Ab or anti-Gpnmb mAb.
Generation of UTX-103 and 3D5 mAb
To generate anti-mouse Gpnmb mAb, 2 rabbits were immunized 4 times with 0.5 mg of recombinant protein mGpnmb-hFc (fused with human IgG-Fc) 3 weeks apart. A week after the third immunization, sera were collected and the titer of anti-mouse Gpnmb evaluated by ELISA using hGpnmb-mFc (fused with mouse IgG-Fc) to eliminate anti-human IgG Ab titer. Rabbit spleen cells were fused with a rabbit myeloma cell line (Epitomics, Burlingame, CA). One clone (UTX-103 mAb IgG1) was chosen and IgG purified from the culture supernatant using Protein A-agarose (Invitrogen).
To generate anti-human Gpnmb mAb, 5 BALB/c mice were each immunized and boosted 3 times with 100 µg of hGpnmb-mFc at 2 week intervals. A week after the last immunization, spleen cells from mice with highest Ab titer were fused with the F/0 myeloma cell line. One IgG1 clone (3D5 mAb) was purified from mouse ascites, using protein-G (Invitrogen) affinity chromatography.
Flow cytometry
For surface expression, pigment cells (1×105) were incubated with UTX-103, 3D5 mAb, or control Ab (each 10 µg/ml). After washing, cells were labeled fluorescently with secondary Ab (PE- or FITC-anti-rabbit IgG or anti-mouse IgG, 5–10 µg/ml). For intracellular staining, cells (1×105) were fixed with 4% paraformaldehyde (PFA) at room temperature for 20 min. After washing, cells were treated with permealization buffer (0.5% Saponin, 0.5% BSA, PBS) for 10 min, and then incubated with anti-Gpnmb mAb (10 µg/ml) in the permealization buffer for 30 min, followed by treatment with secondary Ab (5 µg/ml). Fluorescence intensity of stained cells was analyzed by FACS Calibur (Becton Dickinson, San Jose, CA).
DNA transfection and immunoblotting
Expression vector encoding mouse Gpnmb (2 µg) was transfected into P815 cells (1×106) using Amaxa nucleofector system (Amaxa Inc. Gaithersburg, MD). After culturing for 2 days, P815 cells were cultured in the presence of G418 (1 mg/ml) for 2 weeks and then Gpnmb+ cells were purified by FACS sorting. A plasmid DNA (pmGpnmb or phGpnmb) was delivered into COS-1 cells using Fugene6 (Roche Diagnostics, Indianapolis, IN) (15). At 2–3 days post-transfection, whole cell extracts were prepared from COS-1 cells by lysis with 0.3% Triton X-100/DPBS for 15 min, followed by centrifugation for 20 min at 10,000×g (15). Whole cell extracts were also prepared from other cell lines. Typically, an aliquot (40 µg) of extract was applied to SDS/4–20% gradient PAGE, followed by immunoblotting using UTX-103 or 1E4 (2–10 µg/ml), 3D5 mAb (1 µg/ml), mouse anti-Mel-5 mAb (Clone Ta99, Signet, Deham, MA) (1:10 dilution), rat anti-LAMP-1 mAb (eBioscience, San Diego, CA) (2 µg/ml), or rabbit anti-β-actin polyconal Ab (0.5 µg/ml, Abcam, Cambridge, MA). Color was developed by HRP-secondary Ab (1:10,000 dilution, Jackson ImmunoResearch, West Grove, PA) for 1 h and ECL plus system (Amersham Pharmacia Biotech, Piscataway, NY). Intensity of specific protein bands was measured in the linear range by a densitometric analysis program provided by Image Quant 400 (GE Healthcare, Piscataway, NJ).
Glycosidase treatment
An aliquot (20 µg) of whole cell extracts from melanoma cells was denatured by heating to 100°C for 10 min in the buffer [0.5% SDS, 40mM DTT], and incubated with 50,000 U/ml peptide: N-glycosidase F (New England Biolabs, Ipswich, MA) at 37°C for 1 h. Untreated and treated samples were run in parallel on SDS-PAGE, followed by immunoblotting using anti-Gpnmb mAb.
Confocal fluorescence microscopy
A day prior to staining, melanoma cells were seeded on tissue culture chamber slides (2×105 cells/well) (Lab-Tek, Naperville, IL). Slides were fixed with 4% PFA at room temperature for 20 min. After extensive washing, slides were treated with 5% BSA for 1 h and stained with biotinylated rat anti-LAMP-1 mAb (2 µg/ml), mouse anti-MEL-5 mAb (1:250 dilution) or control IgG plus Alexa 546 streptavidin or anti-mouse IgG (0.5 µg/ml, Invitrogen). After washing, slides were stained with UTX-103 mAb (5 µg/ml) or control IgG, and with anti-rabbit Alexa 488 (0.5 µg/ml, Invitrogen). For staining of dendritic cells, the cells harvested from day 6 culture of bone marrow cells with GM-CSF (16) were doubly stained with UTX-103 and anti-LAMP-1 mAb.
Skin specimens from a biopsy of a healthy subject were snap-frozen, and sectioned (4 µm thickness). The slides were fixed with ethanol, treated with 3% BSA and 10% goat serum (for blocking), and then stained with anti-microphthalmia transcription factor (MITF) mAb (3 µg/ml, clone C5+D5, Invitrogen) and Alexa 546 F(ab')2 fragment of goat anti-mouse IgG (2 µg/ml, Invitrogen) at room temperature for 1 h. After rinsing, slides were again stained with biotinylated 3D5 mAb (10 µg/ml) and Alexa 488-streptavidin (2 µg/ml). Biotinylation of 3D5 mAb was performed using EZ-Link™ NHS-Biotin (PIERCE, Rockford, IL). Control staining was carried out with mouse IgG/Alexa 546 anti-mouse IgG and bitonylated mouse IgG/Alexa 488-streptavidin at the same concentration. Fluorescently stained cells and skin tissues were examined using a Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss Microimaging, Thornwood, NY).
Preparation of vesicle- and melanosome-enriched fractions
These fractions were obtained by sucrose gradient centrifugation, as described previously (17). Briefly, B16F10 cells were homogenized in buffer [0.25 M sucrose, 50 mM sodium phosphate buffer (pH 6.5), 1 mM EGTA, 0.5 mM MgCl2, 100 µM phenylmethylsulfonyl fluoride]. After centrifugation at 700×g for 10 min, the supernatant was recovered and recentrifuged at 11,000×g for 10 min. The pellet was kept for melanosome isolation, and the supernatant used for vesicle isolation. Following centrifugation of the supernatant at 100,000×g for 1 h, the pellet (small granule fractions containing lysosomes) was recovered, resuspended in the homogenizing buffer, and applied to a discontinuous gradient of 0.3–1.8M sucrose at 100,000×g for 1 h. To enrich melanosome-fractions, the pellet was resuspended in the homogenizing buffer and applied to a discontinuous gradient of 1.0–2.0 M sucrose and centrifuged for 90 min at 100,000×g. All fractions were collected separately and an aliquot (5.6% of the total volume) of each fraction was applied to SDS-PAGE and immunoblotting.
