Abstract
The collective dynamics of the nascent polypeptide exit tunnel are investigated with the computationally efficient elastic network model using normal mode analysis. The calculated normal modes are considered individually and in linear combinations with different coefficients mimicking the phase angles between modes, in order to follow the mechanistic motions of tunnel wall residues. The low frequency fluctuations indicate three distinct regions along the tunnel - the entrance, the neck and the exit – each having distinctly different domain motions. Generally the lining of the entrance region moves in the exit direction, with the exit region having significantly larger motions, but in a perpendicular direction, whereas the confined neck region generally has rotational motions. Especially the universally conserved extensions of ribosomal proteins L4 and L22 located at the narrowest and mechanistically strategic region of tunnel undergo generally anti- or non-correlated motions, which may have an important role in nascent polypeptide gating mechanism. These motions appear to be sufficiently robust so as to be unaffected by the presence of a peptide chain in the tunnel.
Keywords: Ribosomal exit tunnel, dynamic domains, collective dynamics, elastic network models, coarse-graining
Introduction
In bacteria, proteins are synthesized at the core of ribosome large subunit 50S at the peptidyl transferase center (PTC). Despite the existence of several gates within the structure,1 the nascent polypeptide chain emerges through the ribosomal tunnel having ∼15Å diameter and ∼100Å long2 (Fig. 1). After the first evidence from low-resolution density maps from electron microscopy3,4 and the monitoring of a nascent polypeptide emerging through an attached polypeptide conducting channel,5 the dynamics of the exit tunnel and the mechanism of nascent polypeptide secretion have been an important subject for the understanding of protein synthesis1,6-10.
Figure 1.
Bacterial ribosome 70S is comprised of the small 30S subunit (dark gray) and the large 50S subunit (light gray) generated from Protein Data bank43 structures 1GIX and 1GIY42. The mRNA (purple) wraps around the neck of the 30S and the cognate A-, P- and E-tRNAs (orange) are located in a cavity between the two subunits. Amino acids are covalently linked at the peptidyl transferase center (PTC) between the amino acids at the CCA ends of the A- and P-tRNAs with nascent polypeptide emerging through the exit tunnel on the lower right (green). Interestingly this tunnel has a highly distinct shape. All molecular graphics are prepared using PyMol45.
The ribosomal tunnel wall is formed mainly from 23S rRNA and several ribosomal proteins, leading to the assumption of its having a ‘non-sticky’ property that allows for the passage of any polypeptide.11 However, at the narrowest part of tunnel not far from the PTC, highly conserved extensions of ribosomal proteins L4 and L22 function as a discriminating gate for specific nascent chains that can cause ribosomal stalling.12,13 In addition, this region is a dynamic area with an open diameter of ∼16-20Å and a closed diameter of ∼10Å conformations imaged with Cryo-electron microscopy (EM) of ligand bound/unbound and L4-L22 mutated structures.1 Furthermore, other mutation studies on the conserved L4 and L22 tips have revealed the erythromycin-resistance of Escherichia Coli,14 and the binding of a macrolide to the tunnel entrance, which is observed to trigger the ‘swing motion’ of the conserved β-hairpin of L22 to block the nascent polypeptide passage7. These findings altogether imply the special importance of L22 in the control and channeling of the nascent polypeptide through the exit tunnel.6,7,15
Along with biological evidence8,10,16 and computational results17 supporting entropic stabilization of α-helical structures in the ribosomal tunnel, experimental data pointed to possible secondary structure folding zones of the tunnel9. However, any tertiary structure would seem not to fit due to simple geometrical restrictions,2 but the deformability of the tunnel is an important issue in this regard. On the other hand, the proteins at the tunnel exit, L22, L23, L24 and L29 are strategically located to interact with membrane proteins,18,19 chaperone trigger factor20,21 and signal recognition particle,22 and point out the possibility that their interactions could affect the tunnel deformations and control the folding process.
