Summary
In addition to hemostasis, platelets mediate inflammation and clearance of bacteria from the bloodstream. As with platelet-platelet interactions, platelet-bacteria interactions involve cytoskeletal rearrangements and release of granular content. Stimulation of the immune Toll-like receptor 2 (TLR2) on the platelet surface, activates phosphoinositide-3-kinase (PI3K) and causes platelet activation and platelet-dependent thrombosis. It remains unknown if platelet activation by immune vs. thrombotic pathways leads to the differential regulation of signal transduction, protein-protein interactions, and α-granule release, and the physiological relevance of these potential differences. We investigated these processes after immune vs. thrombotic platelet stimulation. We examined selected signaling pathways and found that phosphorylation kinetics of Akt, ERK1/2 and p38 differed dramatically between agonists. Next, we investigated platelet protein-protein interactions by mass spectrometry (MS)-based proteomics specifically targeting cytosolic factor XIIIa (FXIIIa) because of its function as a cytoskeleton-crosslinking protein whose binding partners have limited characterization. Four FXIIIa-binding proteins were identified, two of which are novel interactions: FXIIIa-focal adhesion kinase (FAK) and FXIIIagelsolin. The binding of FAK to FXIIIa was found to be altered differentially by immune vs. thrombotic stimulation. Lastly, we studied the effect of thrombin- vs. Pam3CSK4-stimulation on α-granule release and observed differential release patterns for selected granule proteins and decreased fibrin clot formation compared with thrombin. The inhibition of PI3K caused a decrease in protein release after Pam3CSK4- but not after thrombin-stimulation. In summary, stimulation of plates by either thrombotic or immune receptors leads to markedly different signaling responses and granular protein release consistent with differential contribution to coagulation and thrombosis.
Keywords: Toll-like receptor 2, thrombin, signal transduction, mass spectrometry-based proteomics, factor XIIIA
Introduction
Cells of the innate immune system distinguish between pathogen and self by utilizing signals from Toll-like receptors (TLRs). Stimulation of TLRs in vascular cells and monocytes by pathogen-associated molecules results in activation of nuclear factor κB (NF-κB) and production of proinflammatory cytokines (1, 2). TLRs are also expressed on the platelet surface (3, 4). Although platelets are present at sites of inflammation, promote bacterial clearance (5), and cross-talk with other immune cells, their specific role in innate immunity is not well understood.
TLR2, in particular, recognizes bacterial lipopeptides, lipoproteins, and peptidoglycans from gram-positive bacteria (6). The lipopeptide Pam3CSK4, a TLR2/1 agonist, has been shown to induce physiological platelet activation and serotonin release (7). Recent data from our laboratory demonstrate that Pam3CSK4-stimulation of TLR2 leads to P-selectin expression, activation of integrin αIIbβ3, and activation of PI3K/Akt signaling pathway (8).
Thrombin, on the other hand, has been studied extensively in platelets. As one of the major platelet agonists, it activates the protease activated receptor 1 and 4 (PAR1/4) on the platelet surface which leads to activation of PI3K and phospholipase Cβ (PLCβ) pathways. The activation of these pathways leads to an increase in intracellular calcium concentrations, platelet shape change, granule secretion, and platelet aggregation (9). However, many details of these signaling cascades, and the mechanisms of their downstream events remain to be elucidated.
The thrombotic vs. immune stimulation of platelets and their subsequent differential effects on the activation of signaling pathways, on specific protein-protein interactions, and on α-granule release have not been examined. To differentiate these forms of platelet stimulation and define their mechanism of action, we compared platelet stimulation with thrombin vs. Pam3CSK4 and investigated differential changes in (1) the phosphorylation kinetics of Akt, ERK1/2 and p38, (2) FXIIIa-interacting proteins, and (3) the release of several α-granule proteins, as well as the effect of PI3K-inhibition on FXIIIa-interactions and α-granule release.