Cell stimulation
Pigment cells were seeded on chamber slides (2×105 cells/well). 2–3 days after seeding, cells were washed and cultured for indicated time period in the presence of α-MSH (100 nM or varying concentrations), β-MSH (0.2 µM) plus 3-isobutyl-1-methylxanthine (IBMX) (0.1mM), IFN-γ (10 ng/ml) plus phorbol 12-myristate 13-acetate (PMA) (200 unit/ml), or TNF-α (50 ng/ml).
UV irradiation
B16F10 cells (5×105) were resuspended in PBS and irradiated with broadband UVB at different doses, using a bank of four FS40 fluorescent lamps (Philips, Eindhoven, Netherlands) that emit between 280 and 320 nm with a peak at 313 nm (UVC was filtered) (18). B16F10 cells were also irradiated with UVA at different doses, using a tanning lamp (Phillips, HB 404, power 21 mW/cm2) emitting UVA with a maximum at 365 nm. Light intensity was determined by a Waldmann photometer (Waldmann GBH, Schwenningen, Germany).
Adhesion Assays
Adhesion to immobilized Gpnmb-Fc was measured as reported previously (1). PAM212 keraticnocytes (3×105 cells) were radio-labeled with 1 µCi/ml 3H-thymidine (ICN, Costa Mesa, CA) overnight. After harvesting with 0.5 mM EDTA/PBS, cells were assayed for radioactivity using a liquid scintillation counter (usually 0.2–1.5 cpm/cell). During labeling, Gpnmb-Fc or control IgG at indicated concentrations were immobilized to 96-well ELISA plates (in triplicate) by precoating 100 µg/ml goat anti-human Fcγ IgG overnight at 4 °C. Labeled PAM212 cells (2×104/well) were cultured in the Fc fusion protein-coated wells at 37 °C for indicated time period. In some experiments, wells were coated with fibronectin (from human plasma, Invitrogen) (80 µg/ml). After extensive washing, PAM212 cells that adhered to wells were lysed with 1 N NaOH and measured for 3H-cpm. Specific adhesive activity was assessed by cpm left after subtracting with cpm in IgG-coated well.
Results
Gpnmb protein is expressed by melanocytes and melanoma cell lines
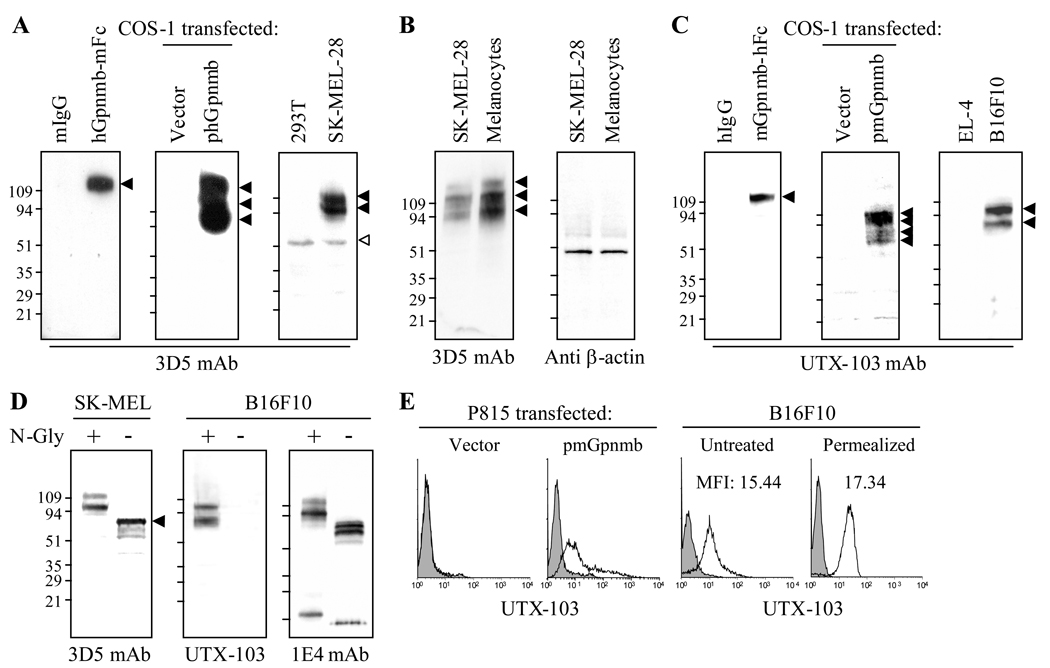
To better examine protein expression of Gpnmb in cells and tissues, we created mAb against human and mouse Gpnmb-Fc (the extracellular domain fused with the Fc portion of IgG). 3D5 mouse anti-human Gpnmb mAb was selected and evaluated for specificity to Gpnmb using immunoblotting (Fig. 1A). 3D5 mAb immunostained human Gpnmb-mFc (fused with mouse IgG) but not the fusion partner mouse IgG; it also stained three protein bands of 90, 120, and 142 KDa in whole cell extracts prepared from COS-1 cells transfected with human Gpnmb gene, but not from those transfected with empty vector. This mAb also stained two major bands of 95 and 120 KDa and a faint band of 142 KDa in SK-MEL-28 melanoma cells (but not 293T cells devoid of endogenous mRNA expression). The 60 KDa band is non-specific because it was also found in protein extracts from 293T cells; the expression level differs among protein samples. We also compared protein expression in SK-MEL-28 melanoma cells vs. human normal melanocytes (Fig. 1B). Melanocytes expressed even greater Gpnmb than SK-MEL-28 cells, with similar β-actin expression levels in both cells.
Figure 1. Protein expression of Gpnmb by melanoma and melanocytes.
