Abstract
The study of micro- or nanocrystalline proteins by magic-angle spinning (MAS) solid-state NMR (SSNMR) gives atomic-resolution insight into structure in cases when single crystals cannot be obtained for diffraction studies. Subtle differences in the local chemical environment around the protein, including the characteristics of the co-solvent and the buffer, determine whether a protein will form single crystals. The impact of these small changes in formulation is also evident in the SSNMR spectra, but leads only to correspondingly subtle changes in the spectra. Here we demonstrate that several formulations of GB1 microcrystals yield very high-quality SSNMR spectra, although only a subset of conditions enable growth of single crystals. We have characterized these polymorphs by X-ray powder diffraction and assigned the SSNMR spectra. Assignments of the 13C and 15N SSNMR chemical shifts confirm that the backbone structure is conserved, indicative of a common protein fold, but sidechain chemical shifts are changed on the surface of the protein, in a manner dependent upon crystal packing and electrostatic interactions with salt in the mother liquor. Our results demonstrate the ability of SSNMR to reveal minor structural differences among crystal polymorphs. This ability has potential practical utility for studying formulation chemistry of industrial and therapeutic proteins, as well as for deriving fundamental insights into the phenomenon of single crystal growth.
Keywords: formulation chemistry, polymorphism, multidimensional, crystal packing, single crystal diffraction, therapeutic proteins
Introduction
Protein crystals offer numerous benefits to industrial and pharmaceutical research over their amorphous soluble or lyophilized counterparts. One benefit of crystallization is improved protein stability, which can be further increased with the introduction of chemical cross-linking.1,2 For example, formulation and cross-linking protocols resulted in increased enzymatic activity and substrate specificity of lipases.1 Pharmaceutical applications of protein crystals benefit from higher concentrations, improved delivery pharmacokinetics, stability and versatility in routes of drug administration.3–5 Insulin, perhaps the most famous therapeutic protein, has been used since 1922 to treat diabetic patients.4,6 Microcrystalline precipitates of insulin provide a controlled release formulation that better mimics the properties of naturally secreted insulin.3,5,7 Despite the great potential of protein crystallization, its application to other therapeutic proteins has so far been limited,1,3,8 in part because many crystallization reagents are not fit for human consumption. Failure to identify effective batch-scale crystallization conditions within this limitation has hindered further use of this method to the expanding number of therapeutic proteins and industrial enzymes.1–3 Therefore improved methods of analysis are likely to accelerate product development, as well as aid in developing a better understanding of the fundamental physical chemistry that underlies crystal growth.
Solid-state NMR (SSNMR) provides a powerful approach to studying the formulation and structure of crystallized proteins, even in cases where single crystals fail to grow. Many formulations of nanocrystals, microcrystals or precipiates from low molecular weight alcohols, polyethylene glycol or salt yield excellent SSNMR spectra.9 Martin and Zilm showed that nanocrystalline preparations of ubiquitin and several other proteins yielded consistently narrow SSNMR linewidths and gave equivalent 13C SSNMR and X-ray powder diffraction spectra to single crystal preparations.9 Micro- or nanocrystalline proteins are easier to prepare by batch crystallization methods and are subject to the same crystal packing effects as single crystals, making SSNMR a sensitive tool to measure changes in hydrogen bonding, ionic interactions and protonation states among crystal formulations.
Specific examples of SSNMR studies to examine crystal formulations include 13C chemical shift analysis of ubiquitin,10,11 peptides12,13 and numerous organic compounds.14–18 One such study by Paulson et al. showed that nanocrystalline sample preparations of ubiquitin yielded three different sets of 1H-15N correlation spectra, resulting from three different polymorphs.10 Subsequently Siedel et al. assigned the C’, CA and CB chemical shifts for two of these polymorphs.11 Both of these studies focused on chemical shift differences among the backbone resonances. Backbone CA shifts report primarily on backbone conformation,19–22 whereas 15N chemical shifts depend upon not only conformation but are especially sensitive to electrostatics and hydrogen bonding.23,24 Carbonyl chemical shifts also show a dependence on hydrogen bonding, as well as backbone conformation.19,25,26 However, the C’, CA and backbone 15N chemical shifts are not the most prominent reporters on the subtle changes among crystal formulations including intermolecular crystal contacts, protonation state of the ionizable sidechains, aromatic ring interactions and salt bridge geometry. Rather, sidechain residues, especially charged sidechains, are involved in all of these events;27 therefore these sites are expected to demonstrate the largest changes in chemical shifts upon minor changes in formulation. Examination of the sidechain resonances would allow for an increased understanding of the structural changes that occur as a result of protein crystal formulations.
In this work, we use SSNMR chemical shifts in combination with X-ray powder diffraction and single crystal diffraction studies to understand the relationship between sample formulation and structural variations of the beta 1 immunoglobulin binding domain of protein G (GB1). Four new polymorphs were discovered for GB1 under similar but distinct formulations. Each of these polymorphs displayed differences in chemical shifts. To further understand these effects single crystal structures were determined in two cases where it was possible to prepare single crystals with similar formulations; one of these structures (PDB ID: 2GI9) was previously reported as part of another study.28 We assigned the complete backbone and sidechain chemical shifts for two of the microcrystalline formulations. Resonance differences among polymorphs correlate to crystal packing, solvent interactions and ionization events.
Experimental Methods
A. Protein Production
GB1 was produced and purified with 50 mM NaPO4 3− buffer (pH=5.5) and 150 mM NaCl using the procedure described in Franks et al.29 Throughout this study it was essential to precisely account for all salts present in the formulation. To ensure that buffers used during purification procedures were fully removed the protein was dialyzed in three steps using 3.5 kDa MWCO SnakeSkin dialysis tubing (Pierce, Rockford, IL, USA) against a total of 12 L of 50 mM NaPO4 3− buffer (pH=5.5). The protein was prepared for X-ray single crystal diffraction, X-ray powder diffraction or SSNMR studies as described below.
B. X-ray Diffraction
Conditions for single crystal growth for GB1 were screened starting from the conditions used by Franks et al.29 and Gallagher et al.,30 with the exception of lower salt concentrations. One trigonal crystal and two orthorhombic crystal morphologies were identified using hanging drop vapor diffusion methods (Table 1). Single crystals were collected and flash-frozen in liquid nitrogen; diffraction data was collected using beamline X12C at Brookhaven National Laboratory (Upton, NY, USA) (Table 1). The structures were solved using molecular replacement based upon the orthorhombic structure of Gallgher et al.(PDB ID: 1PGA),30 with the Amore program within the CCP4 package.31
Table 1.