Materials and methods
Platelet aggregation
Platelets were isolated from healthy volunteers and platelet aggregation was monitored using a PAP-4 platelet aggregometer (Bio/Data Corporation, Horsham, PA) as described previously (10). Briefly, human α-thrombin (0.5 U/mL; Enzyme Research Laboratories, South Bend, IN), Pam3CSK4 (10 μg/mL; Invivogen, San Diego, CA), or ADP (10 μM; Chrono-log Corporation, Havertown, PA) were added, while stirring, to 2×108/mL platelets supplemented with 1 mg/ml fibrinogen, 1 mM CaCl2, and 2 mM MgCl2 for 12 min, 37°C. The peptide Gly-Pro-Arg-Pro (GPRP; Sigma-Aldrich, St. Louis, MO) was added to thrombin-activated samples to inhibit fibrinogen polymerization (11).
Platelet adhesion
Washed platelets were incubated with 10 μM calcein-AM (Invitrogen, Carlsbad, CA) (30 min), then resuspended in HBSS. Subsequently, platelets were stimulated with 0.5 U/mL thrombin, 10 μg/mL Pam3CSK4, 10 μM ADP, or left untreated (10 min). Using a vacuum-based cell adhesion flow chamber (Immunetics, Inc., Boston, MA), platelets were passed at a constant rate over collagen-coated cover slips for 20 min. Cover slips were mounted on glass slides for analysis with a fluorescence microscope (Nikon, Melville, NY; 20X magnification, 505 nm emission).
Activation of platelets and preparation of lysates, releasates
Platelets (2x108/mL) in Tyrodes-buffer were treated with 0.5 U/mL thrombin, 10 μg/mL Pam3CSK4, or 10 μM ADP for 15 min. To resting platelets, prostaglandin E1 (PGE1) (1 μM; Cayman Chemical, Ann Arbor, MI) and apyrase (0.2 U/mL; Sigma-Aldrich) were added. Activation was stopped by adding PGE1/apyrase and pelleting the platelets. Samples were washed with Tyrodes-buffer containing PGE1/ apyrase, and lysed in equal volumes of Tyrodes-buffer and 2x lysis-buffer (2% NP-40, 30 mM Hepes, 150 mM NaCl, 2 mM EDTA, pH 7.4). To remove platelet-derived microvesicles, releasates were centrifuged at 150,000 × g, 90 min, 4°C (12), and tested for the absence of microvesicles by immunoblotting for integrin αIIb (13). Protein concentrations were determined using the DC-protein assay (BioRad, Hercules, CA).
Phosphorylation kinetics and PI3K-inhibition
For kinetics studies, resting platelets and platelets activated for indicated times with thrombin (0.5 U/mL) or Pam3CSK4 (10 μg/mL), were pelleted, washed and lysed. For PI3K-inhibition, platelets were incubated with 25 μM or 50 μM LY294002 (Invivogen) (30 min) prior to platelet activation for 15 min. Lysate and releasate samples were obtained as described above.
Immunoprecipitation of FXIIIA and thrombospondin
Sheep polyclonal anti-FXIIIa (Cedarlane Laboratories, Hornby, ON, Canada) and IgG control antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), or mouse monoclonal anti-thrombospondin and IgG1 control antibodies (LabVision Corporation, Fremont, CA), respectively, were used. After preclearing with protein G+ beads (Santa Cruz Biotechnology), lysates were incubated with either the target-protein-specific or control antibodies. Subsequently, antibodies were precipitated with protein G+ beads and washed with lysis buffer. For 2D gel electrophoresis (2D-GE), precipitates were washed with lysis buffer, followed by 1 mM Hepes/NaCl (pH 7.6), and 1 mM Hepes (pH 7.6) to remove residual salts/detergents.
1D/2D-GE
For 1D-GE, samples were loaded onto 10% Criterion-XT Bis-Tris gels (BioRad). After electrophoresis, gels were either stained with Coomassie Blue Imperial Stain (Pierce, Rockford, IL) or prepared for immunoblotting. For 2D-GE of releasates, samples were prepared using a 2D clean-up kit (GE Healthcare, Piscataway, NJ). Samples (75-125 μg protein) were solubilized in 2D Rehydration/Sample Buffer (BioRad) containing 50 mM dithiothreitol (DTT). For 2D-GE of immunoprecipitates, bead pellets were directly solubilized in the same manner as releasate samples and pelleted. Bio-Lyte 3/10 ampholytes (BioRad) were added to all samples. Immobilized pH gradient (IPG) strips (11 cm; pH 4-7 (for releasates), pH 3-10 (for immunoprecipitates; BioRad) were actively rehydrated with samples overnight in a PROTEAN™ IEF cell (BioRad) followed by isoelectric focusing in the first dimension. Afterwards, proteins in the IPG strips were reduced with DTT and alkylated with iodoacetamide using Equilibrium Buffer I/II (BioRad). IPG strips were placed onto 10% Criterion-XT gels and subjected to separation in the second dimension. Gels were either stained with Coomassie Blue or silver stain (GE Healthcare), or prepared for immunoblotting.