(A) Human Gpnmb-Fc, mouse IgG (2 µg/lane), or whole cell extracts (40 µg/lane) from COS-1 transfected with vector or phGpnmb vector (encoding human Gpnmb), or 293T cells were examined by immunoblotting for immunoreactivity to 3D5 mouse anti-human Gpnmb mAb. (B) Protein expression of Gpnmb in SK-MEL-28 cells was compared with that in primary-cultured human normal melanocytes. Expression of β-actin was also examined. (C) Mouse Gpnmb-Fc, human IgG, or whole cell extracts (40 µg/lane) from COS-1 cells transfected with pmGpnmb (encoding mouse Gpnmb) or empty vector, EL-4 or B16F10 cells were also subjected to immunoblotting using UTX-103 rabbit anti-mouse Gpnmb mAb. Protein bands immunoreactive to 3D5 or UTX-103 mAb are shown by closed arrowheads. Open arrowhead indicates non-specific band. (D) Whole cell extract from SK-MEL-28 or B16F10 was treated with (+)/without (−) N-glycosidase (N-Gly), followed by immunoblotting using anti-human (3D5) or anti-mouse Gpnmb mAb (UTX-103 or 1E4). (E) P815 cells transfected with pmGpnmb or vector alone were examined by flow-cytometry for surface expression of Gpnmb using UTX-103 mAb. B16F10 melanoma cells were left untreated (for surface expression) or permealized (for intracellular expression) and stained with UTX-103 mAb (open histogram) or control IgG (filled in gray). Expression was examined by flow-cytometry.
Previously we created 1E4 rat anti-mouse Gpnmb mAb with low affinity for native Gpnmb protein (but high affinity to the denatured form) (13). To create mAb with high affinity to the native form, we immunized rabbit with Gpnmb-hFc (fused with the human IgG) and selected UTX-103 clone. We then examined its specificity to mouse Gpnmb using immunoblotting (Fig. 1C). This mAb immunostained mouse Gpnmb-Fc but not human IgG. Surprisingly, this mAb stained four bands of 74, 80, 98, and 107 KDa proteins in cell extracts from COS-1 cells transfected with pmGpnmb (but not with vector alone). Immunostaining with UTX-103 mAb revealed 80 and 98 KDa bands in B16F10 melanoma (but not in EL-4 T-lymphoma devoid of Gpnmb mRNA expression); it did not immunostain 107 KDa protein. To examine whether heterogeneity (multiple bands) in molecular weight is due to glycosylation, whole cell extracts prepared from SK-MEL-28 or B16F10 cells were treated with N-glycosidase and the reduction in the molecular weight examined by immunoblotting (Fig. 1D). In SK-MEL-28, N-glycosidase greatly reduced the 2 bands (95 and 120 KDa) to a major band of 78 KDa, with minor bands that migrated faster than it. Surprisingly, N-glycosidase abrogated immunoreactivity of UTX-103 anti-mouse Gpnmb, suggesting that an N-glycan may be involved in the epitope. By contrast, immunoblotting of the same protein sample with 1E4 mAb showed reduction in molecular weight, but it produced 2 bands (80 and 74 KDa), which may be due to another glycosylation (i.e., O-glycosylation). Heterogeneity in molecular weight and differences between mouse and human appears due to different levels of N-glycosylation.
We next examined immunoreactivity of UTX-103 mAb to cell surface Gpnmb (Fig. 1E). P815 mastocytoma cells (lacking Gpnmb expression) transfected with mouse Gpnmb gene or empty vector were incubated with UTX-103 mAb and then labeled fluorescently, followed by flow-cytometric analysis. UTX-103 mAb labeled the surface of P815 cells transfected with Gpnmb but not control cells. We then examined surface expression by melanoma cells. B16F10 cells were untreated (for surface staining) or permealized (for intracellular staining) with saponin prior to staining with UTX-103 mAb. These staining demonstrated that Gpnmb is expressed constitutively on the cell surface, although levels lower than intracellular staining (Fig. 1E). Human SK-MEL-28 cells also expressed Gpnmb on the surface (data not shown). These results confirm specificity of 3D5 and UTX-103 mAb to their corresponding Gpnmb and that UTX-103 rabbit anti-mouse Gpnmb mAb has greater affinity to cell-anchored Gpnmb protein compared to our previously established 1E4 rat anti-Gpnmb mAb (13). No cross-reactivity of these mAb to mouse or human Gpnmb was observed.
Gpnmb+ epidermal cells are melanocytes in human skin
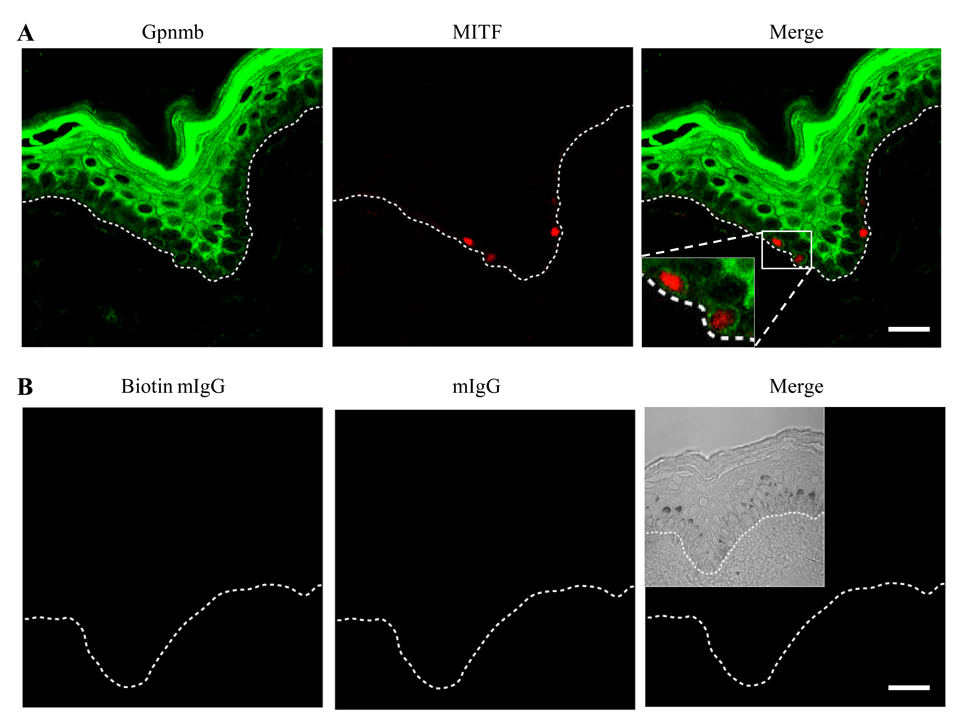
To examine Gpnmb expression by melanocytes in skin, we stained human skin from normal healthy adult volunteers with 3D5 mAb (shown in green fluorescence) and anti-MITF mAb (a marker for melanocytes (19), shown in red), followed by confocal analysis (Fig. 2). 3D5 mAb stained the cytoplasm of almost all epidermal cells (with low intensity in the basal layer and high intensity in the spinous and the granular layer) and of melanocytes sitting on the basement membrane (nuclei was shown in red and the cytoplasm in green). Although Gpnmb expression by Langerhans cells was not shown in this staining, it is noted by flow-cytometric analysis of an epidermal cell suspension stained with 3D5 and anti-CD1a mAb (data not shown). These results indicate that keratinocytes and melanocytes in human skin express Gpnmb. Our study focused on characterization of Gpnmb in melanocytes.