Single Crystal X-ray Diffraction Data
| Orthorhombic | Orthorhombic | Trigonal | |
|---|---|---|---|
| Crystallization conditions |
50% MPDa 20% IPA 150 mM NaCl 25 mM NaCH3CO2 pH=3.8 |
50% MPD 20% IPA 150 mM NaCl 25 mM NaCH3CO2 pH=3.8 |
50% MPD 6% IPA 50 mM NaCl 25 mM NaCH3CO2 pH=4.5 |
| Protein solution | 10 mg/mL of GB1 in 50 mM Na2HPO4 pH=5.5 |
10 mg/mL of GB1 in 50 mM Na2HPO4 pH=5.5 |
10 mg/mL of GB1 in 50 mM Na2HPO4 pH=5.5 |
| Crystal size and growth time |
0.4×0.15×0.15 (mm)3 two weeks |
0.4×0.20×0.15 (mm) 3 two weeks |
0.4×0.20×0.20 (mm) 3 two weeks |
| X-ray source | BNL-X12C | BNL-X12C | BNL-X12C |
| Wavelength | 0.9791 | 0.9791 | 0.9791 |
| Collection temperature |
−150 °C | −150 °C | −150 °C |
| Space group | P212121 | P212121 | P3121 |
| a (Å) | 25.197 | 34.603 | 35.733 |
| b (Å) | 36.384 | 65.769 | 35.733 |
| c (Å) | 50.274 | 132.906 | 75.598 |
| γ | 90 | 90 | 120 |
| Total reflections | 106,995 | 190,218 | 236,393 |
| Number of unique reflections |
16,662 | 31,480 | 25,387 |
| Resolution (Å) | 1.14 | 1.7 | 1.05 |
| Completeness | 96.3 (92.2) | 91.1 (51.7) | 94.5 (64.3) |
| R_merge (I/⌠) | 0.059 | 0.059 | 0.062 |
| High shell | 0.158 | 0.345 | 0.238 |
| Redundancy | 6.4 | 6.0 | 9.3 |
| High shell | 5.8 | 4.0 | 2.9 |
Abbreviations: MPD, 2-methyl-2,4-pentanediol; IPA, 2-propanol
Microcrystalline protein samples were prepared by batch crystallization at 25 °C (Table 2), as described previously.29 The microcrystalline form A conditions are the same as those determined and analyzed by Franks et al.28,29 Microcrystalline conditions for forms D and E were derived from the trigonal and orthorhombic single crystal conditions, respectively, using hanging drop vapor diffusion methods to find the conditions at which microcrystals formed in 2–3 days.
Table 2.
Crystallization Conditions for Microcrystalline Forms of GB1
| Form A | Form B | Form C | Form D | Form E | |
|---|---|---|---|---|---|
| Reservoir solution |
50% MPD 25% IPA |
50% MPD 25% IPA |
50% MPD 25% IPA |
55% MPD 15% IPA 50 mM NaCl in 7.5 mM NaCH3CO2 (pH=4.5) |
55% MPD 25% IPA 150 mM NaCl in 16.7 mM NaCH3CO2 (pH=4.5) |
| Protein solution |
25 mg/mL of GB1 in 50 mM Na2HPO4 (pH=5.5) |
25 mg/mL of GB1 and 5 mM NaCl in 50 mM Na2HPO4 (pH=5.5) |
25 mg/mL of GB1 in 50 mM Tris HCl (pH=5.5) |
10 mg/mL of GB1 in 50 mM Na2HPO4 (pH=5.5) |
10 mg/mL of GB1 in 50 mM Na2HPO4 (pH=5.5) |
GB1 microcrystalline formulations A, D and E were packed, by centrifugation, with excess mother liquor (conditions similar to those used in SSNMR studies) into a 1.5 mm capillary tube for X-ray powder diffraction studies. Powder diffraction patterns were acquired at 25 °C using a Bruker (Bruker AXS, Inc., Madison, WI, USA) General Area Detector Diffraction System (GADDS) equipped with a helium tunnel to reduce air scatter. The program Diffracplus TOPAS (Bruker AXS GMBH, Karlsruhe, Germany) was used for line shape fitting to the trigonal and orthorhombic unit cell parameters using the Pawley method32. Additional experiment details are given in the supporting information.
C. Solid-State NMR Spectroscopy
All SSNMR spectra of microcrystalline forms A, D and E were acquired on a 500 MHz (1H frequency) Varian InfinityPlus spectrometer (Varian Inc., Palo Alto, CA, USA) equipped with a Varian Balun™ 1H-13C-15N 3.2 mm probe at a magic-angle spinning (MAS) rate of 11.11 kHz. The form B spectrum was acquired on a 600 MHz (1H frequency) Varian InfinityPlus spectrometer using a Varian T3 1H-13C-15N 3.2 mm probe and 13.3 kHz of MAS. The form C spectrum was acquired on a 750 MHz (1H frequency) Varian Unity Inova equipped with a Varian Balun™ 1H-13C-15N 3.2 mm probe and a MAS rate of 16.67 kHz. We have previously published several examples of GB1 spectra acquired on all of these spectrometers and demonstrated that the chemical shifts are invariant with respect to these minor differences in relative spinning rates.28,29,33–35
For 2D 15N-13C correlation experiments, band-selective SPECIFIC CP36 was used for polarization transfer between 15N and 13C and DARR37 was employed for 13C-13C mixing. All pulse sequences were implemented with tangent ramp cross polarization38 and 70–80 Hz of TPPM 1H decoupling during acquisition and chemical shift evolution periods.39 All experiments were performed at a nominal temperature set point of 273 K, with ~90 scfh variable temperature flow, resulting in actual sample temperatures of 280±5 K.
Data were processed with NMRPipe40 using backward linear prediction and polynomial baseline correction in the direct dimension. Lorentzian-to-Gausssian apodization and zero filling were applied to all dimensions before Fourier transformation. Chemical shifts were referenced using the downfield resonance of adamantane at 40.48 ppm (as determined from DSS).41 Peak picking and assignment of the spectra were completed with Sparky.42 Additional unique acquisition and processing parameters are included in the figure caption for each spectrum.
Results and Discussion
This study was motivated by observations of variability in the 13C chemical shift spectra of GB1 form A, which correlated with minor variations in the purification and precipitation protocol. For example, the use of NaCl in the purification procedure, if not rigorously removed by dialysis against large volumes of buffer, led to deterioration in spectral quality of GB1 when precipitated according to our previously published protocol.29 For example, prominent splittings in some of the well-resolved signals, such as the I6 CD1 resonance observed at 12.8 ppm. This specific problem was addressed by performing more thorough buffer exchange after purification; the protocol for form A now has been reproduced in a number of laboratories.43–45 To test the hypothesis that residual chloride ions from the purification buffer caused the deterioration in spectral quality of form A, we prepared a protein solution with 25 mg/mL of GB1 and 5 mM of NaCl in 50 mM NaPO4 3− buffer (pH=5.5) and precipitated the protein under the same conditions as those used for form A (Table 2). The SSNMR spectrum acquired on this sample (form B) yielded a spectrum with a similar overall appearance and spectral resolution to the form A spectrum (Figure 1); however, upon closer inspection of the 1D spectrum, it was determined that form B contained two polymorphs, one with chemical shifts consistent with form A and another with chemical shifts not observed in previous GB1 preparations.