Immunoblotting
Proteins transferred onto PVDF membranes (Millipore, Billerica, MA) were probed for the following: anti-FXIIIa, anti-thrombospondin (LabVision); anti-HSP27, anti-fibrinogen β, anti-gelsolin, anti-FAK, anti-platelet basic protein, anti-platelet factor 4, anti-integrin αIIb (Santa Cruz Biotechnology), anti-myosin IIA (Covance Research Products, Berkeley, CA), anti-phospho-Akt(Ser473), anti-Akt, anti-phospho-ERK1/2(Thr202/Tyr204), anti-ERK1/2, anti-phospho-p38(Thr180/Tyr182); and anti-p38 (Cell Signaling Technology, Danvers, MA). Protein bands were quantified by densitometry (VersaDoc Model 3000 Imaging System; Quantity One software version 4.4.1; BioRad).
In-gel digestion of proteins and proteolytic peptide recovery
Gel bands/spots were excised, destained, washed, and dried. Proteins in 1D gel pieces were reduced with DTT, then alkylated with iodoacetamide. Proteins from 2D gels were reduced/alkylated prior to loading of the IPG strip onto the 2D gel. In-gel digestion was performed with Trypsin Gold (Promega, Madison, WI), overnight, 37°C in 50 mM NH4HCO3, 5% acetonitrile (ACN). Tryptic peptides were extracted with 1% trifluoroacetic acid in 50% ACN, desalted with ZipTipC18 pipette tips, and eluted into 0.1% TFA, 50% ACN.
MALDI-TOF MS, mass spectra analysis, protein identification
After co-crystallization of purified proteolytic peptides with matrix (2,5-dihydroxybenzoic acid (Bruker Daltonics, Billerica, MA; 2 mg/ml in 50% ACN, 0.1% TFA)) onto an AnchorChip™ target (Bruker Daltonics), mass spectra were acquired on a Reflex IV MALDI-TOF instrument (Bruker Daltonics) operated in positive-ion reflectron mode, calibrated with Bruker peptide standards. MALDI-TOF mass spectra were analyzed with MoverZ™ software (Proteometrics, LLC, New York, NY). The resulting peak lists were submitted for peptide mass fingerprint (PMF) analysis to Mascot™ (Matrix Science Inc., Boston, MA) against the SwissProt or NCBI non-redundant protein databases, using the following restrictions: human taxonomy; up to 2 missed cleavages allowed with trypsin as the digestion enzyme; cysteine carbamidomethylation as fixed and methionine oxidation as variable modifications; peptide mass error tolerance of ± 0.1-0.3 Da. Under these conditions, a probability-based Mascot™ Mowse score greater than 60 indicated a p-value less than 0.05 and was considered to represent a significant identification.
Confocal microscopy
Activated platelets were placed onto 0.01% poly-L-lysine-coated slides and fixed in methanol:acetone (70:30). Slides were rehydrated, treated with 0.3% H2O2 in methanol, then permeabilized (0.2% Triton X-100). After blocking in Protein block serum-free solution (DakoCytomation, Carpinteria, CA), slides were incubated with antibodies to FXIIIa and FXIIIa-interacting protein of interest (overnight, 4°C). Primary antibodies were the same as those used for immunoblotting, except anti-thrombospondin (Ab-8, LabVision). Slides were incubated with secondary FITC- and TexasRed-conjugated antibodies. Control slides included labeling with non-specific IgGs. Slides were examined by confocal microscopy (Leica TCS SP2 instrument; Leica Microsystems, Exton, PA) using a compensation system to remove spectral overlap (FITC: 488 nm; Texas Red: 595 nm) and Leica Confocal Software (Leica Microsystems). Images were taken at 100X magnification.
