Abstract
There is accumulating evidence that the patterns of altered expression of microRNAs (miRNAs) correlate with distinct types of hematological malignancies. There is also emerging evidence for such miRNAs expression in multiple myeloma (MM), but the identity of the miRNAs that are abnormally expressed in this disease has not been elucidated. In the current study we examined the miRNAs perturbed in CD138 positive MM cell lines and CD138 positive primary MM cells, using microarray analysis and quantitative real-time PCR. The expression levels in these cells were compared to the expression levels in CD138 positive plasma cells isolated from bone marrows of healthy donors. Our data demonstrate that multiple new miRNAs are commonly up-regulated and down-regulated including a new miRNA cluster, miR-193b-365, composed of two paralogs that were significantly up-regulated in MM. The organization and the sequences of the new miR-193b-365 cluster are highly conserved among vertebrates. Given that dysregulation of miR-193b-365 cluster has not been identified in other hematological malignancies examined to date our findings suggest that miR-193b-365 cluster is part of the unique miRNA signature in MM.
Keywords: Multiple Myeloma, microRNA, miR-193b-365 cluster
Introduction
Multiple myeloma (MM) is characterized by proliferation of malignant plasma cells in the bone marrow, secretion of monoclonal protein in the blood and/or urine and skeletal destruction with osteolytic lesions [1]. In MM patients the number of plasma cells are elevated compared to the numbers in normal bone marrow and can be identified by their expression of syndecan-1 (CD138) surface protein. Although increased plasma cell content signifies a common feature in myeloma, the overall biology of MM is quite complex. Therefore elucidation of regulatory circuits altered in myeloma such as microRNAs (miRNAs) will allow us to gain a better understanding into the biology of MM.
miRNAs are recently discovered class of 19–25 nucleotides of small non-coding RNA molecules that are evolutionally conserved and play important regulatory functions by targeting mRNAs for transcriptional or translational repression [2]. Studies in model organisms have revealed that miRNAs are involved in the control of cellular processes, such as differentiation, development, cell growth, and apoptosis, as well as in various human diseases including cancer [3,4]. While several miRNAs have been reported to show differential expression pattern in hematological malignancies and solid tumors very little is known with respect to specific miRNAs that are perturbed in MM. In a recent studyit was shown that several miRNA clusters were perturbed in myeloma using relatively small number (345) of capture probes [3–6]. In the current study using the new locked nucleic acid technology comprising 757 capture probes and quantitative real-time PCR (Q-PCR) we have carried out a comprehensive analysis of miRNAs that are perturbed in MM. These studies allowed us to identify miR-193b-365 as a highly up-regulated cluster in MM. In addition using the same array technology we identified several new commonly up-regulated miRNAs and a smaller set of commonly down-regulated miRNAs in myeloma plasma cells that has not been recognized in previous studies.
MATERIALS AND METHODS
Patient specimens, Cell lines and Cell culture
For microarray and Q-PCR study, we used Trizol™ for RNA isolation from CD138 positive MM cell lines (L363, KMS11, JJN3, RPMI-8226, U266, NCI-H929 and MM.1S), purified CD138 positive plasma cells from newly diagnosed MM patients (10 patients) and purified CD138 positive plasma cells (> 90%) from two healthy donors. L363, KMS11 and JJN3 cells were kindly provided by Dr. Leslie Brents (NIH, Bethesda, MD). MM.1S cells were kindly provided by Drs. Steven T. Rosen and Nancy Krett (Northwestern University, Chicago, IL). U266, NCI-H929 and RPMI-8226 cells were purchased from ATCC. All MM cell lines were maintained in RPMI-1640 containing 10% FBS, and 100 Units/ml penicillin and 100 μg/ml streptomycin (GIBCO). 2.5 μg/ml fungizon (Invitrogen) and 5 μg/ml plasmocin (InvivoGen) were added for MM.1S cells as additional antibiotics. The patient samples were obtained after informed consent and approval from the University of Chicago Institutional Review Board. Plasma cells from patient samples were isolated by CD138 selection using EasySep® Kit (Stem Cell Technologies, BC, Canada). Total RNAs of purified CD138 positive cells from healthy donors were purchased from All Cells, CA, USA.
