Summary
IFN-γ is an important mediator of immunity and inflammation that utilizes the Jak-STAT pathway to activate STAT1. Many functions of IFN-γ have been ascribed to direct STAT1-mediated induction of immune effector genes, but recently it has become clear that key IFN-γ functions are mediated by crossregulation of cellular responses to other cytokines and inflammatory factors. Here we review mechanisms by which IFN-γ and STAT1 regulate signaling by TLRs, inflammatory factors, tissue destructive cytokines, anti-inflammatory cytokines, and cytokines that activate opposing STATs. These signaling mechanisms reveal insights about how IFN-γ regulates macrophage activation, inflammation, tissue remodeling, and Th and Treg differentiation, and how Th1 and Th17 responses are integrated in autoimmune diseases.
Introduction
Since the first description of a type II IFN activity more than three decades ago, much has been learned about the biological effects and signal transduction mechanisms of the sole type II IFN, IFN-γ. IFN-γ is one of the most important endogenous mediators of immunity and inflammation. IFN-γ plays a key role in macrophage activation, inflammation, host defense against intracellular pathogens, Th1 responses, and tumor surveillance/immunoediting. In parallel, IFN-γ exerts regulatory functions to limit tissue damage associated with inflammation and to modulate Th and Treg differentiation. IFN-γ can either augment or suppress autoimmunity and associated pathology in a context- and disease-specific manner.
IFN-γ signals mainly through the Janus kinase (Jak)-signal transducer and activator of transcription (STAT) intracellular signal transduction pathway to achieve transcriptional activation of IFN-γ-inducible genes. The STAT family of transcription factors consists of seven members, all of which are involved in receptor signaling by various cytokines and growth factors. The major STAT protein activated by IFN-γ is STAT1. Many IFN-γ functions are mediated by direct activation of immune effector genes by STAT1, including genes encoding anti-viral proteins, microbicidal molecules, phagocytic receptors, chemokines, cytokines, and antigen presenting molecules. Canonical Jak-STAT signaling mechanisms leading to activation of well-characterized STAT1 target genes have been previously reviewed (Stark, 2007; O'Shea and Murray, 2008), and will not be discussed here. A wide spectrum of IFN-γ activities can not be explained based on activation and direct effector functions of STAT1 target genes. Instead, many key IFN-γ functions are mediated by cross-regulation of cellular responses to other cytokines and inflammatory factors. The capacity of IFN-γ to cross-regulate signaling pathways induced by other endogenous and exogenous factors is less appreciated and underlying mechanisms are more recently described and less understood. The mechanisms and (patho)physiological impact of IFN-γ-mediated cross-regulation of signal transduction will be the main focus of the current review.
IFN-γ-induced Jak-STAT1 signaling
In canonical IFN-γ-Jak-STAT1 signaling (recently reviewed in (Stark, 2007)), ligand engagement of the IFN-γ receptor leads to activation of receptor-associated Jak1 and Jak2 and phosphorylation of a receptor tyrosine residue (Y440) that serves as a docking site for STAT1, which exists in a latent state in the cytoplasm. STAT1 is then activated by phosphorylation of tyrosine 701, translocates to the nucleus, binds to a regulatory DNA element termed gamma-activated sequence (GAS) and stimulates transcription of STAT1 target genes. STAT1 binds to DNA as a dimer comprised of two STAT1 subunits in a parallel configuration, such that amino- and carboxy-termini are aligned (Figure 1). Transcriptional activity of STAT1 is augmented by MAPK-mediated phosphorylation of a serine residue in the carboxy-terminal transcription activation domain, and the amplitude of activation is fine tuned by feedback inhibition mediated by various negative regulators of Jak-STAT signaling such as SOCS1 (O'Shea and Murray, 2008). Recent evidence has highlighted that STAT1 undergoes cycles of activation-inactivation that are coupled with nuclear-cytoplasmic shuttling and regulated by post-translational modifications, including dephosphorylation of tyrosine 701 and acetylation of lysine residues (Figure 1).
Figure 1. IFN-γ-induced STAT1 Activation Cycle Mediated by Tyrosine phosphorylation and lysine acetylation.
Unphosphorylated STAT1 dimers are present in the cytoplasm in an equilibrium state between a parallel or antiparallel conformation of STAT1 monomers. Upon activation of IFNGR signaling, STAT1 is phosphorylated by Jak kinases on tyrosine 701 and phosphorylation stabilizes a parallel conformational state that exhibits DNA binding activity. Phosphorylated STAT1 translocates to nucleus, binds to GAS DNA sequences and activates transcription of STAT1 target genes. Active STAT1 in the nucleus undergoes acetylation on lysines 410 and 413, a process catalyzed by histone acetyltransferase (HAT) CBP. Acetylation flags STAT1 for dephosphorylation by the STAT1 phosphatase TCP45; an antiparallel structure facilitates efficient dephosphorylation and thus deactivation. Dephosphorylated STAT1 recycles back to cytoplasm, where a histone deacetylase (HDAC), HDAC3, deacetylates STAT1 and completes the phosphorylation/acetylation cycle. Deacetylation of STAT1 results in less efficient TCP45-mediated dephosphorylation and thus primes STAT1 for IFNGR-Jak-mediated tyrosine phosphorylation.
Inactivation of nuclear STAT1 occurs rapidly following binding to chromatin and activation of target gene transcription. STAT1 dissociates from DNA and the STAT1 dimer undergoes a conformational change, such that the parallel orientation of STAT1 monomers changes to an antiparallel configuration that exposes phosphotyrosine residues and thus facilitates dephosphorylation of STAT1 by phosphatases (Mao et al., 2005; Mertens et al., 2006; Zhong et al., 2005). Subsequently STAT1 is dephosphorylated by phosphatases such as TCP45, and dephosphorylated STAT1 returns to cytoplasm, where it can potentially serve as the substrate for subsequent rounds of activation and inactivation (Figure 1). There is accumulating evidence that cytoplasmic STATs do not exist predominantly as a monomer (Braunstein et al., 2003; Ota et al., 2004), but instead as a homodimer with the two STAT1 subunits in an anti-parallel configuration (Mao et al., 2005). In this model, STAT1 tyrosine phosphorylation triggers or stabilizes a conformational change of pre-existing STAT1 dimers from antiparallel to parallel configuration and results in increased abundance of parallel dimers with an exposed nuclear localization sequence and high DNA-binding activity (Wenta et al., 2008).
Recent reports suggest that the function of STATs and the transit of STAT1 through the activation-inactivation cycle are regulated by lysine acetylation (Kramer et al., 2006; Kramer et al., 2009; Nie et al., 2009; Tang et al., 2007; Yuan et al., 2005). The acetylation status of several STATs including STAT1, STAT2, and STAT3 is dynamically determined by opposing activities of histone acetyltransferases (HATs) vs. histone deacetylases (HDACs). However, the impact of STAT acetylation on signaling is not well understood, as both positive and negative roles of STAT acetylation on cytokine receptor signaling have been reported. The preponderance of evidence suggests that acetylation of STAT3 is often, although not exclusively, associated with positive regulation of signal transduction (Nie et al., 2009; Wang et al., 2005; Yuan et al., 2005), whereas acetylation of STAT1 is associated with inhibitory effects (Kramer et al., 2006; Kramer et al., 2009). STAT3 acetylation by the HAT CBP has been correlated with increased DNA-binding and transactivation activity (Wang et al., 2005; Yuan et al., 2005) and potentially with its anti-inflammatory properties (Sun et al., 2009). Conversely, deacetylation of STAT3 by the HDAC Sirtuin 1 correlates with decreased STAT3 tyrosine phosphorylation and activity (Nie et al., 2009). Similar to STAT3, STAT1 is also acetylated by CBP (Kramer et al., 2006; Kramer et al., 2009). However, in contrast to STAT3, STAT1 acetylation seems to play a negative role in signaling. It is recently reported that acetylation of STAT1 on lysine residues 410 and 413 in the nucleus results in enhanced interaction with TCP45 and increased dephosphorylation (Kramer et al., 2009). Thus, acetylation “flags” STAT1 for inactivation. The mechanism by which acetylation promotes interaction of STAT1 with TCP45 is not clear. One possibility is that acetylation promotes a change to the anti-parallel configuration of STAT1 subunits that facilitates dephosphorylation by TCP45. In this speculative model, acetylated cytoplasmic STAT1 is refractory to activation because of association with TCP45. De-acetylation of STAT1 that is mediated by HDACs such as HDAC3 (Kramer et al., 2009) thus promotes increased tyrosine phosphorylation and stabilization of the active parallel configuration STAT1 dimer (Figure 1). This requirement for HDAC activity for STAT1 activation could potentially explain the paradoxical observation that HDAC inhibitors suppress STAT1-dependent transcription (Chang et al., 2004; Nusinzon and Horvath, 2003). This acetylation-mediated negative regulatory mechanism can potentially be bypassed by de novo synthesis of STAT1, which is an important mechanism for augmenting long term STAT1 activity.
The role of acetylation in regulating the STAT1 activation cycle opens new avenues for regulation and modulation of STAT1 function and crosstalk with heterologous signaling pathways. For example, the activity of certain STAT HDACs, such as Sirtuin1, is regulated by the overall cellular metabolic state as reflected in the NAD/NADH ratio and can be selectively and therapeutically modulated by small molecule compounds (Finkel et al., 2009). Other, as yet unknown, mechanisms control the translocation of HDACs and HATs to the cytoplasm where they can modify STATs. Despite recent progress, many unanswered questions remain regarding STAT acetylation. One outstanding question is what underlies the differential functional outcomes of acetylation of different STAT molecules. Plausible explanations include different acetylation sites (lysines 410 and 413 of STAT1 versus lysines 679, 685, 707, 709 of STAT3) and different structural changes induced by acetylation. As STAT1 and STAT3 often antagonize each other's functions in many processes including inflammation and tumorigenesis, differential regulation of these STATs by acetylation may represent a mechanism to regulate the balance of STAT function downstream of cytokine receptors.