The different aspects of the ribosome complex dynamics and its constituents have been investigated using various computational approaches such as full-atom molecular dynamics,23 coarse-grained molecular dynamics,24 normal mode analysis using a coarse-grained potential,25,26 elastic network model namely anisotropic network model27,28 and combining experimental observations with computational methods29. However, the ribosomal exit tunnel collective motions have not been yet explored computationally at least to our knowledge.
In this study, we explore the harmonic dynamics of the ribosomal tunnel wall with the coarse-grained elastic network model, using the bacterial ribosome 70S in its complex with mRNA, and three tRNAs alone. The motions are modeled with the anisotropic elastic network model30 based on simple harmonic interactions within a structure-based three-dimensional network from crystallographic data.
Previously the elastic network approach has been applied successfully to extract the slow motions of supramolecular assemblages, including the ribosome27 and the chaperone GroEL-GroES31,32 using coarse-grained model. Even higher levels of coarse-graining, i.e. lower resolutions, result in similar collective dynamics,33-35 which indicates that the macromolecular structures exert exquisite control to limit the motions to the most important functional dynamics, reflecting the highly cohesive nature of these structures. For example the ribosome's computed slowest motions27 overall correspond closely to the ratchet motions as observed in the electron micrograph images of the Frank group36. The principal internal motions within the ribosome during this ratchet motion correspond to the translational motion of the mRNA and tRNAs in the well-known functional direction to facilitate the reading of the genetic code. These prior experiences provide the foundation and justification for the present deeper investigation of the ribosome internal motions.
Our elastic networks calculations point to three distinct regions along the exit tunnel, which we denote as the entrance, the neck and the exit zone, each having distinctively different domain motions in the slow modes of motion. After the normal modes of the elastic network model are obtained for the low frequency spectrum, they are linearly combined with different coefficients to mimic the possible phase angle differences between these modes. Our analysis especially focuses on the narrow gate of the tunnel encompassing the extensions of the ribosomal proteins L4 and L22, and several observations which may play role on the direct control of the nascent polypeptide exit process are reported.
Methods
Elastic Network Modeling
The anisotropic elastic network model30 is a three-dimensional elastic network model that includes the directionality of the residue fluctuations. Our coarse-grained description of the native structure includes representations of amino acids and nucleotides as single geometric points at the actual positions of the Cα and P atoms, respectively.27 Neighboring nodes are linked pairwise with harmonic springs, throughout the network, with the total potential energy calculated to be dependent on the displacements of each point in the structure in the form of
| (1) |
where the Heaviside step function h(x) is 1, if Rij, the distance between the (i,j) pair falls within a cutoff distance rc, but 0 otherwise. For the ribosome protein-RNA complex, rc= 15Å has been found to be suitable for Cα-Cα pairs and 24Å for all other types of pairs 27 since the cutoff distance should be increased for larger node sizes in order to maintain a similar level of cooperativity by adjusting the number of interactions and connectivity in terms of pairwise interactions in the elastic network. This adjustment of the cutoff distance has been demonstrated to be a satisfactory approach when hierarchical levels of coarse-graining are applied to extract the collective motions of folded proteins.33 Similarly, for the available low-resolution ribosome structure, P and Cα atoms represent different sized nodes: a nucleotide and an amino acid, respectively, necessitating the use of different cutoff values 27. ΔRi is the fluctuation in the position vector Ri of residue i (i=1, ‥, N), and the harmonic spring force constant γ is identical (taken as one in this study) for all interactions within the system.
The total potential energy can be also represented in Gaussian form,
| (2) |
where ΔR is the fluctuation in position vector matrix, T is the transpose, and H is the Hessian matrix, or the force constant matrix. After the diagonalization of H, the calculation of 3N-6 normal modes and the respective eigenvalues leads to the collective dynamics of the elastic network at low frequencies, known to be important for biological function.37
Then, mean-square fluctuation of the ith residue averaged over k non-zero normal modes is given by
| (3) |
where λk is the eigenvalue of ith residue with displacement vector uik at kth non-zero normal mode.