Fibrin clot retraction assay
Isolated platelets were resuspended in Tyrodes-buffer at a final concentration of 3×108/mL, with 1 mM CaCl2. Platelets were either stimulated with 0.5 U/mL thrombin or 10 μg/mL Pam3CSK4 ± 1 mg/mL fibrinogen, for 1h. Clots that formed were removed and the volume of buffer remaining was determined. Data is presented as the percent of buffer remaining.
Heterotypic Aggregation Formation and Quantitation through Flow Cytometry
Whole blood (600 μL) was added to 200 μL of 1X PBS and stirred continuously for 10 minutes at 37°C. Samples were left untreated or treated with either 0.5 U/mL thrombin or 10 μL Pam3CSK4. Samples were then incubated with anti-CD41 FITC conjugated antibodies (platelet marker; eBioscience, San Diego, CA) and anti-CD14 PE-Cy7 conjugated antibodies (monocyte marker; eBioscience). Red blood cells (RBC) were then lysed using 1X BD FACS™ Lysing Solution (BD Biosciences, San Jose, CA). Samples were then processed using FACScan Flow Cytometer with CellQuest Pro Software (BD Biosciences). Monocytes were gated for using a combination of light scattering and CD14 PE-Cy7 fluorescence. A large gate was placed around this population to measure an increase in forward light scattering, indicative of heterotypic aggregate formation. 2000 monocytes were acquired. Further discrimination was done to quantitate the percent of platelet-positive monocytes (CD14-positive and CD41-positive).
Preparation of platelets and white blood cells for SEM
After treatment with either 0.5 U/mL thrombin, 10 μg/mL Pam3CSK4, or resting for 10 min, a mixture of platelets, red blood cells, and white cells (isolated through 1-Step ® Polymorphs Method) was fixed with 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) for 1 h. Fixed cells were washed in buffer solution and post-fixed with 1.0% osmium tetroxide (OsO4) in 0.1 M PBS (pH 7.4) for 2 h. Samples were rinsed with distilled water (3×5min), dehydrated serially in 30%, 50%, 70%, 90%, 100% ethanol, then agitated in acetone. After critical point drying, cells were mounted on specimen stub, coated with gold alloy, and examined with a JEOL (JSM-6100) scanning electron microscope (magnification 15,000X).
Statistical analysis
Data are expressed as mean ± standard error. Differences between two groups were assessed by Student's t-test with p < 0.05 being statistically significant.
Results
Platelet aggregation and adhesion studies
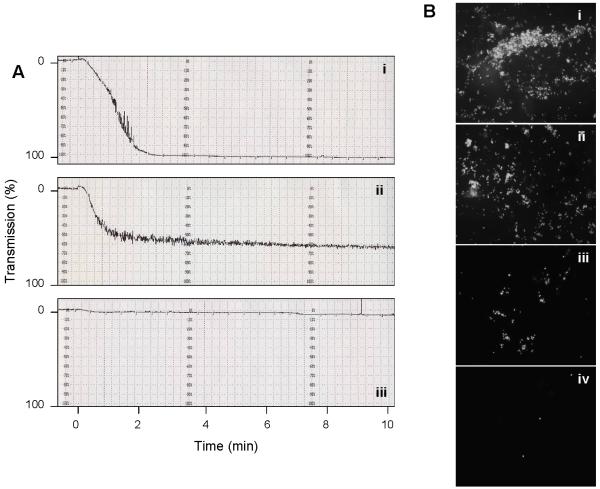
In this study, we compared platelet responses to thrombin and Pam3CSK4 to examine the functional effects of thrombotic vs. immune stimulation. To test the functionality of both agonists, we performed aggregation studies with washed platelets. After 12 minutes, platelet aggregation had progressed to 98 ± 3% for thrombin, 69 ± 5% for Pam3CSK4, and 5 ± 2% for ADP. In Figure 1A, representative platelet aggregation curves for each agonist are shown. The amount of thrombin used (0.5 U/mL/4.43 nM) was below physiological levels (50 nM) (14). ADP (10 μM) was also below its physiologic range (0.05 to > 1 mM) (15). The concentration of Pam3CSK4 used was based on previous work (8).