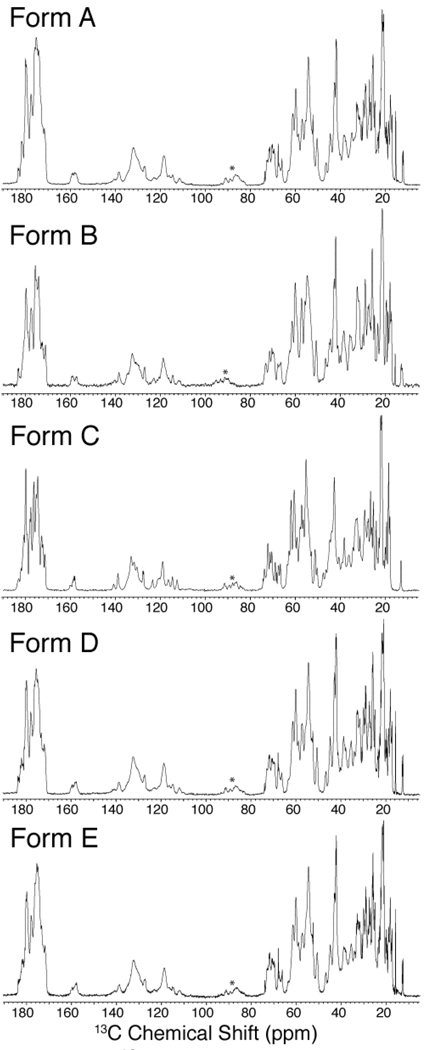
Figure 1. 13C 1D spectra of GB1 microcrystalline forms.
Complete 13C CP-MAS spectra for GB1 forms A, B, C, D and E. Data were processed with 15 Hz of Lorenzian-to-Gaussian apodization. The first order carbonyl spinning side band is indicated with an asterisk. The form A, D and E spectra were acquired at 500 MHz (1H frequency) for 128 scans with 38 ms of acquisition using 11.11 kHz MAS. The form B spectrum was acquired at 600 MHz (1H frequency) for 64 scans with 30 ms of acquisition using 13.3 kHz MAS. The form C spectrum was acquired at 750 MHz (1H frequency) for 256 scans with 30 ms of acquisition using 16.67 kHz MAS.
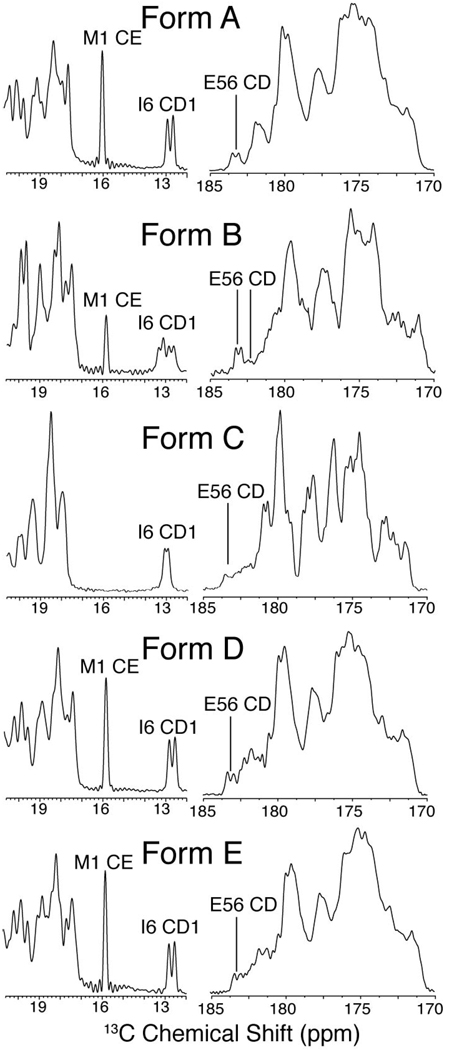
The methyl region of the 13C CP-MAS spectrum of forms A and B (Figure 2), shows the I6 CD1 peak at 12.8 ppm for A and two peaks at 12.8 ppm and 13.3 ppm, with equal intensities, for form B. The M1 CE peak in the form B spectrum is half the intensity observed for the form A sample; this sidechain is observed to be dynamic based on order parameter measurements of form A46 and we therefore attribute the change in signal intensity to different motional properties of form B. Two chemical shifts were also observed in form B for E56 CD (Figure 2); only one of these shifts is consistent with form A. I6 CD1, M1 CE and E56 CD are all located in the exterior of the protein where ion composition and concentration would be most likely to impact observed chemical shifts. Therefore, buffer composition and concentration was postulated as the most likely explanation for the observation of multiple peaks corresponding to the same resonances in form B as both chloride and phosphate anions are present. To further understand the origin of the newly observed resonances in the spectrum of microcrystalline form B, a sample was prepared with chloride as the only anion (form C). The spectrum for form C again gave narrow 13C linewidths (~0.2 ppm, Figure 1) and had unique resonances (Figure 2), such as the I6 CD1 shift at 13.0 ppm, that were not observed in form A or form B. This demonstrates that the chemical shifts of solvent-exposed residues in microcrystalline GB1 are highly sensitive to ions present in the sample. These observations also explain the spectral deterioration of samples prepared with incomplete buffer exchange. We hypothesize that this strong buffer dependence is due to the interactions of the ions with residues on the exterior of the protein, in addition to the formation of crystal contacts.
Figure 2. Enlargements of 13C 1D spectra of GB1 microcrystalline forms.
Methyl (left column) and carbonyl (right column) regions of 13C CP-MAS spectra for GB1 forms A, B, C, D and E are shown. Data were processed with 5 Hz of Gaussian line broadening. Acquisition details are the same as those in Figure 1.
To further understand how the SSNMR chemical shift perturbations relate to the GB1 structure, attempts were made to produce single crystals under identical conditions to those used to create microcrystal form A.29 Despite a large number of crystal trials (in which protein concentration, temperature and precise concentrations of MPD and IPA were varied) none resulted in single crystal growth. Because it was generally observed to be easier to prepare GB1 microcrystalline states to give high quality SSNMR spectra than to prepare new single crystal forms, we adopted the inverse strategy of attempting to identify a single crystal condition that could be conveniently reproduced at batch scale, suitable for SSNMR study. We choose to use conditions with low salt concentrations to permit a greater flexibility in the type of SSNMR probe used (although studies of samples with high conductivity can be performed with new scroll resonator designs47), so GB1 single crystal trails were performed based on a combination of the form A microcrystalline conditions and the Gallagher et al. orthorhombic and trigonal conditions.30 It was observed that the presence of both chloride and acetate ions in the reservoir solution was necessary to produce single crystals (Table 1), which we presume to be due to the fact that each type of ion plays a unique role in stabilizing crystal contacts, favoring propagation of the crystal lattice.