Enhancement of Innate Immune Activation (Priming)
It has been long appreciated that IFN-γ promotes innate immune responses by activating macrophages. One mechanism of IFN-γ-mediated macrophage activation is direct effector gene activation via STAT1 as discussed above. Another way for IFN-γ to achieve strong activation effects is by enhancing macrophage responsiveness to other inflammatory stimuli, such as TLR ligands and TNF; this phenomenon is termed “priming”. Priming of TLR responses by IFN-γ greatly augments TLR-induced expression of inflammatory mediators and immune effectors including multiple cytokines and chemokines, and profoundly affects biological outcomes of innate immunity and inflammation. The mechanisms underlying IFN-γ-mediated priming have been the subject of extensive investigation and it has been suggested that IFN-γ priming enhances TLR-activated signal transduction. For example, IFN-γ priming increases TLR expression, promotes NF-κB activation, and induces transcription factors that are essential for expression of certain TLR responsive genes. However, enhancement of TLR signaling can not explain the full spectrum of activation achieved by IFN-γ priming, and accumulating evidence suggests that inactivation of feedback inhibition pathways by IFN-γ is important for the broad and sustained activation of macrophage effector genes and mechanisms that is characteristic of primed cells. Of note, enhancement of positive signaling and inactivation of feedback inhibition are two complementary mechanisms that reinforce each other to achieve the robust priming effects seen with IFN-γ. The enhancement of positive TLR signaling by IFN-γ has been reviewed elsewhere (Schroder et al., 2006); herein we review recent progress regarding IFN-γ-mediated abrogation of TLR-induced feedback inhibitory loops.
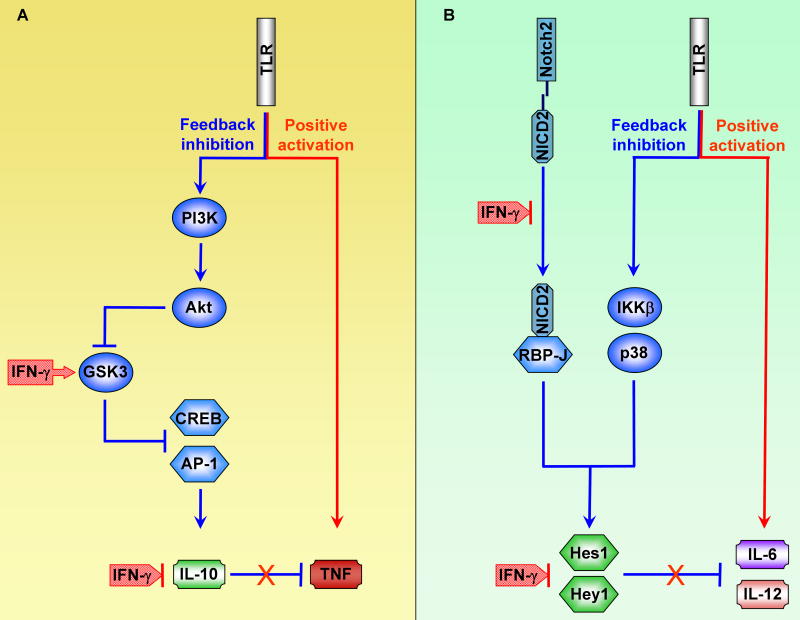
IL-10 is a major anti-inflammatory cytokine induced by TLR signaling and functions to inhibit production of TLR-induced proinflammatory mediators, such as TNF, in a STAT3-dependent manner (Mosser and Zhang, 2008). IFN-γ priming disrupts this IL-10-STAT3 feedback inhibitory loop and thus leads to increased production of the inflammatory cytokines. One mechanism by which IFN-γ suppresses the IL-10-STAT3 axis involves inhibition of TLR-induced Il10 gene expression. IFN-γ suppresses IL-10 production by increasing the activity of GSK3β, a serine/threonine kinase that inhibits the function of AP-1 and CREB, two transcription factors critical for Il10 expression. Upon activation of TLRs, GSK3β is phosphorylated and inactivated by the PI3K/Akt pathway, and inactivation of GSK3β allows Il10 to be expressed. IFN-γ priming overcomes this TLR-induced inhibition of GSK3β and thus restores the capacity of GSK3β to inhibit Il10 expression (Hu et al., 2006) (Figure 2, left panel). IFN-γ-GSK3β-mediated regulation of TLR responses is best characterized with IL-10 as a target. However, given that GSK3β controls the function of CREB and AP-1, key transcription factors involved in expression of many TLR-induced genes, it is likely that IFN-γ regulates expression of a subset of TLR-inducible genes via GSK3 (Ho et al., 2008; Hu et al., 2007). One unanswered question is the mechanism by which IFN-γ activates GSK3β. One potential mechanism is IFN-γ-mediated suppression of TLR-induced PI3K/Akt signaling, with resultant decreased inhibitory phosphorylation of GSK3β (Hu et al., 2006). Alternatively, IFN-γ can inactivate GSK3 phosphatases or promote alternative GSK3 activation via Pyk2 (Tsai et al., 2009). As GSK3 is involved in various signaling pathways including Wnt-β-catenin signaling (Kockeritz et al., 2006), IFN-γ regulation of GSK3β has broader implications for signal transduction crosstalk, such as potential cross-regulation between IFN-γ and Wnt pathways.
Figure 2. IFN-γ Signaling Disrupts TLR-induced Feedback Inhibitory Loops.
(A) IFN-γ enhances TLR-induced TNF production by disrupting an IL-10-mediated inhibitory loop. IFN-γ signaling leads to increased activity of GSK3, which negatively regulates Il10 expression by suppressing activation of transcription factors CREB and AP-1.
(B) IFN-γ enhances TLR-induced IL-6 and IL-12 production by disrupting an inhibitory loop mediated by canonical Notch target genes Hes1 and Hey1. IFN-γ signaling downregulates intracellular NICD2 amounts and thus inhibits expression of Hes1 and Hey1. Hes1 and Hey1 are transcription repressors that negatively regulate Il6 and Il12 gene expression.
In addition to inactivation of the IL-10-STAT3 axis, IFN-γ disrupts another feedback inhibitory loop involving Notch target genes Hes1 and Hey1, which are transcriptional repressors (Hu et al., 2008a). The Notch pathway, whose functions have been predominantly characterized in developmental biology systems, was recently described to modulate macrophage activation and to be regulated by IFN-γ. In macrophages, expression of canonical Notch target genes Hes1 and Hey1 is induced by TLR stimulation. Expression of Notch target genes is synergistically activated by TLR and Notch pathways by cooperation between RBP-J, a master transcription factor downstream of Notch signaling, and the TLR signaling components IKKβ and p38. Following induction by TLRs, transcription repressors Hes1 and Hey1 suppress TLR-induced IL-6 and IL-12 expression, constituting another feedback inhibitory loop that dampens cytokine production (Figure 2B). IFN-γ signaling inhibits expression of Hes1 and Hey1 at least in part by downregulating amounts of NICD2, the intracellular cleaved fragment of Notch2 receptor that binds RBP-J and activates Notch target gene expression. Potential mechanisms by which IFN-γ downregulates NICD2 include modulation of proteases that generate and degrade NICD2, and activation of GSK3 that destabilizes of NICD proteins (Espinosa et al., 2003). Thus, IFN-γ primes for augmented TLR-induced IL-6 and IL-12 production by disrupting an inhibitory loop mediated by Hes1 and Hey1 (Hu et al., 2008a). The above examples suggest that inactivation of feedback inhibitory pathways by IFN-γ is a common mechanism of priming and additional examples are likely to be uncovered. Another notion emerging from these studies is that IFN-γ selectively and differentially regulates expression of subsets of TLR target genes by targeting distinct TLR-induced signaling molecules. This provides an additional mechanism for selective regulation of TLR responses, whose importance has recently been highlighted by Medzhitov and colleagues (Foster et al., 2007; Foster and Medzhitov, 2009).
IFN-γ also directly inhibits signaling pathways downstream of anti-inflammatory cytokines to antagonize their suppressive functions. IFN-γ antagonizes anti-inflammatory effects of IL-10 both by attenuating IL-10 production, as discussed above, and by suppressing IL-10 signaling. Anti-inflammatory action of IL-10 is predominantly mediated by STAT3 and IFN-γ cross-regulates IL-10 signaling by abrogating expression of STAT3 target genes (Herrero et al., 2003). Inhibition of IL-10-STAT3 signaling has significant biological impact as the anti-inflammatory activity of IL-10 is diminished following IFN-γ priming (Herrero et al., 2003). The mechanisms of STAT1-STAT3 cross-regulation are discussed below. TGFβ is another cytokine with important anti-inflammatory function that is subject to the antagonistic action of IFN-γ. IFN-γ induces expression of Smad7, an inhibitory Smad, and thus inhibits TGF-β-induced activation of the activating Smad3 and of TGFβ responsive genes (Ulloa et al., 1999). STAT1 also directly binds Smad3 and inhibits its function (Ghosh et al., 2001). In summary, inhibition of expression and function of anti-inflammatory molecules represents an important mechanism of IFN-γ-mediated priming of enhanced innate immune responses.