As the diagonalization of the (3N × 3N) Hessian matrix (dimension 29,361 × 29,361 in this study) represents a large computational task to derive the displacement vectors of all N = 9,787 residues of the ribosome complex, the computationally efficient software BLZPACK38 using the Lanczos algorithm39 is used to calculate just the first twenty normal modes.
Results and Discussion
The collective motions of the ribosome complex are computed with the coarse-grained elastic network model using the normal mode analysis. Among the several structures available for the ribosome complex,40,41 the structure at 5.5Å resolution42 is one of the most complete including mRNA, three tRNAs and large mobile stalks. Therefore, in this study, as an extension of our earlier work,27 the crystal structure of the ribosome in its complex with tRNAs and mRNA taken from the protein data bank (PDB)43 codes 1GIY (large subunit 50S) and 1GIX (small subunit 30S, three tRNAs and mRNA)42 is modeled, and the collective dynamics around the native structure are computed.
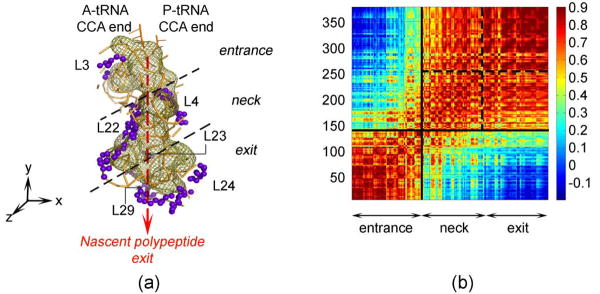
For analysis of the slow mode dynamics, the ribosomal tunnel volume elements have been extracted from the low-resolution crystal structure of the large subunit using the program 3V based on spherical probes searching for solvent accessible areas.2 The exit tunnel conformations are displayed via the coarse-grained residues within 10Å of the tunnel wall (379 residues in total) (Fig. 2(a)). Principal component analysis is used to place the y-axis onto the tunnel nascent polypeptide exit route, and to sort the residue indices accordingly in −y direction. As a result, the residues located around the tunnel have been renumbered starting from the entrance down to the exit. The extracted tunnel, formed mainly of the 23S rRNA with several ribosomal proteins' extensions, has a funnel shape with small secondary openings to solvent observed in the Cryo-EM.1
Figure 2.
(a) Ribosomal tunnel wall (green mesh) extracted with the program 3V2, with the nearby (within 10Å) proteins (purple) and parts of the 23S rRNA nucleotides (orange) shown. The nascent polypeptide, after passing the narrow gate formed of the highly conserved extensions of L4 and L22, emerges through the tunnel. Principal component analysis is performed to place the y-axis in the orientation of the exit tunnel, and the residue indices are sorted accordingly in −y direction. (b) Orientational cross-correlation map for tunnel wall residues averaged over the first 20 modes. The correlations clearly divide the structure into two distinct parts declined by black squares: the entrance and the rest. The region after the narrow gate can be further divided into neck and exit sub-domains separated by the dotted black lines, as shown in (a).
Domains of the exit tunnel observed from slow modes
The ribosomal tunnel, embedded within the large subunit 50S, undergoes translational motions in the slow normal modes, rather than contracting/ expanding motions (based on visual analysis of the first ten slowest modes). From the first twenty cumulative mode directional vectors cross-correlations, two major zones are observed: (i) the entrance including the PTC; (ii) the neck comprising the narrowest region with extensions of proteins L4 and L22, and the broad region near the exit (Fig. 2). Based on the strong orientational-correlation coefficients within residues 250-379, the latter region is also sub-divided into two sub-domains as the neck and exit regions.