Figure 2. In situ expression of Gpnmb in human skin.
Skin biopsy of human healthy adult was snap-frozen, thin-sectioned and stained with 3D5 mAb (green fluorescence) and anti-MITF mAb (red) (A) or corresponding control IgG (biotinylated and unmodified mouse IgG for 3D5 and anti-MITF, respectively) (B). Stained specimen was examined under confocal microscope at a magnification of 63x. Fluorescent images are merged (Merge, a scale bar represents 20 nm). The dotted line indicates location of basement membrane. A phase contrast picture is inserted in the merge of control staining.
Intracellular localization of Gpnmb
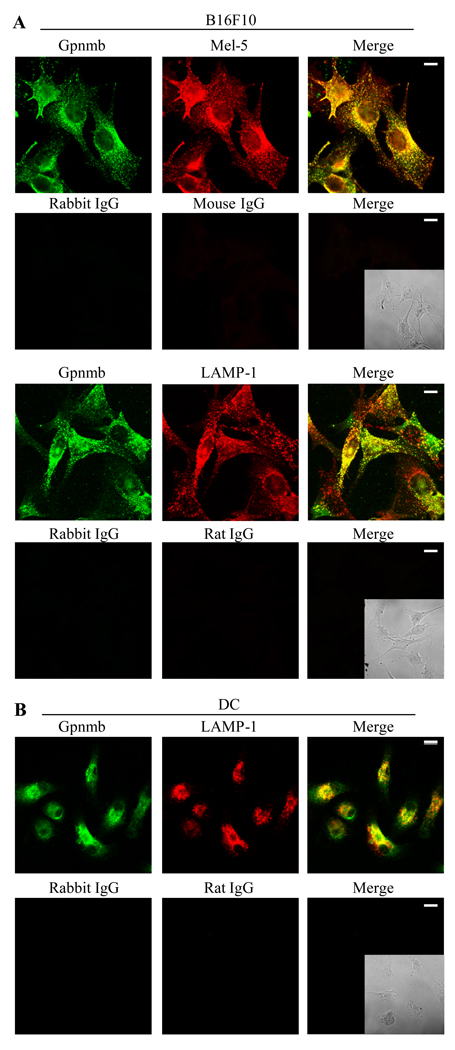
Because Gpnmb structurally resembles Pmel-17 and LAMP-1 and because it contains an endosomal sorting motif in its intracellular domain, we questioned whether intracellular Gpnmb resides within melanosomes and/or lysosomes (Fig. 3). B16F10 cells were fixed, permealized, and doubly-stained with UTX-103 mAb (stained in green fluorescence) and Ab against a melanosome (Mel-5) or lysosome marker (LAMP-1) (both in red). We examined intracellular distribution of Gpnmb, Mel-5, or LAMP-1, using confocal image analysis (Fig. 3A). Anti-Gpnmb-labels accumulated around the nuclei and in intracellular small vesicles, with a distribution pattern similar to that of Mel-5 protein. The majority of Gpnmb+ vesicles were also stained with anti-Mel-5 Ab (in yellow), although some Mel-5+ vesicles (melanosomes) in the periphery did not stain with anti-Gpnmb mAb. By contrast, the distribution of Gpnmb differed from LAMP-1 protein: most LAMP-1+ vesicles did not stain with anti-Gpnmb mAb, but LAMP-1+/Gpnmb+ vesicles were found in the few B16F10 cells. Such heterogeneity may be due to high density of vesicles in the cells. To examine distribution of Gpnmb relative to lysosomes in the absence of melanosomes, we stained dendritic antigen presenting cells (DC) as non-pigment cells (Fig. 3B). As expected from previous studies (20), many lysosomes were seen with high expression of LAMP-1, most also staining with UTX-103 mAb. Altogether, our results indicate that Gpnmb protein resides preferentially in melanosomes; however in the absence of melanosomes, Gpnmb localizes to lysosomes.
Figure 3. Intracellular localization of Gpnmb in melanoma cells and DC.
(A) B16F10 melanoma cells were fixed, permealized, and doubly-stained with UTX-103 (in green fluorescence) and anti-Mel-5 or anti-LAMP-1 mAb (in red), followed by confocal image analysis (Scale bar represents 10 nm). (B) DC generated from bone marrow cells were immunofluorescently stained with UTX-103 mAb and LAMP-1 mAb. Staining with control IgG is shown under staining with specific Ab. Fluorescence images of cells are merged (Merge), in which phase contrast image of stained cells is inserted. Data shown is representative of three independent experiments.
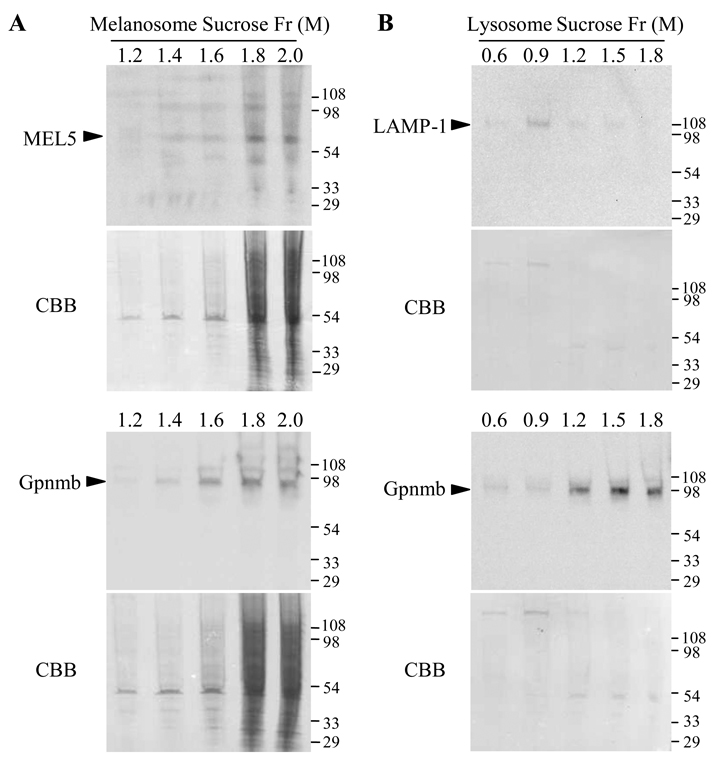
To more precisely examine association of Gpnmb with melanosomes, we enriched melanosomes or lysosomes in B16F10 cells by sucrose-gradient centrifugation and assayed expression of Gpnmb or marker proteins in each fraction (Figs. 4A and B). Immunoblotting showed melanosomal protein Mel-5 to co-migrate with the vast majority of Gpnmb (1.8–2.0 M sucrose fraction) (Fig. 4A). Protein amounts per fraction are shown on blotted membranes stained with coomassie brilliant blue. By contrast, in lysosome-sucrose gradient, Gpnmb protein migrated with 1.2–1.8M sucrose fractions, and it was found minimally in the 0.9M sucrose fraction where LAMP-1 migrated (Fig. 4B). These results strengthen the concept of preferential association of Gpnmb with melanosomes.