Attenuation of tissue destruction
The activating effects of IFN-γ on immunity and inflammation have been extensively studied and are well established. At the same time, IFN-γ possesses crucial homeostatic functions that limit inflammation-associated tissue damage. This enables the host to utilize one mediator, IFN-γ, to regulate the balance between clearance of invading pathogens and limiting collateral damage to the host. IFN-γ plays an important role in limiting tissue damage associated with acute infections and with chronic inflammation in autoimmune diseases such as inflammatory arthritis and experimental allergic encephalomyelitis (EAE). Mechanisms underlying the homeostatic functions of IFN-γ, which include inhibition of gene expression, of migration and differentiation of tissue-destructive cells, and inhibition of signaling by tissue-destructive cytokines, are reviewed in this section.
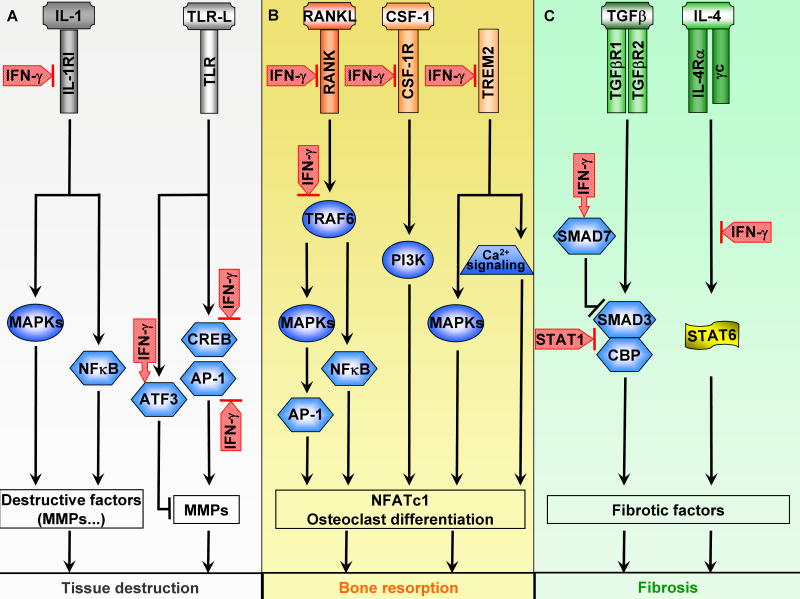
One mechanism by which IFN-γ attenuates tissue destruction is inhibition of expression of genes that encode tissue destructive factors, such as matrix metalloproteinases (MMPs), serine proteases, coagulation factors, complement components, and enzymes involved in prostaglandin metabolism ((Barrios-Rodiles and Chadee, 1998; Ho et al., 2008; Ma et al., 2001; Sanceau et al., 2002; Zhou et al., 2003) and L. Ivashkiv, unpublished data). IFN-γ broadly suppresses expression of multiple MMPs including MMP1, MMP2, MMP3, MMP7, MMP9, and MMP10 induced by various receptors such as TLRs and IL-1R. IFN-γ-mediated suppression of MMPs requires STAT1. However, to date there is no compelling evidence that STAT1 directly suppresses gene expression, including expression of MMP genes. Instead, IFN-γ inhibits receptors and signals that induce MMP expression (Figure 3A). IFN-γ suppresses IL-1-induced MMP expression in macrophages by STAT1-dependent downregulation of IL-1RI (Hu et al., 2005a). Inhibition at this proximal step inactivates all signaling cascades downstream of the IL-1 receptor and results in a global block in macrophage responses to IL-1 (Browning and Ribolini, 1987; Dickensheets and Donnelly, 1997; Hu et al., 2005a). IFN-γ-mediated inhibition of TLR-induced genes targets downstream signaling components and is more selective in inhibiting a subset of approximately 15% of TLR-inducible genes, including MMP genes (Hu et al., 2007). For TLRs, the inhibitory effects of IFN-γ are achieved by superinduction of transcriptional repressors, such as ATF-3 that binds to and inhibits the MMP1 promoter, and by inhibition of AP-1 transcription factors that are required for MMP expression (Ho et al., 2008). This inhibition of AP-1 and downstream target genes is reminiscent of the above-discussed findings that IFN-γ inhibits IL-10 expression in part by inhibiting AP-1 (Hu et al., 2006). IFN-γ suppresses AP-1 activity by several mechanisms, including attenuation of upstream MAPK pathways that induce expression of AP-1 proteins and activate them post-translationally (Hu et al., 2006), suppression of transcription of genes encoding AP-1 components (Hu et al., 2006), downregulation of AP-1 mRNA at the posttranscriptional level (Radzioch and Varesio, 1991), and regulation of AP-1 protein stability (Hu et al., 2007). Destabilization of the AP-1 protein c-Jun by IFN-γ appears to be mediated by GSK3 that phosphorylates c-Jun and creates a binding site for an E3 ubiquitin ligase Fbw7 (Wei et al., 2005). Overall, differential regulation of transcription factors downstream of TLR signaling by IFN-γ (inhibiting AP-1 versus augmenting NF-κB) provides a means to augment inflammatory cytokine production yet to limit expression of tissue destructive factors such as MMPs. Another more universal mechanism of suppression that is independent of upstream signaling involves STAT1-mediated sequestration of the coactivator CBP, which is then not available to activate MMP gene promoters (Ma et al., 2005).
Figure 3. Signaling Mechanisms Associated with IFN-γ-mediated Attenuation of Tissue Destruction.
(A) IFN-γ suppresses inflammatory tissue destruction via regulation of IL-1R and TLR signaling. IFN-γ inhibits IL-1 signaling and subsequent induction of destructive factors in macrophages by downregulating IL-1RI expression. In addition, IFN-γ blocks induction of MMP downstream of TLR signaling by superinducing transcription repressor ATF3 and inhibiting transcription activators CREB and AP-1. IFN-γ inhibits CREB activity by suppressing its serine phosphorylation and inhibits AP-1 by downregulating nuclear protein levels of its subunits.
(B) IFN-γ inhibits osteoclastogenesis and bone resorption via regulation of RANK, CSF-1R, and TREM2 signaling. In osteoclast progenitor cells, IFN-γ suppresses expression as well as signal transduction of RANK, CSF-1R, and TREM2, receptors critical for the process of osteoclastogenesis.
(C) IFN-γ attenuates fibrosis via inhibition of TGFβR and IL-4R signaling. IFN-γ suppresses TGFβR signaling by induction of inhibitory SMAD (SMAD7) and by direct inhibition of SMAD3 by STAT1. IFN-γ inhibits IL-4R signaling by induction of SOCS1 (see Figure 5A for details).
Another way by which IFN-γ exerts homeostatic functions is attenuation of tissue infiltration by neutrophils and monocytes. In several models of human autoimmune disorders such as experimental arthritis and EAE, deficiency of IFN-γ signaling results in increased accumulation of neutrophils and other myeloid cells at sites of inflammation (Ferber et al., 1996; Irmler et al., 2007; Manoury-Schwartz et al., 1997; Vermeire et al., 1997). Several mechanisms may account for the suppressive effects of IFN-γ on inflammatory cell infiltration: 1) IFN-γ attenuates myelopoiesis and granulopoiesis and thus limits availability of infiltrating cells at their source. Several reports have shown that IFN-γ-deficient mice undergo deregulated expansion of macrophages and granulocytes during infections (Matthys et al., 1999; Murray et al., 1998). 2) IFN-γ inhibits expression of chemokines that attract cells to inflammatory sites. One example of such regulation is IFN-γ-mediated inhibition of expression of MCP-2, a major neutrophil chemoattractant in mice (Kelchtermans et al., 2007). 3) IFN-γ alters cellular responsiveness to chemokines. This phenomenon is exemplified by the observation that IFN-γ arrests monocyte migration and alters cellular responses to CCL2 by modulating the activities of signaling molecules Pyk2, Jnk, Rac, and Cdc42 and inhibiting CCL2-induced activation of PAK kinase that regulates cytoskeleton rearrangement and cell polarization (Hu et al., 2008b).
Inflammation often leads to tissue remodeling and bone resorption, processes that are subject to inhibition by IFN-γ. Bone resorption is mediated by myeloid lineage cells called osteoclasts and IFN-γ is a potent inhibitor of osteoclastogenesis (Takahashi et al., 1986; Takayanagi et al., 2005). IFN-γ suppresses osteoclastogenesis in vitro and in vivo by regulating the expression and signaling by two key receptors required for osteoclast generation and differentiation, c-Fms (the receptor for M-CSF, also termed CSF-1) and receptor activator of nuclear factor κB (RANK), a member of the TNF receptor family that binds its cognate ligand RANKL. IFN-γ interferes with RANK signaling by suppressing expression of RANK and by targeting the key adaptor molecule TRAF6 for proteasome-mediated degradation, resulting in diminished activation of downstream signaling events (Takayanagi et al., 2000) (Figure 3B). Similar to IFN-γ, a type I IFN, IFN-β, also inhibits RANK signaling in a STAT1-dependent manner. However, instead of targeting TRAF6, IFN-β inhibits translation of c-Fos, an AP-1 family transcription factor essential for the induction of NFATc1, the master regulator of osteoclastogenesis (Takayanagi et al., 2002). Given that IFN-γ suppresses c-Fos expression in closely-related cell types such as macrophages, it is possible that IFN-γ targets c-Fos in osteoclasts in addition to targeting RANK and TRAF6. One interesting possibility awaiting assessment is the effect of IFN-γ on CREB activation and function in the context of osteoclast differentiation. Given the precedent of inhibition of TLR-induced CREB activity by IFN-γ in macrophages (Hu et al., 2006) and the critical role of CREB in osteoclastogenesis (Sato et al., 2006), inhibition of CREB may contribute to IFN-γ-mediated inhibition of osteoclastogenesis. IFN-γ also inhibits expression of c-Fms, thus conferring resistance to M-CSF stimulation (Baccarini et al., 1992; Inaba et al., 1995) (Figure 3B). Diminished M-CSF responses result in decreased production of osteoclast precursors, and may also explain the suppressive effects of IFN-γ on myelopoiesis.