The exit domain exhibits high amplitude motions compared to the rest of the tunnel, observed from mean-square (ms) fluctuations averaged over the slowest twenty modes (Fig. 3). Collective motions in the first five normal modes show that in general the entrance region moves downwards (along the long y-axis of the tunnel) whereas the residues at the tunnel exit prefer to move in a direction perpendicular to the exit route. Motions of the neck region are relatively more mixed and include twisting, tangential and radial motions (Fig. 3). Since the ribosome is a huge complex, more than one rotational axis exists in each vibrational mode. One interesting finding is that the rotational axis closest to the exit tunnel is generally located at the narrowest gate near the ribosomal proteins L4 and L22 (Fig. 3, third, fourth and fifth modes, axis shown by a red x mark or a red dotted line).
Figure 3.
Mean-square fluctuations of the tunnel wall residues averaged over first twenty modes are displayed with a color coded ribbon presentation of ribosomal tunnel. Using principal component analysis, the y-axis is placed along the nascent polypeptide exit route and the residue indices are sorted in -y direction from entrance to exit. 23S rRNA is shown in ribbon and ribosomal protein extensions in spheres. Colors blue to red indicate increasing mobility (upper left). Displacement vectors of the tunnel wall residues are shown for the first five vibrational modes. Ribosomal proteins L4 and L22's extensions are shown for the individual modes in two alternative conformations in orange and purple. Arrows indicate one of the two alternative directions of oscillation; red x mark (modes 3, 4) and red dotted line (mode 5) locate the global rotational axes.
The inner dot product (i.e. orientational correlation) between the displacement vectors u of ith and jth residues for each of the first twenty normal modes k is calculated for the entrance and exit domains.
| (4) |
The aim is to observe whether the entrance and exit domains remain cohesive in each normal mode k. Figure 4(a)-(b) shows the distribution of the positively correlated (>0.7) residue pair fractions over the first twenty normal modes for the entrance (Fig. 4(a)) and exit zones (Fig. 4(b)). For example in Figure 4(a), in eight normal modes, 90% of residues in the entrance domain fluctuate in same direction. Both entrance and exit regions move as a domain, and a low fraction (0.4) of positive correlation usually corresponds to rotation around an axis (for example in the fourth and fifth modes in Fig. 3). Recent experimental studies have determined three “folding zones” in the ribosomal tunnel, two of which (one close to the PTC and the other at the exit) have an impact on helix formation in a specific nascent polypeptide, with the middle zone having a lower impact.9 Remarkably, our three domains correspond roughly to these folding zones, with our neck region being shorter than the middle zone.
Figure 4.
Distribution of the positively correlated (correlation>0.7 using Eq. (4)) residue pairs fractions over the first twenty normal modes for the entrance (a) and exit zones (b). The same distribution is plotted over 121 generated conformations with Eq. (5) for the entrance (c) and exit regions (d). These plots show that the entrance and exit regions move as distinct cohesive domains in the normal modes as well as in the combined modes.
In contrary to cohesive motions of the entrance and the exit domains, more complex fluctuations are observed for the neck region acting as a transition zone between the two domains. In general, the collective motions of the tunnel wall indicate that the residues around the narrowest gate (Fig. 3) prefer to oscillate in the exit direction, such that the nascent polypeptide is directed (pushed) through the tunnel. Due to their location, the ribosomal proteins L4 and L22 are especially of interest. For example, in first three normal modes, the tips of the L4 (residues Phe61-Ala68) and L22 (residues Leu86-Ala93) move together (with a correlation of ∼0.90), whereas in the fourth and fifth modes their motions are anti-correlated or perpendicular (∼ -0.55 - 0.10 according to residue location).