Figure 4. Fractionation of lysosomes and melanosomes.
Crude fractions containing melanosomes (A) and lysosomes (B) were prepared from B16F10 melanoma cells and then further fractionated by a discontinuous gradient of 1.0–2.0 M and 0.3–1.8 M sucrose, respectively. An aliquot of each fraction was examined for expression of Gpnmb (using 1E4 mAb) and a marker protein (Mel-5 for melanosomes or LAMP-1 for lysosomes) by immunoblotting. Coomasie staining of blotted membranes is shown under the immunoblotting. Blotting images shown in B were acquired by longer exposure than those in A. Second experiment also showed consistent results.
Gpnmb responds to particular extracellular stimuli
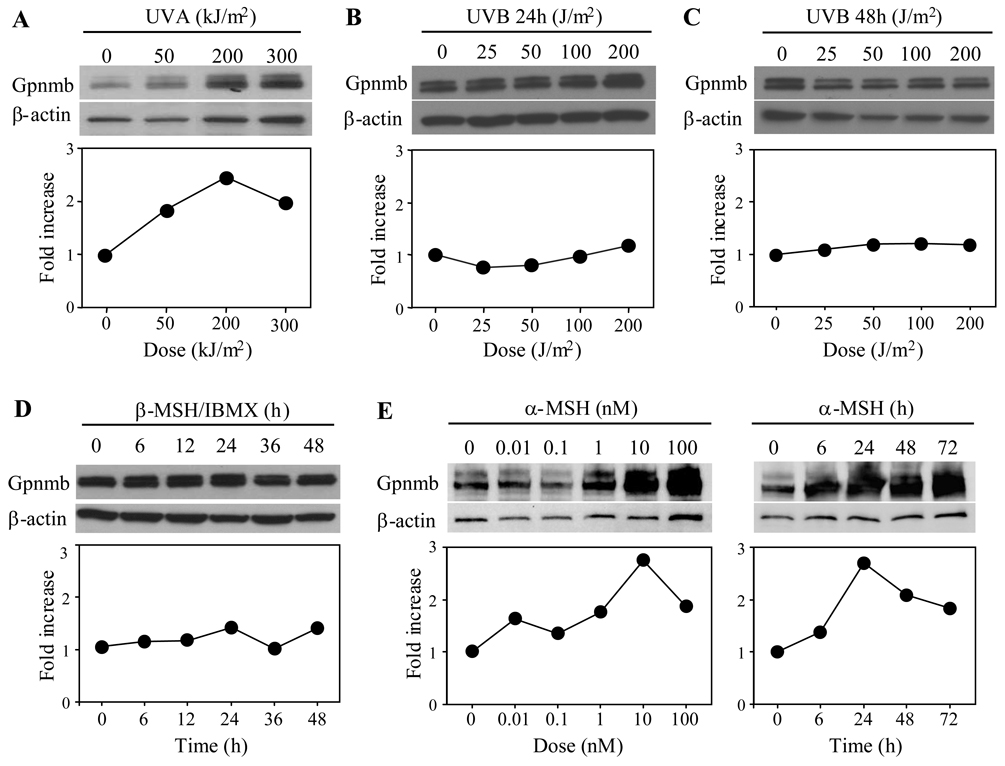
We next assayed the ability of Gpnmb in pigment cells to respond to various extracellular stimuli. B16F10 cells were stimulated with UVA or UVB. A day after stimulation, Gpnmb (with β-actin as control) was assayed by immunoblotting (Fig. 5). UVA upregulated Gpnmb expression in a dose-dependent manner (unexpectedly, β-actin expression was also upregulated to some extent). We thus assessed fold increase by relative expression levels of Gpnmb to β-actin. UVA irradiation (200 KJ/m2) induced a peak response of 2.5-fold greater than control (Fig. 5A). By contrast, UVB irradiation (up to 200 J/m2) had no effect on protein expression (Fig. 5B) and even after culturing for 48 h following irradiation (Fig. 5C). We also examined effects of α-MSH or β-MSH plus IBMX on protein expression (Figs. 5D and E). Gpnmb protein was upregulated by α-MSH (but not β-MSH/IBMX) in a dose-dependent manner up to 8-fold, as compared to basal levels (untreated cells). Correcting for β-actin upregulation, we calculated the increase to be 3-fold.
Figure 5. UVA irradiation upregulates expression of Gpnmb.
B16F10 cells were irradiated with different doses of UVA (A) or UVB (B and C) and cultured for 24 h (A and B) or 48 h (C). B16F10 cells were also cultured with β-MSH (0.2 µM) plus IBMX (0.1mM) (D) for the indicated time or cultured with varying doses of α-MSH for indicated time (E). Whole cell extracts (20 µg/lane) were prepared from treated cells and assayed by immunoblotting for expression of Gpnmb or β-actin. Expression levels shown under the immunoblotting are expressed as fold increase over control that is calculated by dividing experimental relative expression level (intensity of Gpnmb band/that of β-actin) by control expression level (untreated cells). Data shown are representative of three experiments (A and B) and 2 experiments (C through E).