Fibrosis results from aberrant tissue remodeling and excessive connective tissue formation post injury or during chronic inflammation. IFN-γ suppresses fibrosis in several models including viral hepatitis, bleomycin-induced pulmonary fibrosis, and schistosomiasis-induced fibrosis (Wynn, 2004) at least in part by inhibiting signaling by the major pro-fibrotic factors IL-4, IL-13 and TGF-β (Figure 3C). These suppressive effects can be mediated at least in part by the IFN-γ-induced T-bet transcription factor (Aliprantis et al., 2007). Alternatively activated or M2 macrophages have been proposed to play a key role in promoting fibrosis (Mosser and Edwards, 2008; Wynn, 2004), and IFN-γ-mediated diversion of macrophage differentiation away from a wound healing pro-fibrotic M2 phenotype also likely contributes to suppression of fibrosis. Finally, IFN-γ suppresses fibrosis by inhibiting collagen synthesis.
In summary, IFN-γ attenuates tissue destruction by modulating the expression, signaling, and function of tissue-destructive cytokines and their receptors, with resulting suppression of gene expression and of cell recruitment and differentiation. Where studied, these suppressive effects are dependent on STAT1, suggesting indirect regulation mediated by STAT1 target genes such as ATF3. Identification and characterization of STAT1 target genes that regulate tissue-destructive pathways represents a fruitful area for future research.
Regulation of adaptive immunity: Th and Treg differentiation
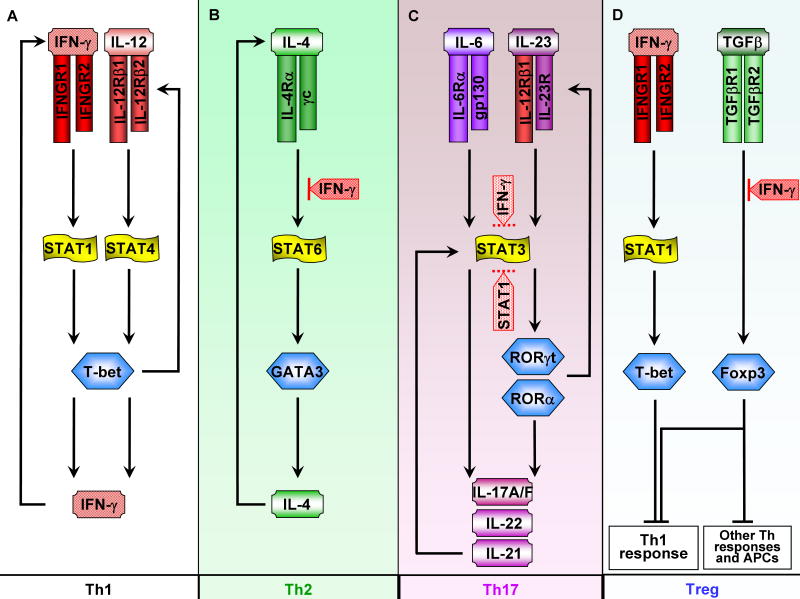
As a major effector cytokine of Th1 immunity, it is no surprise that IFN-γ auto-amplifies Th1 responses (Figure 4A) and cross-inhibits differentiation and function of other Th subsets including Th2 and Th17 cells. This regulation by IFN-γ represents a mechanism for maintaining Th1 lineage commitment and stabilizing Th phenotypes (Szabo et al., 2003). One general theme underlying IFN-γ-mediated cross-inhibition is interference with signal transduction pathways and transcription factors downstream of cytokines that drive differentiation of other Th subtypes. For example, IFN-γ suppresses the IL-4-STAT6 pathway that is required for Th2 differentiation, mediated in part by induction of SOCS1 that inhibits IL-4 receptor signaling (Naka et al., 2001; Yu et al., 2004) (Figure 4B). In addition, IFN-γ-induced Tbet suppresses Th2 differentiation by inhibiting the expression/function of the Th2 transcription factor GATA3 (Hwang et al., 2005; Yu et al., 2004). Another SOCS-independent inhibitory mechanism is posttranscriptional downregulation of IL-4-induced IL-4R gene expression (So et al., 2000).
Figure 4. Signaling Mechanisms Associated with IFN-γ-mediated Regulation of T Cell Differentiation and Function.
(A) In Th1 differentiation, IFN-γ-STAT1 signaling is critical for induction of T-bet and thus for sustaining the positive feedback loop that leads to heightened production of IFN-γ.
(B) IFN-γ blocks Th2 differentiation by inhibiting IL-4-STAT6 signaling.
(C) IFN-γ and STAT1 block Th17 differentiation. IFN-γ-STAT1 signaling can potently inhibit Th17 differentiation but the mechanism of action is not clear. As STAT3 signaling from multiple cytokines including IL-6, IL-23, and IL-21 plays a pivotal role in mediating Th17 differentiation, it is possible that IFN-γ-STAT1 suppresses Th17 by targeting STAT3 (as shown by dotted lines). IFN-γ and STAT1 also inhibit the aryl hydrocarbon nuclear receptor (AHR) important for Th17 differentiation and it is also possible that suppression of TGFβ and IL-1signaling by IFN-γ contributes to inhibition of Th17 differentiation (not depicted).
(D) IFN-γ regulates Treg differentiation and function. IFN-γ can block TGFβ-mediated Treg differentiation. Recently, a more complex role of IFN-γ in regulation of Treg differentiation has emerged. In Foxp3+ Treg cells, IFN-γ upregulates expression of T-bet, which in turns promotes expression of CXCR3 that regulates homing of T-bet+ FoxP3+ Tregs to sites of Th1 inflammation. T-bet also increases suppressive function of Tregs, and T-bet+ FoxP3+ Tregs effectively suppress Th1 inflammation in vivo.
Differentiation of Th17 cells, which is driven IL-6, IL-1, TGF-β, IL-21, and IL-23 (Zhou et al., 2009), is strongly suppressed by IFN-γ in vitro and in vivo. In vitro, treatment with IFN-γ neutralizing antibody during the course of Th17 differentiation leads to increased frequency of Th17 cells, whereas exogenous IFN-γ reduces the Th17 population (Harrington et al., 2005). In vivo, IFN-γ deficient mice exhibit enhanced Th17 responses in several disease models including mycobacterial infection and collagen-induced arthritis (Chu et al., 2007; Cruz et al., 2006; Irmler et al., 2007). Aside from its effects on Th17 development, it was recently reported that IFN-γ inhibits effector functions of Th17 cells (Kelchtermans et al., 2009). Although cross-inhibition of Th17 development by IFN-γ is relatively well-established, the mechanisms are not clear. Inhibition by IFN-γ is likely to be dependent on STAT1, as STAT1-deficient mice mount enhanced Th17 responses, and another STAT1-activating cytokine, IL-27, potently suppresses Th17 development in a STAT1-dependent manner (Stumhofer et al., 2006). The molecules important for Th17 differentiation that are inhibited by IFN-γ have not been unequivocally identified. Possibilities include inhibition of Smad signaling downstream of TGF-β (Tanaka et al., 2008), downregulation of T cell IL-1R expression (X. Hu, unpublished data), and inhibition of the aryl hydrocarbon receptor (Kimura et al., 2008). In addition, STAT1 inhibits STAT3 (see below), which is activated by IL-6, IL-23, and IL-21 and is important for Th17 differentiation; it is possible that STAT1 suppresses Th17 differentiation by targeting STAT3 (Figure 4C).
Th17 responses are important for host defense against extracellular bacteria and yeast, and are characterized by neutrophil infiltration and the potential for severe tissue destruction (McGeachy and Cua, 2008). Thus, counter-regulation of Th17 differentiation by IFN-γ may represent an important pathway to limit tissue inflammation and damage. Emerging evidence suggests a greater complexity in IFN-γ-mediated regulation of Th17 cells than previously appreciated. Many Th cells at sites of inflammation, such as the central nervous system in EAE, co-express IFN-γ and IL-17, and recent evidence supports plasticity in the Th17 lineage, with the potential to evolve into IFN-γ-expressing cells (Lee et al., 2009). Thus, different from Th1 and Th2 cells, the relationship of Th1 and Th17 cells is not limited to cross-inhibition. Instead, there is a potential for ongoing generation and differentiation of Th cells with a changing or mixed effector phenotype. This allows fine tuning of Th1/Th17 effector functions to achieve the most effective host response during the course of infections, and to balance immunity with preservation of tissue integrity.