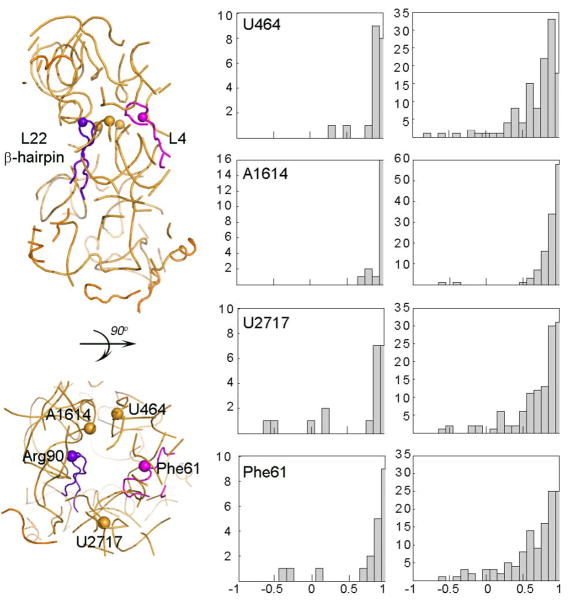
Over most of the first twenty normal modes, the tip of the protein L22 β-hairpin (residue Arg90) is positively correlated (correlation > 0.5) with the neighboring residues across the tunnel wall as partially shown in Figure 5 (middle panel). The bases neighboring Arg90 are U464, A1614 and U2017 (shown as orange spheres) on the 23S rRNA and Phe61 (shown as a magenta sphere) on protein L4 in Figure 5. The bases U464 and A1614 generally move with Arg90, whereas the residues across the tunnel wall, U2017 and Phe61, may move in perpendicular and opposite directions to Arg90 in a few of the slowest normal modes.
Figure 5.
Distribution of the dot products between the displacement vectors of the L22 β-hairpin tip residue Arg90 and proximal residues over the first twenty normal modes (middle panel) and for combined-modes (right panel). These neighboring residues to Arg90 are taken as U464, A1614 and U2017 (shown as orange spheres in left panel) on 23S rRNA and Phe61 (shown as magenta sphere) on protein L4. The x- and y-axis of the histograms refer to the correlation within the range of (-1,1) and to the number of normal or combined modes, respectively.
These motions in the neck region may be meaningful in terms of the gating and signaling mechanism along the polypeptide exit tunnel during protein synthesis. Furthermore, the conformational changes of the L22 β-hairpin for several normal modes (for example the fifth mode in Fig. 3), show that the residues located at the base of the hairpin (Arg84-Leu86 and Ala93-Ile95) are not positively correlated with the tip region of the hairpin (residues Pro87-Arg92). This observation conforms previous experimental results that indicated that the L22 β-hairpin undergoes a ‘swing motion’ around a hinge region (Arg84-Leu86 and Ala93-Ile95).7
The residues at the tunnel exit undergo relatively large harmonic motions at low frequencies since they are located at the periphery of the 50S subunit. This motion may play an important role in polypeptide secretion through the tunnel to the cytoplasm or in protein-protein interactions with accessory membrane and trigger factor proteins.
Dynamics of the tunnel in presence of a nascent polypeptide
A polypeptide chain of 25 residues of random sequence is inserted in the tunnel to investigate its effect on the collective ribosomal tunnel dynamics. Following the geometrical restrictions of the tunnel,2 the coarse-grained polypeptide chain has an extended conformation with a short α-helix, and spans the entrance and narrow neck regions of the ribosomal tunnel (Fig. 6(a)). Based on the first 100 normal modes of the coarse-grained elastic network, there is no significant difference in the cross-correlations of the tunnel residues with and without the nascent polypeptide (Fig. 6(b)). The difference values in residue correlations are all very low, within the range from -0.06 to 0.03. This result together with the mean-square fluctuations averaged over the first 100 modes (not shown) indicates that the ribosome global motions dictate the embedded tunnel dynamics, being virtually unaffected by the presence of the model polypeptide for the important low frequency motions. However, the effect of the nascent polypeptide on the local dynamics might conceivably be more apparent at higher frequencies. Furthermore, various conformations of the polypeptide chain such as more compact chains could possibly have a slightly different effect on the dynamics of the tunnel. However, overall the dominant slow motions appear to be unaffected in the present case, and are likewise probably little affected by different peptide conformations.