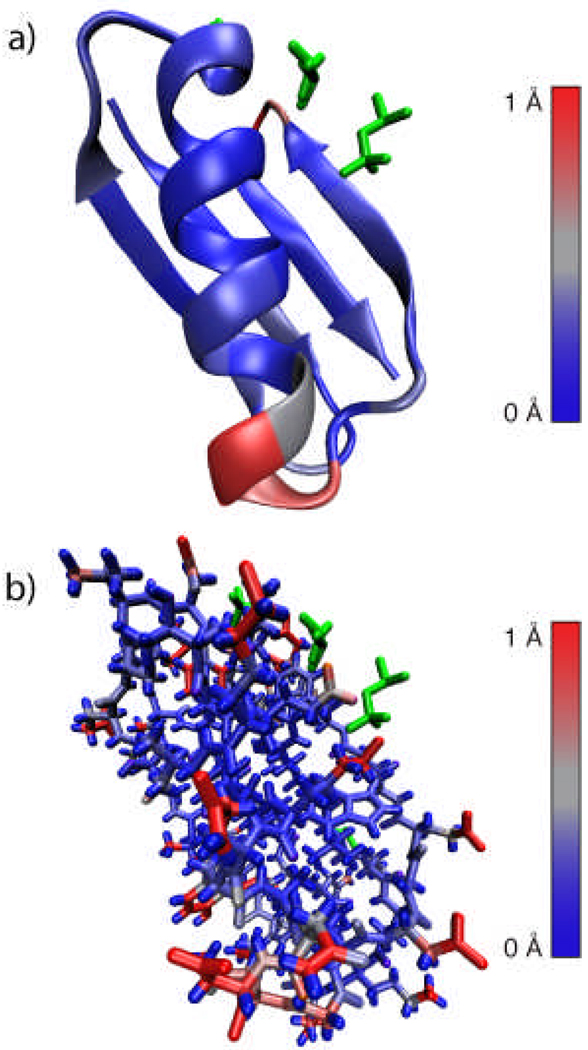
Three high quality, diffracting crystals were produced from these crystal screens. The two orthorhombic crystals diffracted to 1.14 Å and 1.7 Å. The higher resolution data set was solved, as previously reported (PDB ID: 2GI9).28 The second orthorhombic data set was not analyzed in detail because it had the same symmetry but lower resolution than the first data set and the presence of six protein molecules per unit cell made the structure especially difficult to solve. Instead, we focused our efforts on the high quality trigonal crystals we obtained that diffracted to a 1.05 Å resolution. The resulting structure (PDB ID: 2QMT) is shown in Figure 3 with the observed 2-Methyl-2,4-pentanediol, 2-propanol and phosphate molecules (electron density maps of these molecules are given in the supporting information). Comparing orthorhombic28 and trigonal structures, there is little variation (0.47 Å Root Mean Square Deviation (RMSD)) observed for the backbone atoms. Small backbone differences appear in the turn between the beta strand 3 and beta strand 4 (D47, A48, T49) and for N35 through G38 at the start of the helix (Figure 3a, displayed in red). Larger discrepancies are observed for the sidechains (16.41 Å RMSD), in particular for the amino acid residues located in the exterior of the protein (Figure 3b). The differences are greatest among charged sidechains and sidechains that have multiple energetically allowed rotameric states, which are likely to change as a consequence of crystal contacts and salt bridges. For instance, the orthorhombic structure has two intramolecular salt bridges between D47-K50 and E27-K31. The trigonal structure has these same salt bridges, in addition to an additional salt bridge between E15 and K4.
Figure 3. X-ray diffraction crystal structure of GB1.
(a) Structure of trigonal GB1 (PDB ID: 2QMT) displaying secondary structure and location of precipitate molecules and buffer ions. The structure is colored according to differences from the published orthorhombic structure (PDB ID: 2GI9) with conserved atoms shown in blue (0 Å) and structural differences shown in red (1Å or greater). (b) The sidechains indicate greater deviations between the two structures with especially large deviations observed in the polar sidechains (atoms shown in red). Data was acquired at Brookhaven National Laboratory, diffracting to a resolution of 1.05 Å. Figure prepared using VMD.54
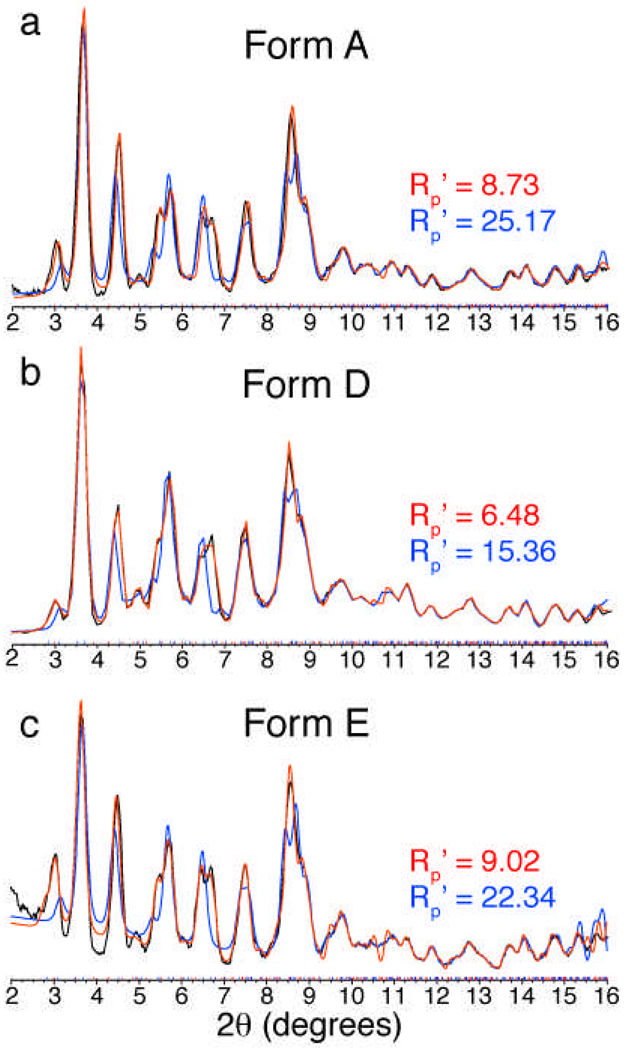
Microcrystalline samples were prepared for the orthorhombic and trigonal crystal forms for SSNMR and X-ray powder diffraction studies. The crystallization conditions used to produce formulation D and formulation E microcrystals (Table 2) were derived from those used for the trigonal and orthorhombic single crystals, respectively. In all cases, minor changes in the conditions were required in order to accommodate batch scale processes; furthermore, the kinetics of mixing under batch precipitation are likely to vary significantly from the vapor diffusion protocol utilized to grow the single crystals. Therefore we performed a direct comparison between the microcrystalline protein obtained by the batch method by acquiring X-ray powder diffraction data for the forms A, D and E. The observed powder diffraction patterns were all highly similar (Figure 4). The experimental data for each form was analyzed by constructing theoretical powder diffraction patterns from the single crystal data of the trigonal and orthorhombic structures. In the course of these fits, the unit cell parameters remained fixed while the background and offset corrections, and line shape parameters were varied. All three microcrystalline samples gave the best fit to the trigonal structure parameters with Rp’ values of 8.73, 6.48 and 9.02 for forms A, D and E, respectively, versus Rp’ values of 25.17, 15.36 and 22.34 when fit to the orthorhombic parameters. This demonstrates that the preparation of GB1 microcrystals yields a trigonal crystal packing arrangement; even the microcrystalline form E, which was derived from the orthorhombic single crystal conditions, produced trigonal microcrystals. It is not surprising that form D, which was derived from the trigonal crystal conditions, fit best to the trigonal structure parameters. However, the fact that all polymorphs prefer the trigonal arrangement indicates that it is the kinetically favored polymorph in this batch process.