Regulatory T cells (Tregs) serve to restrain over-activation of effector T cells and maintain homeostasis. Interest in the role of IFN-γ in Treg development was prompted by the initially paradoxical findings that IFN-γ is protective in models of autoimmune diseases such as EAE (Ferber et al., 1996; Willenborg et al., 1996). Exacerbation of EAE in mice deficient in IFN-γ signaling correlates with reduced numbers and function of Treg cells (Nishibori et al., 2004; Wang et al., 2006). Moreover, adoptive transfer of IFN-γ-treated Treg cells is sufficient to ameliorate EAE symptoms (Nishibori et al., 2004), supporting an essential role of IFN-γ in Treg development, at least in EAE model. However, it is difficult to reconcile the above findings with the observations that Treg development proceeds normally in the absence of IFN-γ signaling under many conditions (Kelchtermans et al., 2005). Recently, the emerging concept of Treg diversity and polarization has shed light on the controversial issue of the involvement of IFN-γ in Treg development (Barnes and Powrie, 2009). Two elegant studies suggest that, similar to effector T cells, Tregs undergo polarization into specialized phenotypes, and that factors important for effector T cell development may also play a critical role in Treg polarization (Koch et al., 2009; Zheng et al., 2009). For example, IRF4, a transcription factor key for differentiation of Th2 cells, is required for differentiation and function of a Treg subset that specifically suppresses Th2 responses (Zheng et al., 2009). In parallel, T-bet, a master regulator of Th1 differentiation, is upregulated by IFN-γ-STAT1 signaling in Foxp3+ Treg cells and Foxp3+T-bet+ cells represent a novel subset of Tregs that selectively dampens Th1 responses (Koch et al., 2009) (Figure 4D). The existence of specialized Treg subsets may help to explain the apparent discrepancy that IFN-γ is necessary for Treg development under certain circumstances but not under others. Interestingly, as a major effector of Th1 responses, IFN-γ promotes differentiation of Foxp3+T-bet+ regulatory T cells that suppress Th1 responses, constituting a negative feedback loop that contributes to homeostatic action of IFN-γ. Overall, recent developments implicate a regulatory role of IFN-γ in modulating many aspects of T cell biology asides from its classic activating role in Th1 responses. In addition to its action on T cells, IFN-γ suppresses early B cell development in the bone marrow and also promotes isotype switching to IgG2a, underscoring its diverse effects on adaptive immunity (Schroder et al., 2004).
Cross-inhibition of opposing STATs (STAT3 and STAT6)
Mechanisms by which IFN-γ and STAT1 regulate the function of receptors that activate distinct signaling pathways were described above. In this section we will review mechanisms by which IFN-γ and STAT1 regulate signaling by cytokines that utilize the Jak-STAT pathway but have different and opposite functions from IFN-γ. Cytokines that oppose each other often activate different STATs that antagonize each other. A good example of antagonistic STATs is STAT1 and STAT3 that are activated by the opposing cytokines IFN-γ and IL-10, respectively. STAT1 and STAT3 oppose each other in many biological processes including macrophage activation that is enhanced by STAT1 and inhibited by STAT3, cell proliferation that is suppressed by STAT1 and promoted by STAT3, and Th differentiation where STAT1 promotes Th1 responses and STAT3 drives Th17 response (O'Shea and Murray, 2008).
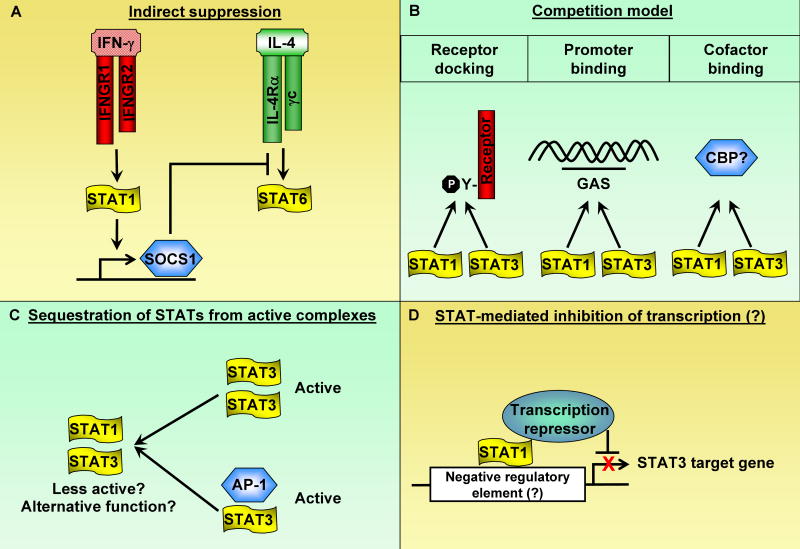
The best established mechanism by which STATs oppose each other is indirect regulation mediated by SOCS proteins that suppress signaling by cytokine receptors by inhibiting receptor-associated Jaks, binding to and blocking STAT docking sites, and targeting receptors for proteosomal degradation (Yoshimura et al., 2007). IFN-γ and STAT1 activate expression of SOCS1, a potent feedback inhibitor of IFN-γ signaling that also cross-inhibits signaling by the type I IFN receptor and the IL-4 receptor (Dickensheets et al., 1999; Fenner et al., 2006; Losman et al., 1999; Naka et al., 2001; Yu et al., 2004; Zimmerer et al., 2007). Thus, SOCS1-mediated inhibition can explain the suppressive properties of IFN-γ on Th2 differentiation (Fig. 5A and Fig. 4). However, SOCS1 does not effectively inhibit signaling by the IL-10 receptor or IL-6-related receptors that utilize gp130, and is not known to inhibit signaling by IL-21 or IL-23. Thus, IFN-γ-mediated antagonism of IL-10 function (Herrero et al., 2003; Lauw et al., 2000; Pajkrt et al., 1997) can not be explained by a SOCS1-dependent mechanism; it also appears likely that regulation of Th17 differentiation by IFN-γ can not be explained solely by induction of SOCS1 or other SOCS proteins. STAT1 also suppresses STAT3 by alternative and more direct mechanisms, as was first suggested by genetic evidence showing increased STAT3 activation in STAT1-deficient cells (Gil et al., 2001; Qing and Stark, 2004; Ramana et al., 2001).
Figure 5. Mechanisms by which STATs can oppose each other.
(A) Antagonizing effects between STATs can be mediated by indirect mechanisms such as induction of inhibitory molecules. An example is depicted here as STAT1 cross-inhibits STAT6 via induction of SOCS1.
(B) Individual STATs compete for receptor docking sites, DNA binding elements, and/or binding cofactors.
(C) A given STAT could bind and sequester other STATs from forming transcriptionally active complexes. Although the function of STAT heterodimers are not clear, it is plausible that heterodimers may be less active than homodimers in gene induction or possess alternative functions distinct from those of homodimers.
(D) Although not experimentally proven, it is conceivable that a given STAT could directly bind to negative regulatory element(s) on the promoter of a gene driven by another STAT and suppress transcription.
Mechanisms by which STAT1 can potentially directly inhibit STAT3 include competition for binding to docking sites on receptors or to target DNA sequences in promoters, competition for binding to other proteins or cofactors, sequestration of STAT3 from active complexes, and direct transcriptional repression of STAT3 target genes (Figure 5). These mechanisms are relevant for cross-inhibition of signaling by other cytokines, but also for establishing the balance of STAT activation downstream of the IFNGR. Thus, STAT1 suppresses IFNGR-mediated activation of STAT3, at least in part by competing for the STAT docking site within the IFNGR cytoplasmic domain. As receptor docking is a prerequisite for activation by tyrosine phosphorylation, the prediction of the competition for docking sites model is that STAT1 suppresses STAT3 tyrosine phosphorylation downstream of IFNGR or other receptors. Several reports using cell lines support this model (Costa-Pereira et al., 2002; Qing and Stark, 2004), but suppression of STAT3 tyrosine phosphorylation by STAT1 appears to be context-dependent, and in primary macrophages it is clear that IFN-γ and STAT1 suppress STAT3 function without suppressing its tyrosine phosphorylation (Herrero et al., 2003). Conceivably, STAT1 could suppress STAT3 function by displacing STAT3 from binding at target gene promoters; in the case of promoter binding by the STAT1β isoform that does not contain a transcription activation domain, such binding would lead to inhibition of transcription. There is, however, very limited evidence to support mechanisms that involve competition for binding to target DNA elements or for recruitment of transcriptional coactivators.
An alternative explanation for how STAT1 can inhibit STAT3 function without suppressing STAT3 tyrosine phosphorylation is sequestration of STAT3 away from active complexes into STAT1:STAT3 heterodimers (Figure 5C). This will result in diminished amounts of STAT3:STAT3 homodimers, such as those activated by IL-10, that are transcriptionally active and functional. It is possible that STAT1:STAT3 heterodimers are less transcriptionally active than STAT3 homodimers, or bind to alternative promoters. Sequestration of STAT3 into STAT1:STAT3 heterodimers is increased in cells that have been primed and express increased amounts of STAT1; near complete sequestration of STAT3 into STAT1:STAT3 heterodimers in primed cells correlates with diminished STAT3 function. Under these conditions of dimerization with excess STAT1, STAT3 can be retained in the cytoplasm, with diminished target gene expression secondary to decreased nuclear translocation (Hu et al., 2005b). In addition to suppressing STAT3 homodimer formation, incorporation of STAT3 into STAT1:STAT3 heterodimers can result in diminished formation of other active STAT3-containing complexes, such as STAT3-Jun complexes important for activation of specific target genes (Ivanov et al., 2001). Interestingly, this sequestration model by which STAT1 inhibits transcription factors extends to inhibition of RUNX2 and NF-κB by STAT1 binding and subsequent trapping of these transcription factors in the cytoplasm (Kim et al., 2003; Kramer et al., 2006). Finally, it is possible that STAT1 can bind to STAT3 target genes and directly suppress transcription by recruiting transcriptional repressors (Figure 5D). An interesting area for future investigation will be to determine whether STAT1 can indeed directly repress gene transcription, in contrast to the indirect mechanisms that have been described previously and reviewed here. It will also be important to determine mechanisms by which IFN-γ and STAT1 inhibit STAT3-mediated IL-6, IL-21 and IL-23 function during Th17 differentiation (Figure 4C).