Figure 6.

(a) A polypeptide (in green) of 25 residues with a random sequence modeled to investigate its possible effect on the ribosomal tunnel dynamics. The location of ribosomal proteins L4 and L22 (purple), and the exit route of the nascent polypeptide (light green arrow) are indicated in the ribbon presentation of the tunnel wall. (b) The cross-correlation plot based on the first 100 normal modes gives the difference between the motions of the tunnel residues with and without the nascent polypeptide in the tunnel, with all differences observed to be extremely small.
Linearly combined normal modes and resulting dynamics
In sum, biological systems would undergo a superposition of harmonic mode motions with different phase angles. To obtain some further insight into the dynamics of the ribosomal tunnel area, we have combined linearly the first five non-zero normal modes with different coefficients (ai= -1, 0, 1; i=1,…, 5), randomly mimicking phase angles multiplied with the eigenvector sets ui as,
| (5) |
leading to k=35 eigenvector sets but to (35-1)/2 distinct combined modes.
In a harmonic oscillation, the displacement along the kth normal mode coordinate with time t is given by Ak cos(ωkt + εk), where Ak, ωk and εk are the corresponding amplitude, vibrational frequency and phase angle, respectively. Our motivation in combining linearly the displacement vectors of the first five normal modes without considering the effect of the vibrational frequencies and amplitudes is based on the fact that their values are close to one another (ωk=1, 5 ratio: 1: 1.07: 1.30: 1.40: 1.56). In order to include the effect of the phase angle, in the extreme cases, the minimum, the equilibrium and the maximum values of the cosine term (-1, 0, 1) have been considered in the calculations, leading to the equation (5).
With the above assumptions, it is then possible to visualize the combined modes and the resulting ribosomal tunnel conformations that could give insight into the polypeptide channeling mechanism during protein synthesis. The current approach is similar to a previous study that generated different receptor conformations for ligand docking.44 In addition, combining only a few normal modes (5 normal modes in this study) provides some variety and a continuous distribution of motions when compared to a smaller number of normal modes (20 normal modes in this study) (Fig. 5, right and middle panels).
In Figure 7, several randomly selected combined modes are shown and coefficients of the eigenvector sets are given in brackets in the order of from the first to fifth modes. For example in the combination {a1-5=1, 1, 1, 1, 1} all five normal modes contribute positively. In this pseudo-motion, through the narrowest region of the ribosomal tunnel, a rotation axis passes about which all residues rotate. As noted above for the normal modes, the closest rotation axis to the tunnel is located near the narrow gate in these pseudo-motions.
Figure 7.
Displacement vectors of tunnel wall residues calculated from different combinations of first five normal modes (pseudo-motions) computed to explore the possible range of overall motions. For example {1, 1, 1, 0, 0} indicate that only first three normal modes contribute positively to the resulting motion. Coloring is as indicated in Figure 3.
In the combination {a1-5=1, 1, 1, 0, 0} where only first three modes are summed, the β-hairpin of protein L22 oscillates in a tangential motion while the tip of L4 moves in the y direction, such as explicitly directing a nascent polypeptide through the tunnel. Another interesting motion is observed for pseudo-motion {a1-5= -1, 0, 1, 1, 0} where the L22 β-hairpin is displaced in a tangential direction with respect to tunnel wall and L4 oscillates perpendicularly to L22 as in a gate opening.