MiRNA microarray experiments and Data analysis
MiRNA expression was determined using the miRCURY™ LNA (Locked Nucleic Acid) microarray (Exiqon, Vedbaek, Denmark). All miRNAs were isolated by Trizol™ and miRNeasy Mini Kit (Qiagen) was used to eliminate organic solvents. The quality of the RNA was verified by a 2100 Bioanalyzer (Agilent Technologies, CA, USA). The miRCURY™ LNA array used in this study contained 757 annotated human miRNA capture probes spotted in four subgrids, listed in the miRBase 11.0 version of the Exiqon array. After the hybridization the slides were scanned by the Agilent G2565BA Microarray Scanner System (Agilent Technologies) and image analysis was carried out using the ImaGene 8.0 software (BioDiscovery, CA, USA). The slides were pre-processed in ImaGene to ensure uniformity of hybridization before being subject to data analysis. Background correction was performed to remove nonbiological contributions to the measured signal [7]. The quantified signals were normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm [8] excluding spots that were flagged due to poor spot morphology. Hy3 and Hy5 labeling dyes were used to label the samples and the common reference pool respectively. The common reference pool represents a pool of all samples used in the miRNA studywhich contained two hundred nano grams of each sample labeled with Hy5. The common reference pool is specifically used in two-color cDNA microarray experiments for adjustment of experimental variation between slides. This approach named common reference design enabled us to compare directly between samples (Hy3) since a common factor is made (Hy5) [9,10]. The normalized Hy3-and Hy5-signals were presented as four (Hy3 (sample)/Hy5 (common reference pool)) ratios per probe ID. The median of the four ratios per probe ID was calculated and log2– transformed. Prior to the data analysis of the normalized signals two specific criteria were evaluated. The criteria for deciding that a miRNA (probe ID) had failed on a particular array, is that 3 or more of the 4 replicated measures of this miRNA were flagged (ie. Poor spot morphology). Further, all capture probes with both Hy3- and Hy5-median signals lower than 1.5 X the median signal intensity of all probes on the given slide is excluded during further analysis.
After the extraction of normalized data, a candidate list of miRNAs was generated based on the filtering criteria on variation across all samples being a standard deviation (SD) >0.50 [11]. All genes SD>0.5 were selected as candidate genes. The cut-off SD > 0.5 is an arbitrary criteria corresponding to a regulation > 2 fold change when comparing the lowest and highest log2– transformed ratio per miRNA. This arbitrary cut-off approach is used when a group of samples do not fulfill at least 3 biological replicates used for statistical approaches. The clustering was performed on log2 (Hy3 (sample)/Hy5 (common reference pool)) ratios which passed the filtering criteria on variation across samples; SD > 0.50 [11].
Quantitative real-time PCR
Reverse transcription (RT) and real-time PCR for miRNAs expression were performed using TaqMan MicroRNA Assays (Applied Biosystems), TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems), and TaqMan® Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems). TaqMan MicroRNA Assays, namely: hsa-miR-193b (Part No. 4395478), hsa-miR-365 (Part No. 4373194) and RNU6B (Part No. 4373381) were used with amplification efficiencies of 100% for real-time PCR. RT and real-time PCR were carried out according to the manufacturer’s instructions. Briefly, cDNAs were synthesized from total RNA using gene-specific primers. Reverse transcriptase reactions contained 5 ng of RNA samples, 7 μl of RT master mix including 0.15 μl of 100 mM dNTP mix, 1 μl of 50 U/μl MultiScribe™ Reverse Transcriptase, 1.5 μl of 10X Reverse Transcriptase Buffer, 0.19 μl of 20 U/μl RNase Inhibitor and 4.16 μl of Nuclease-free water, and 3 μl of stem-loop RT primer. The 15 μl of reactions were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and then held at 4°C. Real-time PCR was performed using a Bio-Rad MiniOpticon Real-Time PCR Detection System (Bio-Rad, CA, USA). The 20 μl of PCR included 1 ul of TaqMan MicroRNA Assay, 1.33 μl of RT product, 10 μl of TaqMan 2X Universal PCR Master Mix, No AmpErase UNG and 7.67 μl of Nuclease-free water. Reactions were incubated in optical tubes at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. All TaqMan MicroRNA Assays were designed with same conditions. All experiments were done in triplicate and data were calculated using the comparative Ct (2−ΔΔCt) method. U6 snRNA (RNU6B) was used as an internal control to normalize RNA input in the Q-PCR assay.