Role in autoimmune diseases
Autoimmune diseases are characterized by the development of autoimmunity against self antigens, together with an effector phase characterized by chronic inflammation and attendant tissue damage. Many autoimmune diseases, such as rheumatoid arthritis, multiple sclerosis, inflammatory bowel disease, psoriasis and lupus nephritis are characterized by the presence of activated macrophages at sites of inflammation and disease. These macrophages exhibit an “M1” classically activated phenotype and are believed to be key players in pathogenesis via production of cytokines such as TNF, IL-1 and IL-6 (Mosser and Edwards, 2008). Thus, based on its macrophage-activating properties, IFN-γ has been considered an attractive candidate pathogenic cytokine in autoimmune diseases. Several mouse models of autoimmune diseases, such as collagen-induced arthritis (CIA) and EAE, were originally thought to be predominantly Th1-mediated, further supporting the notion that IFN-γ is pathogenic. However, consistent with the pleiotropic activating and suppressive functions of IFN-γ described above, it is now clear that IFN-γ has both promoting and suppressive effects in autoimmune diseases. Most strikingly, IFN-γ suppresses Th17-mediated autoimmunity in mice, and can have both augmenting and suppressive effects on autoimmunity and on the effector inflammatory phase of autoimmune diseases (Kelchtermans et al., 2008), depending on the specific disease and the timing, location and intensity of IFN-γ action.
It is now clear based on genetic evidence that EAE and CIA are Th17-mediated disease models. In EAE, genetic ablation of IFN-γ or the IFNGR results in increased morbidity and mortality (Ferber et al., 1996; Willenborg et al., 1996). Exacerbated disease in the absence of IFN-γ signaling is associated with massive central nervous system infiltrates composed of neutrophils and macrophages (Ferber et al., 1996; Willenborg et al., 1996). In CIA, deficiency of IFNGR leads to accelerated onset and increased incidence of disease (Manoury-Schwartz et al., 1997; Vermeire et al., 1997). Joint lesions of IFNGR knockout mice in CIA are characterized by increased infiltration of neutrophils and macrophages, with increased tissue destruction and bone erosion (Manoury-Schwartz et al., 1997; Vermeire et al., 1997). In both EAE and CIA, the protective role of IFN-γ has been attributed to its suppression of Th17 responses, and this notion is supported by evidence that IL-17 antibodies attenuate arthritis in IFN-γ-deficient animals in two different models (Chu et al., 2007; Kelchtermans et al., 2009). However, as discussed above, attenuation of disease by IFN-γ is also likely mediated by additional protective mechanisms such as suppression of production of chemokines, cytokines, and tissue-destructive enzymes (Guedez et al., 2001; Kelchtermans et al., 2007; Willenborg et al., 1999), infiltration of inflammatory cells, and differentiation of osteoclasts; modulation of Treg function may also be important and IFN-γ-induced Treg subsets may specifically attenuate Th1-mediated pathology while allowing Th17-mediated pathology to progress.
Although IFN-γ is clearly protective in EAE and CIA, it is overly simplistic to conclude that IFN-γ plays a protective role in multiple sclerosis and rheumatoid arthritis based upon its role in these two acute neutrophil-dominated models of chronic human autoimmune diseases that exhibit a more complex and often different pathology. Indeed, administration of IFN-γ induces exacerbations of MS in humans (Panitch et al., 1987), and IFN-γ is pathogenic in other models of RA, such as proteoglycan-induced arthritis, and in CIA when complete Freund's adjuvant (CFA) is not used during disease induction (Finnegan et al., 2002; Matthys et al., 1999). Even in CIA induced using standard CFA-utilizing protocols, exogenous IFN-γ can exacerbate disease depending on whether it is provided locally or systemically, and on timing of administration (Boissier et al., 1995). More recent work (Kroenke et al., 2008; Luger et al., 2008; Steinman, 2008) indicates that both Th1 and Th17 cells can contribute to pathogenesis of EAE and experimental allergic uveitis (EAU). The predominant pathogenic Th cell type is determined by the methods used to induce disease, especially by the use of adjuvants such as CFA that contain various TLR ligands. Th17-mediated disease was characterized by neutrophil-rich infiltrates, whereas Th1 disease had predominant macrophage infiltrates, which is more characteristic of MS, RA and many human autoimmune diseases. Thus, a more balanced role for Th1 cells and IFN-γ in autoimmune diseases is emerging, with a mixed picture where Th1 and Th17 cells can coexist and contribute to pathology. This mixed picture is consistent with lineage plasticity and co-expression of IFN-γ and IL-17 by certain Th cells as discussed above, and is supported by data showing co-expression of IFN-γ and IL-17 in various models and diseases, including RA, systemic lupus erythematosus (SLE), EAE, Crohn's disease and psoriasis. One recent study shows that IFN-γ actually contributes to induction of Th17 cell migration and differentiation in the context of psoriasis, suggesting that IFN-γ may play a positive role in Th17 responses (Kryczek et al., 2008). Overall, a large body of work highlights the complex interplay between Th1 cells/IFN-γ and Th17 cells in vivo and suggests that IFN-γ could differentially regulate Th17 responses under different disease conditions.
A pathogenic role of Th1 cells and IFN-γ in autoimmune diseases raises the question of mechanisms by which IFN-γ contributes to pathogenesis. Given the above discussion, a good candidate mechanism is IFN-γ-mediated activation of macrophages and other cell types at sites of inflammation, and thus augmentation of the effector inflammatory component of autoimmune diseases. In this scenario, the activating and priming functions of IFN-γ that lead to increased inflammatory cytokine production and abrogate homeostatic mechanisms contribute to disease pathology. Indeed, we and others have provided evidence supporting IFN-γ-mediated priming of macrophages in human RA and mouse models of lupus nephritis (Hu et al., 2002; Wang et al., 2008). In support of a role for IFN-γ in augmenting inflammation in autoimmune diseases, local administration or tissue-specific transgene-mediated expression of IFN-γ at inflammatory sites exacerbates disease in arthritis and autoimmune diabetes models. Additional support for a role for IFN-γ in the effector phase of autoimmune disease is provided by genetic evidence showing that deletion of the Ifng gene ameliorates nephritis in the MRL/lpr model of SLE where nephritis is dependent on pathogenic macrophages (Baccala et al., 2005). Importantly, autoimmunity did not appear to be diminished in IFN-γ-deficient animals, supporting the idea that IFN-γ can boost inflammation and tissue destruction in the kidney independently of the autoimmune process. However, there is also evidence that IFN-γ can suppress the inflammatory effector phase of autoimmunity. The clearest example may be the increased severity of arthritis in IFN-γ-deficient mice in the K/BxN model that is induced by passive transfer of auto-antibodies and does not depend on acquired immunity (Wu et al., 2007). Conversely, systemic administration of exogenous IFN-γ suppressed K/BxN arthritis. The mechanism by which IFN-γ suppresses K/BxN arthritis is inhibition of neutrophil infiltration of joints, although it is possible that direct attenuation of tissue destruction and osteoclastogenesis could also play a role.
The complex role of IFN-γ in autoimmune diseases has important therapeutic implications. A detailed understanding of key pathogenic processes will be required to determine whether blocking endogenous IFN-γ or administering exogenous IFN-γ may be efficacious, and at which point in the disease process. It will be equally important to understand the interplay between Th1 and Th17 responses in specific autoimmune diseases. Blockade of solely IFN-γ or Th17 cytokines may result only in partial therapeutic efficacy and a shift to a different pathology. In diseases where both Th1 and Th17 cells work together, blocking both may be required for effective therapy. Indeed, the striking beneficial effects antibodies against IL-12 p40 in diseases such as Crohn's disease and psoriasis may be explained by attenuation of both Th1 and Th17 responses (Ghosh et al., 2006; Nestle et al., 2009). It will be interesting to see the effects of IL-12 p40 blockade in autoimmune diseases such as MS and RA.