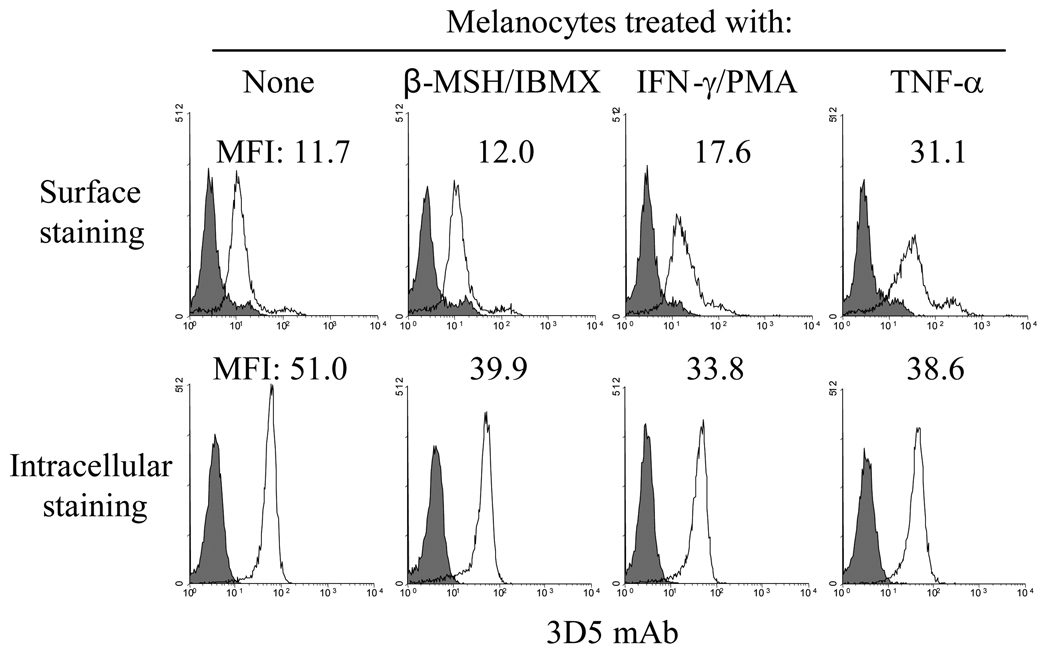
We next compared surface vs. intracellular expression of Gpnmb in human melanocytes. Consistent with immunoblotting data (Fig. 5D), β-MSH/IBMX had no effect on surface expression by melanocytes; if at all it produced some down-regulation in intracellular fractions (Fig. 6). By contrast, the pro-inflammatory cytokines IFN-γ and TNF-α, which can modulate melanin synthesis or melanocyte proliferation (21), upregulated surface expression; control mean fluorescence intensity (MFI) increased from 11.7 to 17.6 and 31.1, respectively. This is likely due to transport to the cell surface rather than de novo protein synthesis because there was no change (a decrement if at all) in intracellular expression. Interestingly, none of the cytokines tested failed to change surface expression of Gpnmb in mouse B16F10 or human SK-MEL-28 melanoma cells (data not shown). We were unable to assay UVA-dependent upregulation in human melanocytes because they were highly toxic to UVA radiation.
Figure 6. Cytokine-dependent regulation of Gpnmb surface expression.
Human melanocytes were cultured in the presence of different agents. After culturing for 1 day, cells were stained with 3D5 mAb (open histograms) or control IgG (closed histograms). Staining was examined by flow-cytometry. Mean fluorescence intensity (MFI) of staining by 3D5 mAb is shown in the histograms. Second experiment showed similar results.
PAM212 keratinocytes adhered to Gpnmb in a RGD-dependent manner
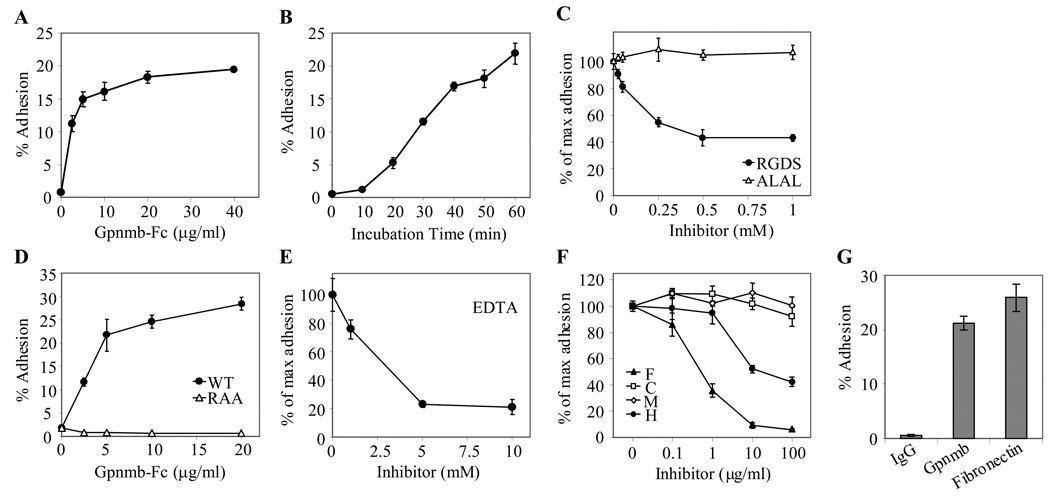
Previously, we showed that a human endothelial line SVEC adheres to immobilized DC-HIL in a RGD-dependent manner (1). This led us to examine whether Gpnmb can mediate adhesion to keratinocytes. Using PAM212 keratinocytes (which do not express Gpnmb) as a surrogate, we radio-labeled these cells and incubated them in 96-ELISA wells immobilized with Gpnmb-Fc. PAM212 cells adhered to immobilized Gpnmb in a dose-dependent manner (but not to control IgG) (Fig. 7A): At its maximum, 20% of input PAM212 cells adhered. At a constant amount of Gpnmb-Fc (20 µg), we showed that increasing incubation time promoted the adhesion up to 25% (Fig. 7B). We next assessed whether the RGD motif of Gpnmb was required for adhesion (Fig. 7C). Addition of a RGDS tetramer peptide inhibited adhesion by up to 40% of the maximum adhesion activity. However, the RAA mutant (RGD was mutated to RAA) lost the adhesive activity completely (Fig. 7D). Adhesion was calcium-dependent as shown by its abrogation by EDTA (Fig. 7E). Finally, we examined a role for polysaccharides in the adhesion (Fig. 7F) because we showed previously that Gpnmb binds to heparin/heparan sulfate (1). PAM212 cells were allowed to adhere Gpnmb-Fc in the presence of different doses of polysaccharides. Fucoidan was the most effective inhibitor, followed by heparin; chondroitin sulfate and mannan had no effect. This profile is different from adhesion of SVEC to Gpnmb, suggesting that a disparate mechanism may be involved for PAM212 keratinocytes. We also compared adhesion of PAM212 cells to Gpnmb vs. fibronectin, an extracellular matrix glycoprotein that binds to integrins (Fig. 7G). The molecular weight of fibronectin (220 KDa) is about twice as large as similar to Gpnmb-Fc (120KDa) (22) and these two proteins contain only one RGD motif (23). To make a fair comparison, we coated ELISA wells with 40 µg/ml for Gpnmb-Fc or 80 µg/ml for fibronectin. Gpnmb-Fc exhibited 20% adhesion, similar to that of fibronectin (80 µg/ml).
Figure 7. PAM212 keratinocytes adhere to immobilized Gpnmb in a RGD-dependent manner.