RESULTS AND DISCUSSION
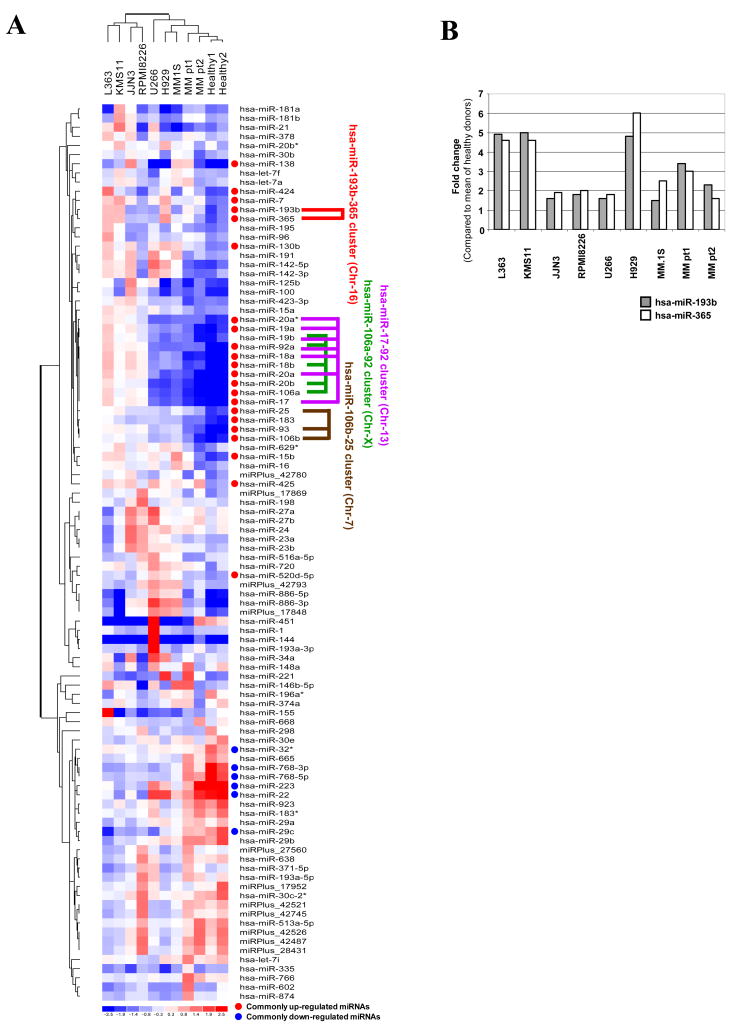
In order to identify commonly-perturbed miRNAs in myeloma cells we first carried out global miRNAs expression using 7 myeloma cell lines, purified plasma cells from 2 MM patients and 2 healthy donors, which lead to the identification of 22 commonly up-regulated and 6 commonly down-regulated miRNAs in myeloma cells (MM patient samples and MM cell lines) compared to the healthy donor samples (Tables I and II). The heat map and unsupervised hierarchical clustering of miRNAs shown in Figure 1A allowed us to identify 4 clusters, which were located in distinct chromosomes. Of the four miRNA clusters, miR-17-92, miR-106b-25 and miR-106a-92 were also identified in another study whereas miR-193b-365 is a novel cluster that was not examined in that study or any other study in the context of myeloma [6]. The miR-193b-365 cluster we have identified here has not been previously implicated in hematological malignancies. However, miR-193b and miR-365 are highly perturbed in endometrioid adenocarcinoma and breast cancer respectively [12,13]. Therefore, our report provides the first evidence of up-regulation of miR-193b and miR-365 in hematological malignancies. With respect to the other three clusters overexpression of miR-17–92 cluster was reported in several hematological malignancies including Burkitt lymphoma, mantle cell lymphoma, diffuse large B-cell lymphoma, acute lymphocytic leukemia, MLL-rearrangement leukemias and most recently in myeloma [6,14–16], in addition to various solid tumors including lung cancer [17,18]. On the other hand miR-106b-25 cluster was up-regulated mostly in solid tumors such as in gastric cancers [19]. miR-17–92 and miR-106b-25 clusters have been suggested to function as oncogenic miRNAs over the MYC/E2F1/TGFβ network [20]. miR-106a-92 cluster has been reported to be overexpressed in leukemia as well as in certain solid tumors such as colon and prostate [17,21,22].