Over the 121 combined modes, correlations between the tip of L22 β-hairpin (Arg90) and the neighboring residues across the tunnel wall namely U464, A1614 and U2017 on the 23S rRNA and Phe61 on the protein L4 are calculated. The closest base to Arg90, A1614 is generally positively correlated with the tip of the hairpin in the combined modes (Fig. 5, right panel). On the other hand, U464, U2017 and Phe61 are all positively correlated and in some pseudo-modes move in an anti-correlated way and in a perpendicular direction to Arg90, different from the first twenty normal modes (Fig. 5, middle panel). The variety of displacements are shown in the pseudo-motions {a1-5= 1, 1, 1, 0, 0} and {a1-5= -1, 0, 1, 1, 0} in Figure 7. The perpendicular and anti-correlated motions seem to have an importance in the opening/closing motion of the tunnel entrance observed in various Cryo-EM images,1 that could channel the nascent polypeptide.
Domain motions of the entrance and exit regions are investigated for all of the 121 combined modes. In Figure 4(c)-(d), the distributions of the positively correlated (>0.7) residue pairs fractions over the combined modes for entrance (Fig. 4(c)) and exit zones (Fig. 4(d)) are shown. As in the normal modes, these zones move as cohesive domains while small fractions correspond to rotational movements about an axis such as in the pseudo-modes {1, 1, 1, 1, 1} and {0, 1, 1, 1, 1} displayed in Figure 7.
Conclusions
It is interesting to contrast the motions in the ribosome tunnel with the motions of other structures having cavities. The GroEL-GroES structure has a large interior cavity, and as a result it exhibits extremely diverse motions – shearing and stretching in various directions.31 These are the most diverse motions that we have ever observed for a protein structure. These diverse motions present a strong contrast to those reported here for the ribosome tunnel. The motions of the ribosome tunnel appear to relate much more specifically to its function. The motions of the entrance region of the tunnel moving dominantly in the exit direction clearly relate to the functional processing motions of the ribosome. The rotational motion at the neck and the mixed rotational and perpendicular motions at the exit region would lead to a dispersion of the growing peptide chain.
We have also placed a polypeptide chain of 25 residues including a short α-helix spanning the entrance and neck regions of the ribosomal tunnel to investigate its possible effects on the tunnel dynamics. The result is that there appears to be no significant change in the global motions observed for both the ribosome and the tunnel region including the slowest 100 normal modes. This is consistent with the ribosome controlling its own motions, independently of the growing peptide chain, a behavior that may be required for its robust function.
Understanding ribosomal tunnel dynamics is essential for understanding the mechanism of protein synthesis and in extensions to develop new therapeutics for bacterial infections. Our results suggest that during the harmonic collective dynamics of the ribosome.tRNA3.mRNA complex, the ribosomal tunnel goes through different conformations dividing the tunnel into three distinct regions - the entrance together with the PTC, the neck including the narrowest region of tunnel and the exit domain. The entrance domain motions are generally in the direction of nascent polypeptide exit. However, at the narrowest part of tunnel, extensions of ribosomal proteins L4 and L22 usually undergo uncorrelated or anti-correlated motions, which may have relevance for the opening/closing motion of this particular region observed with Cryo-EM1 and the polypeptide gating mechanism7,12,13. And, another interesting observation is the location of rotational axes usually passing through the narrowest part of the tunnel, for the low frequency motions. Finally the exit region comprising the ribosomal proteins L22, L23, L24 and L29 is highly mobile compared to other parts of tunnel and in some cases, undergoes rotational motion around an axis passing through the tunnel.
Acknowledgments
We gratefully acknowledge the support of NIH grant R01GM073095. PD and OK acknowledge partial support by TUBITAK Project 104M247 and EU-FP6-ACC-2004-SSA-2 contract no. 517991. OK gratefully acknowledges the support of the TUBITAK-BIDEB 2214 Fellowship.
Footnotes
This work was performed in Bogazici University and Iowa State University.