Acknowledgments
X.H. is supported by Within Our Reach rheumatoid arthritis research grant from the American College of Rheumatology and L.B.I. is supported by grants from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliprantis AO, Wang J, Fathman JW, Lemaire R, Dorfman DM, Lafyatis R, Glimcher LH. Transcription factor T-bet regulates skin sclerosis through its function in innate immunity and via IL-13. Proc Natl Acad Sci U S A. 2007;104:2827–2830. doi: 10.1073/pnas.0700021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala R, Kono DH, Theofilopoulos AN. Interferons as pathogenic effectors in autoimmunity. Immunol Rev. 2005;204:9–26. doi: 10.1111/j.0105-2896.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- Baccarini M, Dello Sbarba P, Buscher D, Bartocci A, Stanley ER. IFN-gamma/lipopolysaccharide activation of macrophages is associated with protein kinase C-dependent down-modulation of the colony-stimulating factor-1 receptor. J Immunol. 1992;149:2656–2661. [PubMed] [Google Scholar]
- Barnes MJ, Powrie F. Hybrid Treg cells: steel frames and plastic exteriors. Nat Immunol. 2009;10:563–564. doi: 10.1038/ni0609-563. [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Chadee K. Novel regulation of cyclooxygenase-2 expression and prostaglandin E2 production by IFN-gamma in human macrophages. J Immunol. 1998;161:2441–2448. [PubMed] [Google Scholar]
- Boissier MC, Chiocchia G, Bessis N, Hajnal J, Garotta G, Nicoletti F, Fournier C. Biphasic effect of interferon-gamma in murine collagen-induced arthritis. Eur J Immunol. 1995;25:1184–1190. doi: 10.1002/eji.1830250508. [DOI] [PubMed] [Google Scholar]
- Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- Browning JL, Ribolini A. Interferon blocks interleukin 1-induced prostaglandin release from human peripheral monocytes. J Immunol. 1987;138:2857–2863. [PubMed] [Google Scholar]
- Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marie I, Levy DE. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CQ, Swart D, Alcorn D, Tocker J, Elkon KB. Interferon-gamma regulates susceptibility to collagen-induced arthritis through suppression of interleukin-17. Arthritis Rheum. 2007;56:1145–1151. doi: 10.1002/art.22453. [DOI] [PubMed] [Google Scholar]
- Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is'harc H, Gesualdo I, Newman SJ, Kerr IM, Poli V. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci U S A. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz A, Khader SA, Torrado E, Fraga A, Pearl JE, Pedrosa J, Cooper AM, Castro AG. Cutting edge: IFN-gamma regulates the induction and expansion of IL-17-producing CD4 T cells during mycobacterial infection. J Immunol. 2006;177:1416–1420. doi: 10.4049/jimmunol.177.3.1416. [DOI] [PubMed] [Google Scholar]
- Dickensheets HL, Donnelly RP. IFN-gamma and IL-10 inhibit induction of IL-1 receptor type I and type II gene expression by IL-4 and IL-13 in human monocytes. J Immunol. 1997;159:6226–6233. [PubMed] [Google Scholar]
- Dickensheets HL, Venkataraman C, Schindler U, Donnelly RP. Interferons inhibit activation of STAT6 by interleukin 4 in human monocytes by inducing SOCS-1 gene expression. Proc Natl Acad Sci U S A. 1999;96:10800–10805. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa L, Ingles-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, Schreiber RD, Sheehan K, Hilton DJ, Alexander WS, Hertzog PJ. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- Ferber IA, Brocke S, Taylor-Edwards C, Ridgway W, Dinisco C, Steinman L, Dalton D, Fathman CG. Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE) J Immunol. 1996;156:5–7. [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan A, Grusby MJ, Kaplan CD, O'Neill SK, Eibel H, Koreny T, Czipri M, Mikecz K, Zhang J. IL-4 and IL-12 regulate proteoglycan-induced arthritis through Stat-dependent mechanisms. J Immunol. 2002;169:3345–3352. doi: 10.4049/jimmunol.169.6.3345. [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009;130:7–15. doi: 10.1016/j.clim.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Yuan W, Mori Y, Chen S, Varga J. Antagonistic regulation of type I collagen gene expression by interferon-gamma and transforming growth factor-beta. Integration at the level of p300/CBP transcriptional coactivators. J Biol Chem. 2001;276:11041–11048. doi: 10.1074/jbc.M004709200. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Chaudhary R, Carpani M, Playford R. Interfering with interferons in inflammatory bowel disease. Gut. 2006;55:1071–1073. doi: 10.1136/gut.2005.090134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil MP, Bohn E, O'Guin AK, Ramana CV, Levine B, Stark GR, Virgin HW, Schreiber RD. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci U S A. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedez YB, Whittington KB, Clayton JL, Joosten LA, van de Loo FA, van den Berg WB, Rosloniec EF. Genetic ablation of interferon-gamma up-regulates interleukin-1beta expression and enables the elicitation of collagen-induced arthritis in a nonsusceptible mouse strain. Arthritis Rheum. 2001;44:2413–2424. doi: 10.1002/1529-0131(200110)44:10<2413::aid-art406>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- Herrero C, Hu X, Li WP, Samuels S, Sharif MN, Kotenko S, Ivashkiv LB. Reprogramming of IL-10 activity and signaling by IFN-gamma. J Immunol. 2003;171:5034–5041. doi: 10.4049/jimmunol.171.10.5034. [DOI] [PubMed] [Google Scholar]
- Ho HH, Antoniv TT, Ji JD, Ivashkiv LB. Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-gamma via superinduction of ATF-3 and suppression of AP-1. J Immunol. 2008;181:5089–5097. doi: 10.4049/jimmunol.181.7.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Chen J, Wang L, Ivashkiv LB. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J Leukoc Biol. 2007;82:237–243. doi: 10.1189/jlb.1206763. [DOI] [PubMed] [Google Scholar]
- Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, Tateya T, Kang YJ, Han J, Gessler M, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008a;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Herrero C, Li WP, Antoniv TT, Falck-Pedersen E, Koch AE, Woods JM, Haines GK, Ivashkiv LB. Sensitization of IFN-gamma Jak-STAT signaling during macrophage activation. Nat Immunol. 2002;3:859–866. doi: 10.1038/ni828. [DOI] [PubMed] [Google Scholar]
- Hu X, Ho HH, Lou O, Hidaka C, Ivashkiv LB. Homeostatic role of interferons conferred by inhibition of IL-1-mediated inflammation and tissue destruction. J Immunol. 2005a;175:131–138. doi: 10.4049/jimmunol.175.1.131. [DOI] [PubMed] [Google Scholar]
- Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, Ivashkiv LB. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24:563–574. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Hu X, Park-Min KH, Ho HH, Ivashkiv LB. IFN-gamma-primed macrophages exhibit increased CCR2-dependent migration and altered IFN-gamma responses mediated by Stat1. J Immunol. 2005b;175:3637–3647. doi: 10.4049/jimmunol.175.6.3637. [DOI] [PubMed] [Google Scholar]
- Hu Y, Hu X, Boumsell L, Ivashkiv LB. IFN-gamma and STAT1 arrest monocyte migration and modulate RAC/CDC42 pathways. J Immunol. 2008b;180:8057–8065. doi: 10.4049/jimmunol.180.12.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Inaba T, Gotoda T, Harada K, Shimada M, Ohsuga J, Ishibashi S, Yazaki Y, Yamada N. Induction of sustained expression of proto-oncogene c-fms by platelet-derived growth factor, epidermal growth factor, and basic fibroblast growth factor, and its suppression by interferon-gamma and macrophage colony-stimulating factor in human aortic medial smooth muscle cells. J Clin Invest. 1995;95:1133–1139. doi: 10.1172/JCI117761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler IM, Gajda M, Brauer R. Exacerbation of antigen-induced arthritis in IFN-gamma-deficient mice as a result of unrestricted IL-17 response. J Immunol. 2007;179:6228–6236. doi: 10.4049/jimmunol.179.9.6228. [DOI] [PubMed] [Google Scholar]
- Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, Horvath CM, Ronai Z. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7:517–528. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- Kelchtermans H, Billiau A, Matthys P. How interferon-gamma keeps autoimmune diseases in check. Trends Immunol. 2008;29:479–486. doi: 10.1016/j.it.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Kelchtermans H, De Klerck B, Mitera T, Van Balen M, Bullens D, Billiau A, Leclercq G, Matthys P. Defective CD4+CD25+ regulatory T cell functioning in collagen-induced arthritis: an important factor in pathogenesis, counter-regulated by endogenous IFN-gamma. Arthritis Res Ther. 2005;7:R402–415. doi: 10.1186/ar1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelchtermans H, Schurgers E, Geboes L, Mitera T, Van Damme J, van Snick J, Uyttenhove C, Matthys P. Effector mechanisms of interleukin-17 in collagen-induced arthritis in the absence of interferon-gamma and counteraction by interferon-gamma. Arthritis Res Ther. 2009;11:R122. doi: 10.1186/ar2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelchtermans H, Struyf S, De Klerck B, Mitera T, Alen M, Geboes L, Van Balen M, Dillen C, Put W, Gysemans C, et al. Protective role of IFN-gamma in collagen-induced arthritis conferred by inhibition of mycobacteria-induced granulocyte chemotactic protein-2 production. J Leukoc Biol. 2007;81:1044–1053. doi: 10.1189/jlb.0806486. [DOI] [PubMed] [Google Scholar]
- Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockeritz L, Doble B, Patel S, Woodgett JR. Glycogen synthase kinase-3--an overview of an over-achieving protein kinase. Curr Drug Targets. 2006;7:1377–1388. doi: 10.2174/1389450110607011377. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, Grez M, Pfitzner E, Heinzel T. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, Stauber RH, Bohmer FD, Heinzel T. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke MA, Carlson TJ, Andjelkovic AV, Segal BM. IL-12- and IL-23-modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. J Exp Med. 2008;205:1535–1541. doi: 10.1084/jem.20080159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Bruce AT, Gudjonsson JE, Johnston A, Aphale A, Vatan L, Szeliga W, Wang Y, Liu Y, Welling TH, et al. Induction of IL-17+ T cell trafficking and development by IFN-gamma: mechanism and pathological relevance in psoriasis. J Immunol. 2008;181:4733–4741. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauw FN, Pajkrt D, Hack CE, Kurimoto M, van Deventer SJ, van der Poll T. Proinflammatory effects of IL-10 during human endotoxemia. J Immunol. 2000;165:2783–2789. doi: 10.4049/jimmunol.165.5.2783. [DOI] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Chen XP, Hilton D, Rothman P. Cutting edge: SOCS-1 is a potent inhibitor of IL-4 signal transduction. J Immunol. 1999;162:3770–3774. [PMC free article] [PubMed] [Google Scholar]