Figure 4. X-ray powder diffraction data of GB1 microcrystalline forms.
Powder diffraction patterns (black) and theoretical fits to the trigonal (shown in red) and orthorhombic (blue) structures’ unit cell parameters are shown for microcrystalline forms A (a), B (b) and C (c). Figure prepared using Diffracplus TOPAS (Bruker AXS GMBH, Karlsruhe, Germany).
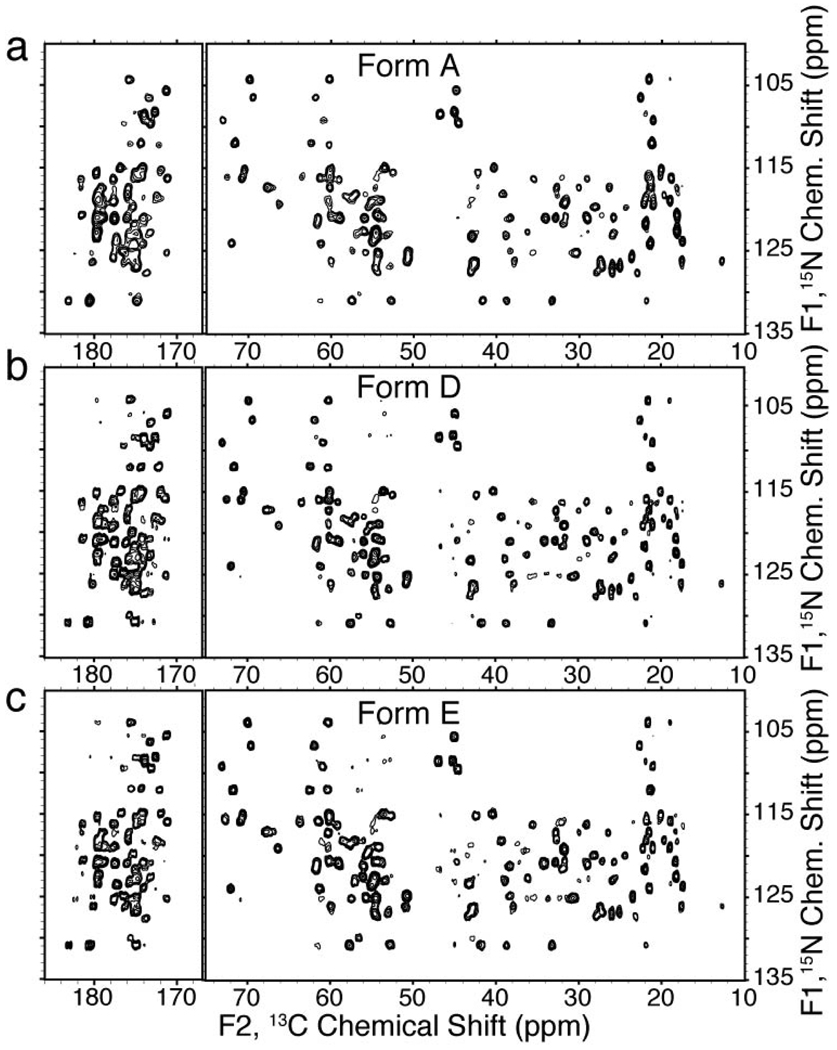
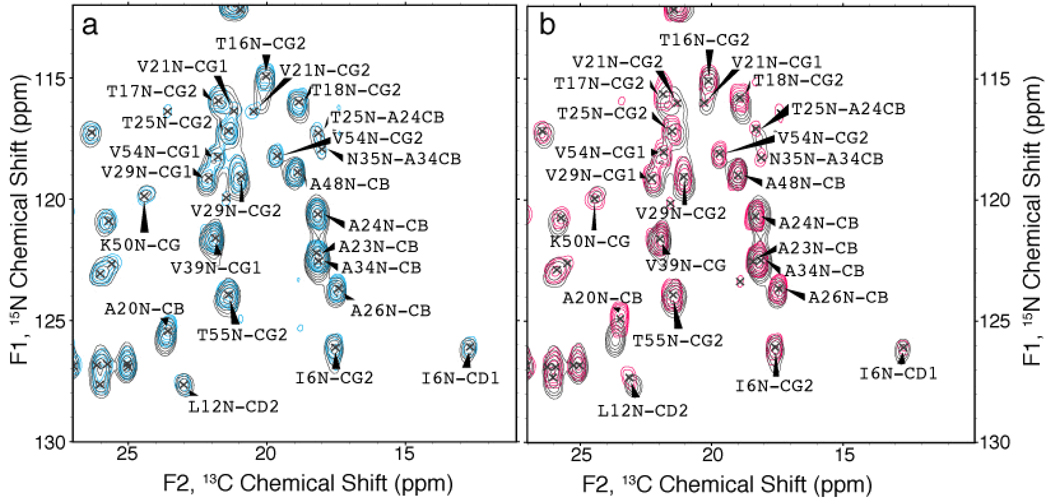
The SSNMR analysis of forms D and E yielded spectra with very few overlapped resonances and narrow linewidths, ~0.2 ppm for 13C (displayed in Figure 1 and Figure 2) and less than 0.8 ppm for 15N. In both of these forms, a single resonance was observed for every 13C and 15N atom, consistent with a single homologous structure. N(CA)CX spectra of both these forms are displayed in Figure 5 along with the spectrum acquired on form A, highlighting the overall similarities. The majority of the chemical shifts are identical among the three forms, but several differences are observed for the backbone amide and aliphatic sidechain shifts. The methyl region of the N(CA)CX contains the greatest number of resonance variation (Figure 6). Many small (~0.5 to 1.0 ppm) differences are observed for the backbone amide shifts between form A and form E, the most significant being for A20 with a difference of ~1 ppm (Figure 6a). The form A and form D spectra have a greater amount of similarities, although differences are observed for the A20 N, A34 N and V21 CG shifts in the methyl region of the spectra (Figure 6b). The initial SSNMR spectra of formulations D and E microcrystals revealed that the chemical shifts were unique to these forms with numerous differences observed among the form A, D and E spectra. This evidence suggests that there are multiple microcrystalline structures and crystal lattice arrangements, in which the protein is thermodynamically stable.