Table I.
Fold increase in expression of commonly up-regulated miRNAs in MM compared to samples from healthy donors.
| microRNAs | L363 | KMS11 | JJN3 | RPMI8226 | U266 | H929 | MM.1S | MM pt1 | MM pt2 | SD |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-138 | 4.8 | 6.8 | 23.7 | 7.9 | 1.7 | 1.8 | 15.5 | 14.9 | 3.2 | 1.63 |
| hsa-miR-106a | 17.0 | 11.1 | 13.6 | 12.4 | 4.0 | 3.5 | 4.2 | 2.7 | 1.7 | 1.49 |
| hsa-miR-20a | 15.8 | 10.4 | 12.5 | 11.7 | 5.3 | 3.2 | 3.9 | 3.1 | 1.6 | 1.45 |
| hsa-miR-17 | 16.1 | 11.3 | 13.2 | 12.2 | 4.4 | 3.5 | 4.1 | 2.7 | 2.1 | 1.44 |
| hsa-miR-18a | 13.7 | 8.9 | 11.8 | 9.4 | 3.8 | 5.0 | 4.1 | 1.8 | 2.0 | 1.37 |
| hsa-miR-20b | 13.2 | 10.2 | 9.3 | 9.3 | 2.9 | 2.7 | 3.5 | 2.5 | 1.8 | 1.34 |
| hsa-miR-18b | 11.7 | 7.5 | 10.0 | 7.9 | 3.4 | 4.1 | 3.6 | 1.9 | 2.3 | 1.25 |
| hsa-miR-93 | 7.7 | 8.1 | 6.5 | 7.6 | 4.5 | 5.3 | 5.7 | 2.8 | 2.3 | 1.13 |
| hsa-miR-92a | 9.3 | 6.8 | 7.1 | 6.6 | 2.6 | 2.4 | 2.4 | 2.6 | 2.4 | 1.11 |
| hsa-miR-19a | 8.0 | 5.4 | 6.4 | 4.5 | 2.8 | 2.4 | 2.8 | 2.0 | 1.1 | 1.08 |
| hsa-miR-424 | 8.9 | 2.7 | 2.0 | 1.1 | 2.3 | 3.8 | 1.3 | 3.3 | 2.0 | 0.97 |
| hsa-miR-15b | 4.9 | 5.0 | 3.2 | 3.5 | 4.5 | 3.3 | 7.8 | 3.7 | 1.6 | 0.97 |
| hsa-miR-106b | 5.0 | 5.5 | 4.7 | 4.9 | 3.2 | 3.6 | 4.3 | 2.6 | 1.2 | 0.96 |
| hsa-miR-193b | 4.9 | 5.0 | 1.6 | 1.8 | 1.6 | 4.8 | 1.5 | 3.4 | 2.3 | 0.88 |
| hsa-miR-365 | 4.6 | 4.6 | 1.9 | 2.0 | 1.8 | 6.0 | 2.5 | 3.0 | 1.6 | 0.87 |
| hsa-miR-20a* | 5.3 | 4.8 | 4.0 | 3.2 | 1.4 | 1.5 | 1.7 | 1.7 | 1.7 | 0.87 |
| hsa-miR-7 | 3.6 | 4.8 | 2.9 | 1.5 | 2.8 | 4.6 | 2.2 | 2.1 | 2.8 | 0.78 |
| hsa-miR-183 | 4.5 | 3.5 | 2.8 | 3.2 | 3.3 | 3.0 | 2.9 | 1.8 | 1.9 | 0.72 |
| hsa-miR-130b | 3.4 | 2.1 | 3.1 | 1.4 | 2.8 | 3.8 | 3.2 | 2.0 | 1.7 | 0.70 |
| hsa-miR-25 | 3.6 | 4.1 | 2.8 | 2.8 | 2.7 | 2.3 | 3.4 | 2.6 | 2.1 | 0.68 |
| hsa-miR-425 | 2.8 | 2.3 | 2.9 | 1.6 | 3.3 | 2.0 | 2.4 | 2.0 | 3.6 | 0.63 |
| hsa-miR-520d-5p | 1.5 | 1.7 | 1.2 | 1.9 | 3.0 | 2.6 | 2.0 | 1.3 | 1.4 | 0.52 |
Fold increase in expression of miRNA common to myeloma were derived by comparing to expression in CD138 positive plasma cells from two healthy donors. The miRNAs shown in bold face are ones that were newly identified by the current study whereas the ones that are underlined were identified by the current study as well as by a recent report [6]. SD values were calculated from four technical replicates. SD >0.50.