(A) Dose-dependent adhesion: 3H-thymdine-labeled PAM212 cells were allowed to adhere immobilized Gpnmb-Fc at varying doses. Adherent cells were lysed and measured for 3H-cpm. Adhesive activity is expressed as percent 3H-cpm relative to total input cpm. (B) Incubation time: At different time points after incubation of PAM212 cells with immobilized Gpnmb-Fc (20 µg/ml), adhesion to Gpnmb was assayed by 3H-cpm. (C) RGD-dependency: Adhesion of PAM212 cells to Gpnmb was blocked by addition of RGDS or ALAL (tetramer peptide) at varying doses. Adhesive activity is shown as % of maximum adhesion (without inhibitors). (D) RGD-deficient Gpnmb: PAM212 cells were incubated in 96-ELISA wells precoated with wild-type Gpnmb-Fc or RAA mutant (20 µg/ml) for 1 h. Adhesion is expressed by % adhesion of input PAM212 cells. (E and F) Effect of inhibitors on adhesion: Adhesion assay was performed in the presence of EDTA (E) or polysaccharides (F) (F, fucoidan; C, chondroitin sulfate; M, mannan; and H, heparin) at different concentrations. (G) Comparison with adhesion to fibronectin: 3H–labeled PAM212 cells were incubated for 60 min in ELISA wells precoated with IgG, Gpnmb-Fc (each 40 µg/ml) or fironectin (80 µg/ml) and adhesion measured. All data are representative of at least three independent experiments.
Finally, we examined inhibition of Gpnmb-mediated adhesion by UTX-103 mAb or Gpnmb-Fc protein (each 40 µg/ml). Continuous presence of these inhibitors during adhesion assay inhibited just 10–20%, compared to controls (rabbit IgG or human Fc protein) (data not shown). This poor ability may be due either to inability of UTX-103 mAb to block access of integrins on PAM212 cells (the mAb epitope may be distal from the RGD motif) or to low molecular ratio of the RGD sequence on Gpnmb-Fc to integrins on PAM212 cells.
Discussion
Gpnmb mRNA and protein were shown to be expressed by human melanoma cells (2), but their subcellular localization and function in these cells were not identified. Recently green fluorescent protein (GFP)-fused Gpnmb was shown to localize to endosome-like vesicles in non-pigmented COS-7 and HEK293 cells (24). Our present study in non-transfected melanoma cells show that Gpnmb primarily resides intracellularly mostly within melanosomes, but with a minor portion on the cell surface where it may serve as a receptor for integrins regulating cell migration and proliferation (25,26). Other melanosome proteins like Pmel-17, tyrosinase, and tyrosinase-related protein-1 (Tyrp1) can be expressed transiently on the cell surface (11,12,27–29). Importantly, Gpnmb-mediated adhesion may be regulated by pro-inflammatory cytokines that upregulate the surface expression.
In a DC line, Gpnmb accumulated within a single large vesicle adjacent to the nucleus (perinuclear lysosome) and also in smaller vesicles (other lysosomes) scattered in the periphery (1). Localization study using UTX-103 mAb also confirmed accumulation in lysosomes of normal DC prepared from bone marrow cells. By contrast, in B16F10 melanoma cells, Gpnmb localized primarily in melanosomes and to a lesser degree in some lysosomes. This difference in lysosome- vs. melanosome-localization may be due to the specific presence of the adaptor protein (AP) complex for endosomal sorting system in melanocytes (26,30). For example, Pmel-17 is escorted by AP1 and/or AP2 from early endosomes or trans-Golgi to the stage II melanosomes (31). We assume that Gpnmb is also captured similarly by the AP complex and sorted from endosomes to melanosomes. In the absence of such AP, Gpnmb may very well be transported to lysosomes and cell surfaces (e.g. DC) (32,33).
E-cadherin is the major molecule mediating melanocyte adhesion to keratinocytes (34); α2, α3, and β1 integrins also play a minor but significant role in this process (35). In fact, adhesion of B16F10 to PAM212 cells was inhibited by up to 25% following addition of RGDS peptide (data not shown), consistent with a previous report showing anti-integrin Ab that inhibits adhesion of melanocytes to keratinocytes by 20% (34). Thus, interaction of Gpnmb with integrins may be an additional mechanism mediating adhesion of melanocytes to keratinocytes. However, we showed Gpnmb-mediated adhesion to be as strong as fibronectin-mediated adhesion, which is an extracellular matrix also utilizing a RGD-binding domain (36).
Melanin is synthesized in melanosomes and stored there prior to transfer to keratinocytes in response to external stimuli (26,37). We examined the possibility that Gpnmb may be involved in transfer of melanosomes from melanocytes to keratinocytes by labeling B16F10 melanoma with CSFE and co-culturing these cells with PAM212 cells in the presence or absence of UTX-103 mAb or Gpnmb-Fc protein. Either inhibitor failed to inhibit transfer. Although the biological significance of Gpmb is still unknown, we speculate that Gpnmb may regulate melanocyte function in response to stimuli transducing ITAM-based tyrosine phosphorylation signals. In fact, our recent studies indicate that cross-linking of Gpnmb by mAb induces tyrosine phosphorylation in DC (manuscript in preparation).
The human homolog of Gpnmb has been implicated in the development of the iris and retinal pigment epithelium (24), susceptibility to intraocular pressure elevation (3), and anti-melanoma immunity as a tumor-associated antigen akin to MART-1 and Pmel-17/gp100 (38). These observations suggest an important function of Gpnmb in pigment cells. Our demonstration, Gpnmb adheres to PAM212 keratinocytes in a RGD-dependent fashion, suggests a role for integrin-mediated binding between melanocytes and keratinocytes that may be involved in development of melanocytes, melanin synthesis, and generation of melanoma (26,35). Moreover, Gpnmb-mediated adhesion may be achieved bidirectionally because keratinocytes and melanocytes express Gpnmb and integrins, respectively.
Acknowledgments
We are grateful to Irene Dougherty for technical assistance and Susan Milberger for administrative assistance. This work was supported by grants from the National Institutes of Health (A164927-01) and VA merit review.