Figure 5. N(CA)CX SSNMR spectra of GB1 microcrystalline forms.
(a) N(CA)CX 2D spectra of form A, (b) form D and (c) form E. All data was acquired at 500 MHz (1H frequency), 0 °C with 11.11 kHz MAS. The form A spectrum was acquired for 14.5 hours with 50 ms DARR mixing, 46 ms of acquisition and 14.4 ms of total t1 evolution (dwell time of 22.5 µs, 1280 rows TPPI); data is shown with 25 Hz (F2) and 15 Hz (F1) Lorenzian-to-Gaussian apodization applied, forward linear prediction in the F1 (15N) dimension and zero filling to 8192 (F2)×4096 (F1) points. The form D and E spectra were acquired for 9.4 hours each with 31 ms of acquisition and 11.5 ms of total t1 evolution (dwell time of 45 µs, 512 rows TPPI). In the form D spectrum, 45 ms of DARR mixing was utilized and in the form E spectrum, 27 ms of DARR mixing was used. Data is shown with 15 Hz (F2) and 5 Hz (F1) line broadening applied, forward linear prediction in the F1 (15N) dimension and zero filled to 8192 (F2)×4096 (F1) points.
Figure 6. Expansion of N(CA)CX spectra of GB1 forms.
Expansion of the methyl region of N(CA)CX spectra for (a) form D (cyan) overlaid with form A (gray) and (b) form E (pink) overlaid with form A (gray) is shown on the right. Numerous chemical shift differences among the microcrystalline formulations are displayed. Data and processing parameters are the same as those used in figure 5.
The backbone and sidechain 15N and 13C chemical shifts for microcrystalline forms D and E were assigned, and are reported for the first time here for comparison with previously reported form A chemical shifts.29 Among forms A, D and E, the majority of the chemical shifts were consistent within 0.2 ppm. All of the CA and C’ chemical shifts were consistent among all the forms, indicating a conserved backbone conformation. Several backbone amide and sidechain sites displayed chemical shift deviations of more than 0.4 ppm for 13C resonances and 0.5 ppm for 15N resonances. These values are two times the standard deviation observed between spectra acquired on samples of the same formulation. The complete list of resonances with significant deviations between microcrystalline forms (Table 3) shows only small variations, with no clear trend among the forms. Each microcrystalline polymorph gives a unique set of chemical shifts, arising from the unique electrostatic and crystal packing properties for a given formulation.
Table 3.
Chemical Shift Differences Among Microcrystalline Polymorphs
| Form D-Form A | |
|---|---|
| Resonance | Δδ (ppm) |
| L7 CD | −0.76 |
| V21 CG | 0.50 |
| V39 CG | 3.74 |
| K31 CB | −1.0 |
| Form E- Form A | |
| Resonance | Δδ (ppm) |
| L12 N | −0.61 |
| T17 N | −0.73 |
| T18 N | −0.69 |
| A20 N | −1.17 |
| V21 N | −0.50 |
| T49 N | −0.52 |
| L7 CD | −0.81 |
| Y33 CB | 0.62 |
| V39 CD | 3.78 |
| K10 NZ | 0.80 |
| K13 NZ | 0.68 |
| K28 NZ | 0.88 |
| E42 CG | 0.42 |
| D47 CG | −3.17 |
| Form D- Form E | |
| Resonance | Δδ (ppm) |
| L12 N | 0.65 |
| T17 N | 0.66 |
| A20 N | 0.95 |
| V21 N | 0.60 |
| V21 CG | 0.46 |
| Y33 CB | −0.75 |
| F52 CB | −0.52 |
| E15 CG | 0.65 |
| K31 CB | −1.09 |
| D47 CG | 2.97 |
The chemical shift differences among microcrystalline polymorphs were mapped onto the GB1 trigonal crystal structure to determine the locations of these perturbations (Figure 7). The trigonal structure was chosen since the powder diffraction data for all of the microcrystalline forms had the best agreement with this structure. Differences of greater than 0.5 ppm are shown in red and conserved regions are in blue. Forms D and A show the least amount of differences with only four differing residues having a total RMSD greater than 0.5 ppm (Figure 7a). The majority of the differences were observed for the methyl-bearing sidechains (L7 CD, V21 CG and V39 CG). It is well know that the CA shifts of beta-branched amino acid residues show a strong dependence upon chi1 in addition to phi and psi.48–51 As no change in CA is observed, these chemical shift differences must result from crystal packing and ionic strength. The sidechains of L7, V21 and V39 are on the exterior of the protein, subjecting them to changes in solvent interactions and crystal contacts as a result of formulation changes.
Figure 7. Chemical shift differences among microcrystalline forms A, D and E.
The average RMSD chemical shift between (a) forms A and D, (b) forms A and E and (c) forms D and E is shown mapped onto the GB1 trigonal crystal structure (PDB ID: 2QMT). Differences range from conserved (blue, 0 ppm) to large (red, 0.5 ppm or greater) values. The average RMSD of the backbone and sidechain (excluding aromatic sidechains) chemical shifts are plotted. Figure prepared using VMD.54
Form E has the largest chemical shift differences relative to form A. (Table 3). The greatest difference is observed for the D47 CG resonance where there is a 3 ppm upfield shift relative to form A. In forms A and D the D47 CG resonance is downfield at 179.8 ppm, consistent with a deprotonated carboxylic acid. In form E the D47 CG resonance is at 176.6 ppm, indicating a protonated aspartic acid residue.52 This would be expected since the form E sample was prepared at a lower pH (pH=4.5 verses pH=5.5). Solution NMR studies by Khare et al.52 and Lindman et al.53 have found D47 to have a pKa ranging from 3.1–3.4 for wild type and mutant GB1, respectively. This suggests that the local electrostatic environment has altered the pKa of D47 in form E. In the trigonal crystal structure, D47 creates a salt bridge with K50. A 0.46 ppm resonance change is observed for K50 NZ, in addition to chemical shift perturbations of the backbone amides (T17-V21) in the turn between beta sheet 2 and the helix (Figure 7b). We attribute these chemical shift differences to interactions with a phosphate ion located in this region in the crystal structure (Figure 3). Perturbations are also observed for V39 CG and L21 N, which are in close proximity to each other, and K28, K13 and K10 NZ and E42 CG; all of these residues are in solvent exposed regions of the protein where intermolecular contacts and buffer ions effects would be accentuated.