Table II.
Fold decrease in expression of commonly down-regulated miRNAs in MM compared to samples from healthy donors.
| microRNAs | L363 | KMS11 | JJN3 | RPMI8226 | U266 | H929 | MM.1S | MM pt1 | MM pt2 | SD |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-223 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.2 | 0.1 | 0.2 | 0.9 | 1.61 |
| hsa-miR-22 | 0.2 | 0.1 | 0.2 | 0.1 | 0.7 | 0.6 | 0.3 | 0.4 | 0.8 | 1.40 |
| hsa-miR-768-3p | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 | 0.2 | 0.2 | 0.5 | 0.3 | 1.28 |
| hsa-miR-29c | 0.1 | 0.2 | 0.2 | 0.2 | 0.1 | 0.3 | 0.4 | 0.6 | 0.6 | 1.19 |
| hsa-miR-32* | 0.5 | 0.5 | 0.4 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 | 0.6 | 1.19 |
| hsa-miR-768-5p | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 | 0.1 | 0.2 | 0.5 | 0.4 | 1.16 |
Fold decrease in expression of miRNA common to myeloma were derived by comparing to expression in CD138 positive plasma cells from two healthy donors. The miRNAs shown are ones that were newly identified by the current study. Previously no miRNAs have been shown to be down-regulated. SD values were calculated from four technical replicates. SD >0.50.
Figure 1.
Differential expression of miRNAs in multiple myeloma. (A) Heat map and unsupervised hierarchical clustering. The clustering was performed on log2 (Hy3 (sample)/Hy5 (common reference pool)) ratios which passed the filtering criteria on variation across samples; standard deviation >0.50. miRNAs that are commonly up-regulated and are commonly down-regulated in CD138 positive myeloma cells compared to CD138 positive cells from healthy donors are indicated in addition to identified miRNA clusters. (B) Expression of the hsa-miR-193b-365 cluster in MM cells (CD138) compared to CD138 positive cells from healthy donor. The data were plotted after conversion of log ratios to fold changes. The mean value of two healthy samples was used as a control.
The fold increase in expression of this new cluster, miR-193b and miR-365, ranged from 2–6 fold depending on the myeloma sample compared to CD138 plasma cells isolated from the 2 healthy donors (Figure 1B). To further confirm that up-regulation of this cluster was common in myeloma, we carried out Q-PCR using RNA isolated from a total of 10 (CD138 purified) patient samples along with RNA isolated from 2 healthy donors. As shown in Figure 2 by Q-PCR we observed 2–12 and 2–7 fold increase in expression of miR-193b and miR-365 respectively depending on the patient samples. In addition to this novel cluster we have identified several new miRNAs that are up-regulated and a smaller set of miRNAs that are down-regulated in myeloma cells using the same version 11.0 array in our studies (Tables I and II).