References
- 1.Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 2.Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 3.Milligan RA, Unwin PN. Location of exit for nascent protein in 80S ribosome. Nature. 1986;319:693–695. doi: 10.1038/319693a0. [DOI] [PubMed] [Google Scholar]
- 4.Yonath A, Leonard KR, Wittmann HG. A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction. Science. 1987;236:813–816. doi: 10.1126/science.3576200. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 6.Agmon I, Auerbach T, Baram D, Bartels H, Bashan A, Berisio R, Fucini P, Hansen HAS, Harms J, Kessler M, Peretz M, Schluenzen F, Yonath A, Zarivach R. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes. Eur J Biochem. 2003;270:2543–2556. doi: 10.1046/j.1432-1033.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 7.Berisio R, Schluenzen F, Harms J, Bashan A, Auerbach T, Baram D, Yonath A. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat Struct Biol. 2003;10:366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- 8.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 11.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 12.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 13.Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 14.Chittum HS, Champney WS. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 16.Kosolapov AL, Tu L, Wang J, Deutsch C. Structure acquisition of the T1 domain of Kv1.3 during biogenesis. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Ziv G, Haran G, Thirumalai D. Ribosome exit tunnel can entropically stabilize {alpha}-helices. Proc Natl Acad Sci USA. 2005;102:18956–18961. doi: 10.1073/pnas.0508234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 19.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, III, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 21.Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- 22.Gu SQ, Peske F, Weiden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tung CS, Sanbonmatsu KY. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophys J. 2004;87:2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trylska J, Tozzini V, McCammon JA. Exploring global motions and correlations in the ribosome. Biophys J. 2005;89:1455–1463. doi: 10.1529/biophysj.104.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tama F, Valle M, Frank J, Brooks CL., III Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trylska J, Konecny R, Tama F, Brooks CL, III, McCammon JA. Ribosome motions modulate electrostatic properties. Biopolymers. 2004;74:423–431. doi: 10.1002/bip.20093. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Rader AJ, Bahar I, Jernigan RL. Global ribosome motions revealed with elastic network model. J Struct Biol. 2004;147:302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Jernigan RL. Comparison of tRNA motions in the free and ribosomal bound structures. Biophys J. 2005;89:3399–3409. doi: 10.1529/biophysj.105.064840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 30.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keskin O, Bahar I, Flatow D, Covell DG, Jernigan RL. Molecular mechanisms of chaperonin GroEL-GroES function. Biochemistry. 2002;41:491–501. doi: 10.1021/bi011393x. [DOI] [PubMed] [Google Scholar]
- 32.Chennubhotla C, Bahar I. Markov propagation of allosteric effects in biomolecular systems: application to GroEL-GroES. Mol Sys Biol. 2006;2:36. doi: 10.1038/msb4100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doruker P, Jernigan RL, Bahar I. Dynamics of large proteins through hierarchical levels of coarse-grained structures. J Comp Chem. 2002;23:119–127. doi: 10.1002/jcc.1160. [DOI] [PubMed] [Google Scholar]
- 34.Ming D, Kong Y, Lambert MA, Huang Z, Ma J. How to describe protein motion without amino-acid sequence and atomic coordinates. Proc Natl Acad Sci USA. 2002;99:8620–8625. doi: 10.1073/pnas.082148899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurkcuoglu O, Jernigan RL, Doruker P. Mixed levels of coarse-graining of large proteins using elastic network model succeeds in extracting the slowest motions. Polymer. 2004;45:649–657. [Google Scholar]
- 36.Frank J, Agrawal K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 37.Tama F, Sanejouand YH. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Marques O, Sanejouand YH. Hinge-bending motion in citrate synthase arising from normal mode calculations. Proteins. 1995;23:557–560. doi: 10.1002/prot.340230410. [DOI] [PubMed] [Google Scholar]
- 39.Grimes RG, Lewis JG, Simon HD. A shifted block Lanczos algorithm for solving sparse symmetric eigenvalue problems. SIAM J Matrix Anal Appl. 1994;15:228–272. [Google Scholar]
- 40.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 41.Schurwith BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 42.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 43.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavasotto CN, Kovacs JA, Abagyan R. Representing receptor flexibility in ligand docking through relevant normal modes. J Amer Chem Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 45.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]