Although few chemical shift differences are observed between forms D and E, the presence of any perturbation confirms the existence of two unique polymorphic structures. These samples have very similar formulations yet the changes to the D47 CG and the backbone amide resonances are unique to the formulation E polymorph and resonance changes observed for K31 CB and V21 CG in form D are not observed in form E. Again these observations demonstrate that SSNMR chemical shifts are sensitive to differences in electrostatic interactions caused by minor formulation changes to the GB1 microcrystals.
Conclusions
We have demonstrated the use of SSNMR chemical shifts to examine subtle structural differences among crystalline polymorphs of GB1. The microcrystalline preparations of GB1 can be quickly prepared using batch method in several formulations, although only a very small subset of these formulations yielded diffraction-quality single crystals. In the instances were single crystals formed, the resulting high-resolution data displayed co-solvent and buffer ion interactions with the protein. The SSNMR spectra of all microcrystalline forms yield very narrow signals, enabling observation of ~0.2 ppm changes in backbone and sidechain chemical shifts. Using solely the chemical shifts, we were able to identify four new polymorphs of GB1 (forms B, C, D and E), all dependent upon microcrystalline formulation. From a practical standpoint, identifying highly reproducible conditions for GB1 microcrystallization has proved invaluable in a number of detailed studies of GB1 structure and dynamics, as well as the use of GB1 as an excellent model protein for NMR methods development.
X-ray powder diffraction data for these polymorphs show qualitative similarities, which we attribute to the trigonal form being the kinetically preferred product in the course of rapid microcrystal formation; we did not observe any microcrystalline forms that were consistent with orthorhombic space groups. Beyond this qualitative observation, it was not possible to extract more detailed, site-specific conformational data from the powder diffraction experiments. In contrast, using straightforward 2D 13C-13C and 15N-13C chemical shift correlation spectra in the solid state, detailed analysis in a site-specific manner throughout the protein was possible. The observed chemical shift differences among forms were minimal along the peptide backbone, where the stable fold of GB1 is conserved across all known structures to within ~0.5 Å RMSD (backbone). However, sidechain conformations, protonation states, electrostatics and intermolecular interactions differ significantly; these phenomena are reflected in the changes of sidechain chemical shifts, especially among ionizable residues and/or those involved in crystal contacts. For instance, methyl 13C chemical shift differences were observed for residues on the exterior of protein, L7, V21 and V39, while their CA shifts were conserved, indicating that these changes result from differences in crystal packing and/or solvent interactions and not changes in rotameric states. In addition, among charged sidechains such as lysine, aspartic acid and glutamic acid, chemical shifts in some cases were greatly perturbed, which we attribute to differences in protonation state, solvent interactions and salt bridges, that in turn depend strongly upon salt concentration and type.
These subtle issues of formulation chemistry, as demonstrated here for GB1, are known in general to impact protein stability and enzymatic reaction rates. The results here illustrate that SSNMR methods can be applied to study structural effects due to crystal formulation changes of therapeutic proteins and industrial enzymes. These capabilities are useful for analysis of structural factors that correlate with the increases in stability, concentration and substrate specificity that protein crystallization provides. Moreover, understanding the issues that affect the quality of GB1 microcrystals enables its general use as a model protein for solid-state NMR, as well as fundamental studies of protein structure and dynamics.
Supplementary Material
Acknowledgements
This research was supported by the National Science Foundation (CAREER Award MCB 0347824 to C.M.R.) and National Institutes of Health (NIGMS/Roadmap Initiative R01GM075937 to C.M.R.). H.L.F.S. was supported by a Drickamer Fellowship. The authors thank Sara A. Stellfox for determining microcrystalline conditions for new GB1 formulations, Merideth Burkhart for assistance in the early stages of this work, and the School of Chemical Sciences NMR and George L. Clark X-Ray Facilities, University of Illinois for technical support.
Footnotes
Supporting Information Available: Solid-state NMR chemical assignment procedure and table of assignments from formulation D and E microcrystalline samples, graphical comparison and discussion of assigned SSNMR chemical shifts, electron density maps of GB1 trigonal single crystal data (displaying precipitate molecules) and additional details of X-ray powder diffraction experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Margolin AL, Navia MA. Angew. Chem. Int. Edit. 2001;40:2205–2222. doi: 10.1002/1521-3773(20010618)40:12<2204::aid-anie2204>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 2.Jen A, Merkle HP. Pharm. Res. 2001;18:1483–1488. doi: 10.1023/a:1013057825942. [DOI] [PubMed] [Google Scholar]
- 3.Basu SK, Govardhan CP, Jung CW, Margolin AL. Expert Opin. Biol. Ther. 2004;4:301–317. doi: 10.1517/14712598.4.3.301. [DOI] [PubMed] [Google Scholar]
- 4.Beals J, Shanafelt A. Drug Discovery Today:Technol. 2006;3:87–94. doi: 10.1016/j.ddtec.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Merkle HP, Jen A. Nat. Biotechnol. 2002;20:789–790. doi: 10.1038/nbt0802-789. [DOI] [PubMed] [Google Scholar]