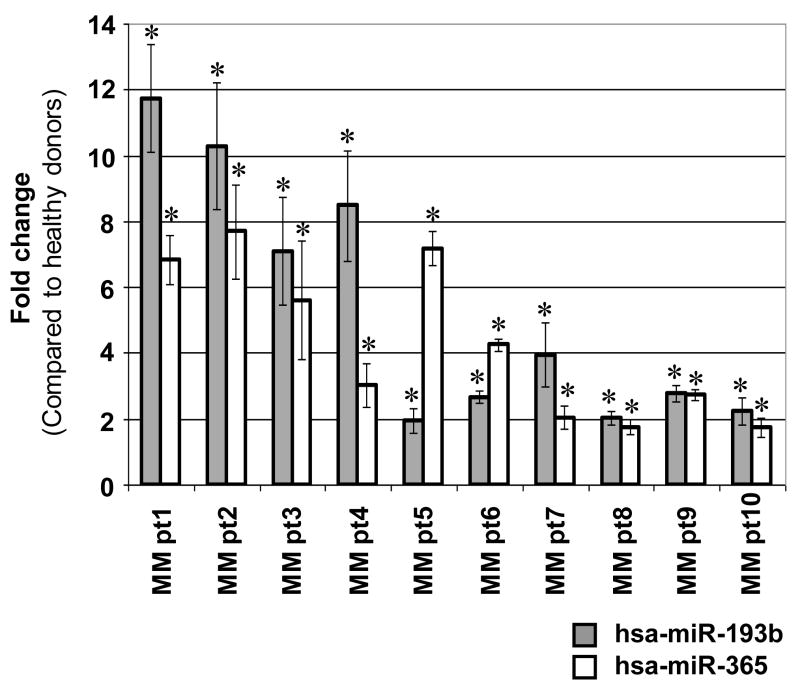
Figure 2.
Expression of hsa-miR-193b and hsa-miR-365 in purified CD138 positive cells from 10 MM patients determined by Q-PCR. The data are shown as fold change in expression of hsa-miR-193b and hsa-miR-365 compared to expression levels observed in purified CD138 positive cells of two pooled healthy donors. Q-PCR was done in triplicate and data were calculated using the comparative Ct (2−ΔΔCt) method. U6 snRNA was used as an internal control. All Q-PCR data were statistically significant (*P<0.05). The P values of hsa-miR-193b and hsa-miR-365 for each myeloma patient sample were 0.008 and 0.005 (MM pt1), 0.014 and 0.015 (MM pt2), 0.023 and 0.047 (MM pt3), 0.016 and 0.032 (MM pt4), 0.046 and 0.002 (MM pt5), 0.004 and 0.001 (MM pt6), 0.034 and 0.036 (MM pt7), 0.014 and 0.029 (MM pt8), 0.007 and 0.003 (MM pt9), 0.038 and 0.044 (MM pt10) respectively. The mean signal intensities for hsa-miR-193b in myeloma patient samples and healthy donors were 429 and 157 respectively. The mean signal intensities for hsa-miR-365 in myeloma patient samples and healthy donors were 351 and 135 respectively.
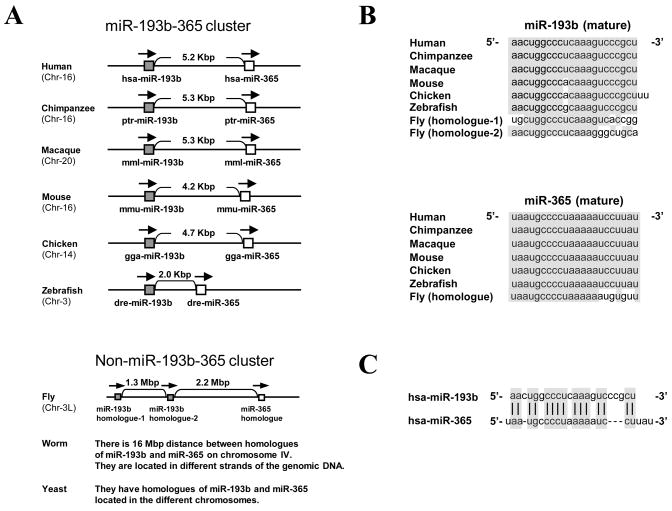
In order to obtain further insight into the organization and sequence conservation of miR-193b-365 cluster we performed cluster and nucleotide sequence alignment of multiple species respectively. The organization of miR-193b-365 cluster was highly conserved among vertebrates such as Human, Chimpanzee, Macaque, Mouse, Chicken and Zebrafish (Figure 3A). It was not conserved in Fly, Worm and Yeast suggesting that organization of this cluster evolved recently and is important in vertebrates. Furthermore, we investigated sequence alignment of mature miR-193b and miR-365 and their homologues among vertebrates and flies which revealed that the miR-193b sequence was highly conserved with the exception of one nucleotide and miR-365 sequence was perfectly conserved among vertebrates but not in Drosophila (Figure 3B). In order to obtain insight into whether hsa-miR-193b and hsa-miR-365 are paralogs that may act in concert to target the same mRNA or mRNA family, we next searched similarity of mature hsa-miR-193b and hsa-miR-365 sequences using ClustalW2 analysis program. This analysis showed a 57.7% similarity between the two sequences suggesting that miR-193b and miR-365 potentially can cooperate to target some of mRNA families in order to synergize the phenotypic outcome (Figure 3C).