References
- 1.Shikano S, Bonkobara M, Zukas PK, Ariizumi K. Molecular cloning of a dendritic cell-associated transmembrane protein, DC-HIL, that promotes RGD-dependent adhesion of endothelial cells through recognition of heparan sulfate proteoglycans. J Biol Chem. 2001;276:8125–8134. doi: 10.1074/jbc.M008539200. [DOI] [PubMed] [Google Scholar]
- 2.Weterman MA, Ajubi N, van Dinter I, et al. nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. Int J Cancer. 1995;60:73–81. doi: 10.1002/ijc.2910600111. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MG, Smith RS, Hawes NL, et al. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 4.Safadi FF, Xu J, Smock SL, et al. Cloning and characterization of osteoactivin, a novel cDNA expressed in osteoblasts. J Cell Biol. 2001;84:12–26. doi: 10.1002/jcb.1259. [DOI] [PubMed] [Google Scholar]
- 5.Metz RL, Patel PS, Hameed M, et al. Role of human HGFIN/nmb in breast cancer. Breast Cancer Res. 2007;9:R58. doi: 10.1186/bcr1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 7.Bycroft M, Bateman A, Clarke J, et al. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J. 1999;18:297–305. doi: 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting CP, Hofmann K, Bork P. A latrophilin/CL-1-like GPS domain in polycystin-1. Curr Biol. 1999;9:R585–R588. doi: 10.1016/s0960-9822(99)80379-0. [DOI] [PubMed] [Google Scholar]
- 9.Strzelecka A, Kwiatkowska K, Sobota A. Tyrosine phosphorylation and Fc gamma receptor-mediated phagocytosis. FEBS Lett. 1997;400:11–14. doi: 10.1016/s0014-5793(96)01359-2. [DOI] [PubMed] [Google Scholar]
- 10.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 11.Le Borgne R, Planque N, Martin P, et al. The AP-3-dependent targeting of the melanosomal glycoprotein QNR-71 requires a di-leucine-based sorting signal. J Cell Sci. 2001;114:2831–2841. doi: 10.1242/jcs.114.15.2831. [DOI] [PubMed] [Google Scholar]
- 12.Theos AC, Berson JF, Theos SC, et al. Dual loss of ER export and endocytic signals with altered melanosome morphology in the silver mutation of Pmel17. Mol Biol Cell. 2006;17:3598–3612. doi: 10.1091/mbc.E06-01-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung JS, Sato K, Dougherty I, et al. DC-HIL is a negative regulator of T lymphocyte activation. Blood. 2007;109:4320–4327. doi: 10.1182/blood-2006-11-053769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung JS, Dougherty I, Cruz PD, Jr, Ariizumi K. Syndecan-4 mediates the coinhibitory function of DC-HIL on T cell activation. J Immunol. 2007;179:5778–5784. doi: 10.4049/jimmunol.179.9.5778. [DOI] [PubMed] [Google Scholar]
- 15.Sato K, Shikano S, Xia G, et al. Selective expression of vacuolar H+-ATPase subunit d2 by particular subsets of dendritic cells among leukocytes. Mol Immunol. 2006;43:1443–1453. doi: 10.1016/j.molimm.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 16.Inaba K, Inaba M, Romani N, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araki K, Horikawa T, Chakraborty AK, et al. Small Gtpase rab3A is associated with melanosomes in melanoma cells. Pigment Cell Res. 2000;13:332–336. doi: 10.1034/j.1600-0749.2000.130505.x. [DOI] [PubMed] [Google Scholar]
- 18.Bonkobara M, Yagihara H, Yudate T, et al. Ultraviolet B Radiation Upregulates Expression of Dectin-2 on Epidermal Langerhans Cells by Activating the Gene Promoter. Photochem Photobiol. 2005;81:944–948. doi: 10.1562/2004-10-21-RC-349. [DOI] [PubMed] [Google Scholar]
- 19.King R, Weilbaecher KN, McGill G, et al. Microphthalmia transcription factor. A sensitive and specific melanocyte marker for MelanomaDiagnosis. Am J Pathol. 1999;155:731–738. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geuze HJ. The role of endosomes and lysosomes in MHC class II functioning. Immunol Today. 1998;19:282–287. doi: 10.1016/s0167-5699(98)01269-9. [DOI] [PubMed] [Google Scholar]
- 21.Krasagakis K, Garbe C, Zouboulis CC, Orfanos CE. Growth control of melanoma cells and melanocytes by cytokines. Recent Results Cancer Res. 1995;139:169–182. doi: 10.1007/978-3-642-78771-3_12. [DOI] [PubMed] [Google Scholar]
- 22.Engvall E, Ruoslahti E. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int J Cancer. 1977;20:1–5. doi: 10.1002/ijc.2910200102. [DOI] [PubMed] [Google Scholar]
- 23.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 24.Bachner D, Schroder D, Gross G. mRNA expression of the murine glycoprotein (transmembrane) nmb (Gpnmb) gene is linked to the developing retinal pigment epithelium and iris. Brain Res Gene Expr Patterns. 2002;1:159–165. doi: 10.1016/s1567-133x(02)00012-1. [DOI] [PubMed] [Google Scholar]
- 25.De LM, Pellegrini G, Zambruno G, Marchisio PC. Role of integrins in cell adhesion and polarity in normal keratinocytes and human skin pathologies. J Dermatol. 1994;21:821–828. doi: 10.1111/j.1346-8138.1994.tb03296.x. [DOI] [PubMed] [Google Scholar]
- 26.Schallreuter KU, Kothari S, Chavan B, Spencer JD. Regulation of melanogenesis--controversies and new concepts. Exp Dermatol. 2008;17:395–404. doi: 10.1111/j.1600-0625.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 27.Calvo PA, Frank DW, Bieler BM, et al. A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. J Biol Chem. 1999;274:12780–12789. doi: 10.1074/jbc.274.18.12780. [DOI] [PubMed] [Google Scholar]
- 28.Simmen T, Schmidt A, Hunziker W, Beermann F. The tyrosinase tail mediates sorting to the lysosomal compartment in MDCK cells via a di-leucine and a tyrosine-based signal. J Cell Sci. 1999;112(Pt 1):45–43. doi: 10.1242/jcs.112.1.45. [DOI] [PubMed] [Google Scholar]
- 29.Lepage S, Lapointe R. Melanosomal targeting sequences from gp100 are essential for MHC class II-restricted endogenous epitope presentation and mobilization to endosomal compartments. Cancer Res. 2006;66:2423–2432. doi: 10.1158/0008-5472.CAN-05-2516. [DOI] [PubMed] [Google Scholar]
- 30.Hearing VJ. Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 31.Valencia JC, Watabe H, Chi A, et al. Sorting of Pmel17 to melanosomes through the plasma membrane by AP1 and AP2: evidence for the polarized nature of melanocytes. J Cell Sci. 2006;119:1080–1091. doi: 10.1242/jcs.02804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raposo G, Marks MS. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harper DC, Theos AC, Herman KE, et al. Premelanosome amyloid-like fibrils are composed of only golgi-processed forms of Pmel17 that have been proteolytically processed in endosomes. J Biol Chem. 2008;283:2307–2322. doi: 10.1074/jbc.M708007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang A, Eller MS, Hara M, et al. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. J Cell Science. 1994;107:983–992. doi: 10.1242/jcs.107.4.983. [DOI] [PubMed] [Google Scholar]
- 35.Haass NK, Smalley KS, Li L, Herlyn M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18:150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole EA. Extracellular matrix and keratinocyte migration. Clin Exp Dermatol. 2001;26:525–530. doi: 10.1046/j.1365-2230.2001.00891.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 38.Ahn JH, Lee Y, Jeon C, et al. Identification of the genes differentially expressed in human dendritic cell subsets by cDNA subtraction and microarray analysis. Blood. 2002;100:1742–1754. [PubMed] [Google Scholar]