- 6.Bliss M. Bull. Hist. Med. 1982;56:554–568. [PubMed] [Google Scholar]
- 7.Hallas-Moller K, Peterson D, Schlichtkrull J. Science. 1952;116:394–398. doi: 10.1126/science.116.3015.394. [DOI] [PubMed] [Google Scholar]
- 8.Govardhan C, Khalaf N, Jung CW, Simeone B, Higbie A, Qu S, Chemmalil L, Pechenov S, Basu SK, Margolin AL. Pharm. Res. 2005;22:1461–1470. doi: 10.1007/s11095-005-6021-x. [DOI] [PubMed] [Google Scholar]
- 9.Martin RW, Zilm KW. J. Magn. Reson. 2003;165:162–174. doi: 10.1016/s1090-7807(03)00253-2. [DOI] [PubMed] [Google Scholar]
- 10.Paulson EK, Morcombe CR, Gaponenko V, Dancheck B, Byrd RA, Zilm KW. J. Am. Chem. Soc. 2003;125:15831–15836. doi: 10.1021/ja037315+. [DOI] [PubMed] [Google Scholar]
- 11.Seidel K, Etzkorn M, Heise H, Becker S, Baldus M. Chembiochem. 2005;6:1638–1647. doi: 10.1002/cbic.200500085. [DOI] [PubMed] [Google Scholar]
- 12.Kamihira M, Naito A, Nishimura K, Tuzi S, Saito H. J. Phys. Chem. B. 1998;102:2826–2834. [Google Scholar]
- 13.Kameda T, McGeorge G, Orendt AM, Grant DM. J. Biomol. NMR. 2004;29:281–288. doi: 10.1023/B:JNMR.0000032557.71691.a2. [DOI] [PubMed] [Google Scholar]
- 14.Zell MT, Padden BE, Grant DJW, Chapeau MC, Prakash I, Munson EJ. J. Am. Chem. Soc. 1999;121:1372–1378. [Google Scholar]
- 15.Middleton DA, Le Duff CS, Peng X, Reid DG, Saunders D. J. Am. Chem. Soc. 2000;122:1161–1170. [Google Scholar]
- 16.Harper JK, Grant DM. J. Am. Chem. Soc. 2000;122:3708–3714. [Google Scholar]
- 17.Byrn SR, Pfeiffer RR, Stephenson G, Grant DJW, Gleason WB. Chem. Mater. 1994;6:1148–1158. [Google Scholar]
- 18.Harris RK. J. Pharm. Pharmacol. 2007;59:225–239. doi: 10.1211/jpp.59.2.0009. [DOI] [PubMed] [Google Scholar]
- 19.Wishart DS, Sykes BD. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 20.Cornilescu G, Delaglio F, Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Spera S, Bax A. J. Am. Chem. Soc. 1991;113:5490–5492. [Google Scholar]
- 22.Heller J, Laws DD, Tomaselli M, King DS, Wemmer DE, Pines A, Havlin RH, Oldfield E. J. Am. Chem. Soc. 1997;119:7827–7831. [Google Scholar]
- 23.Wylie BJ, Sperling LJ, Frericks HL, Shah GJ, Franks WT, Rienstra CM. J. Am. Chem. Soc. 2007;129:5318–5319. doi: 10.1021/ja0701199. [DOI] [PubMed] [Google Scholar]
- 24.Gu ZT, Ebisawa K, McDermott A. Solid State Nucl. Magn. Reson. 1996;7:161–172. doi: 10.1016/s0926-2040(96)01259-3. [DOI] [PubMed] [Google Scholar]
- 25.Gu ZT, Zambrano R, McDermott A. J. Am. Chem. Soc. 1994;116:6368–6372. [Google Scholar]
- 26.Lipsitz RS, Tjandra N. J. Am. Chem. Soc. 2001;123:11065–11066. doi: 10.1021/ja016854g. [DOI] [PubMed] [Google Scholar]
- 27.Oki H, Matsuura Y, Komatsu H, Chernov AA. Acta Crystallogr. D. 1999;55:114–121. doi: 10.1107/S0907444998008713. [DOI] [PubMed] [Google Scholar]
- 28.Franks WT, Wylie BJ, Stellfox SA, Rienstra CM. J. Am. Chem. Soc. 2006;128:3154–3155. doi: 10.1021/ja058292x. [DOI] [PubMed] [Google Scholar]
- 29.Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. J. Am. Chem. Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 30.Gallagher T, Alexander P, Bryan P, Gilliland GL. Biochemistry. 1994;33:4721–4729. [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, N. Acta. Cryst. 1994;D50:760–763. [Google Scholar]
- 32.Pawley GS. J. Appl. Crystallogr. 1981;14:357–361. [Google Scholar]
- 33.Wylie BJ, Franks WT, Graesser DT, Rienstra CM. J. Am. Chem. Soc. 2005;127:11946–11947. doi: 10.1021/ja053862e. [DOI] [PubMed] [Google Scholar]
- 34.Wylie BJ, Franks WT, Rienstra CM. J. Phys. Chem. B. 2006;110:10926–10936. doi: 10.1021/jp060507h. [DOI] [PubMed] [Google Scholar]
- 35.Zhou DH, Kloepper KD, Winter KA, Rienstra CM. J. Biomol. NMR. 2006;34:245–257. doi: 10.1007/s10858-006-0026-6. [DOI] [PubMed] [Google Scholar]
- 36.Baldus M, Geurts DG, Meier BH. Solid State Nucl. Magn. Reson. 1998;11:157–168. doi: 10.1016/s0926-2040(98)00036-8. [DOI] [PubMed] [Google Scholar]
- 37.Takegoshi K, Nakamura S, Terao T. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 38.Hediger S, Meier BH, Ernst RR. Chem. Phys. Lett. 1993;213:627–635. [Google Scholar]
- 39.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Morcombe CR, Zilm KW. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 42.Goddard TD, Kneller DG. 3.106 ed. San Francisco: University of California; [Google Scholar]
- 43.Nadaud PS, Helmus JJ, Hofer N, Jaroniec CP. J. Am. Chem. Soc. 2007;129:7502–7504. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- 44.Polenova T. Personal communication. Newark. DE: University of Delaware; 2007. [Google Scholar]
- 45.Ladizhandsky V. Personal communication. Guelph, ON, Canada: University of Guelph; 2007. [Google Scholar]
- 46.Wylie BJ, Sperling LJ. Urbana, IL: University of Illinois; 2007. Unpublished work. [Google Scholar]
- 47.Stringer JA, Bronnimann CE, Mullen CG, Zhou DH, Stellfox SA, Li Y, Williams EH, Rienstra CM. J. Magn. Reson. 2005;173:40–48. doi: 10.1016/j.jmr.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 48.Pearson JG, Le HB, Sanders LK, Godbout N, Havlin RH, Oldfield E. J. Am. Chem. Soc. 1997;119:11941–11950. [Google Scholar]
- 49.Straus SK, Bremi T, Ernst RR. J. Biomol. NMR. 1997;10:119–128. doi: 10.1023/a:1008280716360. [DOI] [PubMed] [Google Scholar]
- 50.Havlin RH, Le HB, Laws DD, deDios AC, Oldfield E. J. Am. Chem. Soc. 1997;119:11951–11958. [Google Scholar]
- 51.Villegas ME, Vila JA, Scheraga HA. J. Biomol. NMR. 2007;37:137–146. doi: 10.1007/s10858-006-9118-6. [DOI] [PubMed] [Google Scholar]
- 52.Khare D, Alexander P, Antosiewicz J, Bryan P, Gilson M, Orban J. Biochemistry. 1997;36:3580–3589. doi: 10.1021/bi9630927. [DOI] [PubMed] [Google Scholar]
- 53.Lindman S, Linse S, Mulder FAA, Andre I. Biophys. J. 2007;92:257–266. doi: 10.1529/biophysj.106.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.