Figure 3.
Organization and sequences of miR-193b-365 cluster is highly conserved among vertebrates. (A) Conservation of the miR-193b-365 cluster among vertebrates. Using miRBase (version 12.0) (http://microrna.sanger.ac.uk/sequences/index.shtml), FlyBase (http://flybase.org/), WormBase (http://www.wormbase.org/) and Saccharomyces Genome Database (http://www.yeastgenome.org/), conservation of the miR-193b-365 cluster was investigated among organisms. (B) Sequence alignment of mature miR-193b and miR-365 and their homologues among vertebrates and fly. Using miRBase (version 12.0) and FlyBase, mature miR-193b and miR-365 sequences and their homologues were confirmed among vertebrates and fly. (C) miR-193b-365 cluster composed of two paralogs; miR-193b and miR-365. Similarity of mature miR-193b and miR-365 sequences was searched using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) program.
In the current study we were successful in identifying a common set of miRNAs that are perturbed in myeloma by considering microarray and Q-PCR data obtained from every sample examined without discarding any data. This approach allowed us to identify a cluster that has not been previously recognized in hematological malignancies. Furthermore we were able to identify multiple new miRNAs and confirm several other clusters that have been identified recently in myeloma. Although the new miRNAs identified by this study that is not part of the miR-193b-365 cluster need to be further tested using a large number of primary myeloma samples, it provides us the basis for future studies to understand the role/s played by specific miRNAs in regulating gene transcription in MM.
Acknowledgments
This work is supported in part by National Institutes of Health Grant RO1CA98550 (AW) and Leukemia Lymphoma Society SCOR (AW) grants. LCP was supported by NIH grant CA77816. We thank Jodie Ulaszek, Hui Liu, Jungjin Kim, John Alexandrou (Exiqon), and Dr. Michael Hansen (Exiqon) for their technical assistance.
References
- 1.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–98. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Dalmay T. MicroRNAs and cancer. J Intern Med. 2008;263:366–75. doi: 10.1111/j.1365-2796.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 4.Lee YS, Dutta A. MicroRNAs in Cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrie CH. microRNA expression in lymphoid malignancies: new hope for diagnosis and therapy? J Cell Mol Med. 2008;12:1432–44. doi: 10.1111/j.1582-4934.2008.00399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 8.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang CW, Hsiao CF, Chou CK. Evaluation of experimental designs for two-color cDNA microarrays. J Comput Biol. 2005;12:1202–20. doi: 10.1089/cmb.2005.12.1202. [DOI] [PubMed] [Google Scholar]
- 10.Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet. 2002;3:579–88. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- 11.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–5. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 13.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 2007;98:1482–90. doi: 10.1111/j.1349-7006.2007.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinaldi A, Poretti G, Kwee I, Zucca E, Catapano CV, Tibiletti MG, et al. Concomitant MYC and microRNA cluster miR-17–92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma. 2007;48:410–2. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 16.Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva WA, Jr, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–40. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 19.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1- regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor β signaling. Cancer Res. 2008;68:8191–4. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 21.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106–363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699– 707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]