- Luger D, Silver PB, Tang J, Cua D, Chen Z, Iwakura Y, Bowman EP, Sgambellone NM, Chan CC, Caspi RR. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J Exp Med. 2008;205:799–810. doi: 10.1084/jem.20071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Chang MJ, Shah RC, Benveniste EN. Interferon-gamma-activated STAT-1alpha suppresses MMP-9 gene transcription by sequestration of the coactivators CBP/p300. J Leukoc Biol. 2005;78:515–523. doi: 10.1189/jlb.0205112. [DOI] [PubMed] [Google Scholar]
- Ma Z, Qin H, Benveniste EN. Transcriptional suppression of matrix metalloproteinase-9 gene expression by IFN-gamma and IFN-beta: critical role of STAT-1alpha. J Immunol. 2001;167:5150–5159. doi: 10.4049/jimmunol.167.9.5150. [DOI] [PubMed] [Google Scholar]
- Manoury-Schwartz B, Chiocchia G, Bessis N, Abehsira-Amar O, Batteux F, Muller S, Huang S, Boissier MC, Fournier C. High susceptibility to collagen-induced arthritis in mice lacking IFN-gamma receptors. J Immunol. 1997;158:5501–5506. [PubMed] [Google Scholar]
- Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr, Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Matthys P, Vermeire K, Mitera T, Heremans H, Huang S, Schols D, De Wolf-Peeters C, Billiau A. Enhanced autoimmune arthritis in IFN-gamma receptor-deficient mice is conditioned by mycobacteria in Freund's adjuvant and by increased expansion of Mac-1+ myeloid cells. J Immunol. 1999;163:3503–3510. [PubMed] [Google Scholar]
- McGeachy MJ, Cua DJ. Th17 cell differentiation: the long and winding road. Immunity. 2008;28:445–453. doi: 10.1016/j.immuni.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE., Jr Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20:3372–3381. doi: 10.1101/gad.1485406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Young RA, Daley GQ. Hematopoietic remodeling in interferon-gamma-deficient mice infected with mycobacteria. Blood. 1998;91:2914–2924. [PubMed] [Google Scholar]
- Naka T, Tsutsui H, Fujimoto M, Kawazoe Y, Kohzaki H, Morita Y, Nakagawa R, Narazaki M, Adachi K, Yoshimoto T, et al. SOCS-1/SSI-1-deficient NKT cells participate in severe hepatitis through dysregulated cross-talk inhibition of IFN-gamma and IL-4 signaling in vivo. Immunity. 2001;14:535–545. doi: 10.1016/s1074-7613(01)00132-7. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibori T, Tanabe Y, Su L, David M. Impaired development of CD4+ CD25+ regulatory T cells in the absence of STAT1: increased susceptibility to autoimmune disease. J Exp Med. 2004;199:25–34. doi: 10.1084/jem.20020509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci U S A. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208–215. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- Pajkrt D, Camoglio L, Tiel-van Buul MC, de Bruin K, Cutler DL, Affrime MB, Rikken G, van der Poll T, ten Cate JW, van Deventer SJ. Attenuation of proinflammatory response by recombinant human IL-10 in human endotoxemia: effect of timing of recombinant human IL-10 administration. J Immunol. 1997;158:3971–3977. [PubMed] [Google Scholar]
- Panitch HS, Hirsch RL, Haley AS, Johnson KP. Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet. 1987;1:893–895. doi: 10.1016/s0140-6736(87)92863-7. [DOI] [PubMed] [Google Scholar]
- Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- Radzioch D, Varesio L. c-fos mRNA expression in macrophages is downregulated by interferon-gamma at the posttranscriptional level. Mol Cell Biol. 1991;11:2718–2722. doi: 10.1128/mcb.11.5.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Stat1-independent regulation of gene expression in response to IFN- gamma. Proc Natl Acad Sci U S A. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanceau J, Boyd DD, Seiki M, Bauvois B. Interferons inhibit tumor necrosis factor-alpha-mediated matrix metalloproteinase-9 activation via interferon regulatory factor-1 binding competition with NF-kappa B. J Biol Chem. 2002;277:35766–35775. doi: 10.1074/jbc.M202959200. [DOI] [PubMed] [Google Scholar]
- Sato K, Suematsu A, Nakashima T, Takemoto-Kimura S, Aoki K, Morishita Y, Asahara H, Ohya K, Yamaguchi A, Takai T, et al. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med. 2006;12:1410–1416. doi: 10.1038/nm1515. [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Schroder K, Sweet MJ, Hume DA. Signal integration between IFNgamma and TLR signalling pathways in macrophages. Immunobiology. 2006;211:511–524. doi: 10.1016/j.imbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- So EY, Park HH, Lee CE. IFN-gamma and IFN-alpha posttranscriptionally down-regulate the IL-4-induced IL-4 receptor gene expression. J Immunol. 2000;165:5472–5479. doi: 10.4049/jimmunol.165.10.5472. [DOI] [PubMed] [Google Scholar]
- Stark GR. How cells respond to interferons revisited: from early history to current complexity. Cytokine Growth Factor Rev. 2007;18:419–423. doi: 10.1016/j.cytogfr.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman L. A rush to judgment on Th17. J Exp Med. 2008;205:1517–1522. doi: 10.1084/jem.20072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM, Villarino AV, Huang Q, Yoshimura A, Sehy D, et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat Immunol. 2006;7:937–945. doi: 10.1038/ni1376. [DOI] [PubMed] [Google Scholar]
- Sun Y, Chin YE, Weisiger E, Malter C, Tawara I, Toubai T, Gatza E, Mascagni P, Dinarello CA, Reddy P. Cutting edge: Negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J Immunol. 2009;182:5899–5903. doi: 10.4049/jimmunol.0804388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Mundy GR, Roodman GD. Recombinant human interferon-gamma inhibits formation of human osteoclast-like cells. J Immunol. 1986;137:3544–3549. [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Ogasawara K, Hida S, Chiba T, Murata S, Sato K, Takaoka A, Yokochi T, Oda H, Tanaka K, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Sato K, Takaoka A, Taniguchi T. Interplay between interferon and other cytokine systems in bone metabolism. Immunol Rev. 2005;208:181–193. doi: 10.1111/j.0105-2896.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Ichiyama K, Hashimoto M, Yoshida H, Takimoto T, Takaesu G, Torisu T, Hanada T, Yasukawa H, Fukuyama S, et al. Loss of suppressor of cytokine signaling 1 in helper T cells leads to defective Th17 differentiation by enhancing antagonistic effects of IFN-gamma on STAT3 and Smads. J Immunol. 2008;180:3746–3756. doi: 10.4049/jimmunol.180.6.3746. [DOI] [PubMed] [Google Scholar]
- Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kai JI, Huang WC, Wang CY, Wang Y, Chen CL, Fang YT, Lin YS, Anderson R, Chen SH, et al. Glycogen synthase kinase-3beta facilitates IFN-gamma-induced STAT1 activation by regulating Src homology-2 domain-containing phosphatase 2. J Immunol. 2009;183:856–864. doi: 10.4049/jimmunol.0804033. [DOI] [PubMed] [Google Scholar]
- Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- Vermeire K, Heremans H, Vandeputte M, Huang S, Billiau A, Matthys P. Accelerated collagen-induced arthritis in IFN-gamma receptor-deficient mice. J Immunol. 1997;158:5507–5513. [PubMed] [Google Scholar]
- Wang L, Tassiulas I, Park-Min KH, Reid AC, Gil-Henn H, Schlessinger J, Baron R, Zhang JJ, Ivashkiv LB. ‘Tuning’ of type I interferon-induced Jak-STAT1 signaling by calcium-dependent kinases in macrophages. Nat Immunol. 2008;9:186–193. doi: 10.1038/ni1548. [DOI] [PubMed] [Google Scholar]
- Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hong J, Sun W, Xu G, Li N, Chen X, Liu A, Xu L, Sun B, Zhang JZ. Role of IFN-gamma in induction of Foxp3 and conversion of CD4+ CD25- T cells to CD4+ Tregs. J Clin Invest. 2006;116:2434–2441. doi: 10.1172/JCI25826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Wenta N, Strauss H, Meyer S, Vinkemeier U. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc Natl Acad Sci U S A. 2008;105:9238–9243. doi: 10.1073/pnas.0802130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenborg DO, Fordham S, Bernard CC, Cowden WB, Ramshaw IA. IFN-gamma plays a critical down-regulatory role in the induction and effector phase of myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. J Immunol. 1996;157:3223–3227. [PubMed] [Google Scholar]
- Willenborg DO, Fordham SA, Staykova MA, Ramshaw IA, Cowden WB. IFN-gamma is critical to the control of murine autoimmune encephalomyelitis and regulates both in the periphery and in the target tissue: a possible role for nitric oxide. J Immunol. 1999;163:5278–5286. [PubMed] [Google Scholar]
- Wu HJ, Sawaya H, Binstadt B, Brickelmaier M, Blasius A, Gorelik L, Mahmood U, Weissleder R, Carulli J, Benoist C, Mathis D. Inflammatory arthritis can be reined in by CpG-induced DC-NK cell cross talk. J Exp Med. 2007;204:1911–1922. doi: 10.1084/jem.20070285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- Yu CR, Mahdi RM, Ebong S, Vistica BP, Chen J, Guo Y, Gery I, Egwuagu CE. Cell proliferation and STAT6 pathways are negatively regulated in T cells by STAT1 and suppressors of cytokine signaling. J Immunol. 2004;173:737–746. doi: 10.4049/jimmunol.173.2.737. [DOI] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Zhou M, Zhang Y, Ardans JA, Wahl LM. Interferon-gamma differentially regulates monocyte matrix metalloproteinase-1 and -9 through tumor necrosis factor-alpha and caspase 8. J Biol Chem. 2003;278:45406–45413. doi: 10.1074/jbc.M309075200. [DOI] [PubMed] [Google Scholar]
- Zimmerer JM, Lesinski GB, Kondadasula SV, Karpa VI, Lehman A, Raychaudhury A, Becknell B, Carson WE., 3rd IFN-alpha-induced signal transduction, gene expression, and antitumor activity of immune effector cells are negatively regulated by suppressor of cytokine signaling proteins. J Immunol. 2007;178:4832–4845. doi: 10.4049/jimmunol.178.8.4832. [DOI] [PubMed] [Google Scholar]