Figure 1. Platelet aggregation curves and platelet adhesion to collagen.
(A) Washed platelets were activated under stirring conditions with (i) 0.5 U/mL thrombin, (ii) 10 μg/mL Pam3CSK4 and (iii) 10 μM ADP, respectively. Platelet aggregation was measured for 12 min by light transmission in an aggregometer and repeated four times. A representative curve for each agonist is shown where aggregation had progressed to 100% (i), 66% (ii) and 6% (iii), respectively, at the 12-min time point. (B) Platelet adhesion to a collagen-coated cover slip was measured in a flow chamber for washed platelets activated for 10 min with 0.5 U/mL thrombin (i), 10 μg/mL Pam3CSK4 (ii), 10 μM ADP (iii), and for unstimulated platelets (iv), respectively, demonstrating agonist-dependent differences in the induction of platelet adhesion. Images were taken at 20X magnification.
To further investigate the functionality of thrombin vs. Pam3CSK4, we measured platelet adhesion to collagen-coated slides under flow conditions (Figure 1B). Stimulation with thrombin (Figure 1Bi) or Pam3CSK4 (Figure 1Bii) resulted in significant platelet adhesion, whereas stimulation with ADP (Figure 1Biii) induced less platelet adhesion. Unactivated platelets did not adhere to the collagen surface (Figure 1Biv). Taken together, Pam3CSK4 appears to activate platelets significantly more than the weak agonist, ADP.
Differential induction of signaling pathways after platelet activation with thrombin or Pam3CSK4
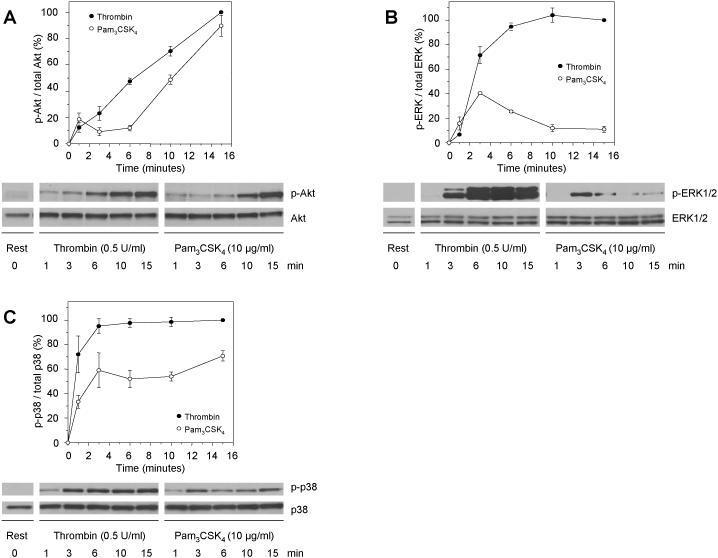
To elucidate whether thrombin and Pam3CSK4 activate signaling pathways differentially downstream of their respective receptors, we examined the phosphorylation kinetics of three major signaling proteins. Previous data from our group showed that Pam3CSK4-stimulation of TLR2 activates the PI3K/Akt signaling pathway (8). Here, we measured the phosphorylation kinetics of Akt(Ser473) as a downstream target of PI3K after platelet activation with thrombin or Pam3CSK4 for up to 15 minutes (Figure 2A). Interestingly, at early time points of activation, each agonist led to distinct phosphorylation kinetics of Akt, whereas at 15 minutes the extent of Akt-phosphorylation became comparable. We also tested ERK1/2- and p38-phosphorylation as they have been shown to be involved in platelet signaling after thrombin stimulation (16, 17). ERK1/2-phosphorylation peaked around 3 minutes after Pam3CSK4-activation and then abated, while thrombin-activation caused a quick increase, which remained constant after 10 minutes (Figure 2B). For p38, thrombin caused an almost immediate increase in phosphorylation, reaching a plateau after 3 minutes, while Pam3CSK4 led to only 60-70% of phosphorylation relative to thrombin (Figure 2C).
Figure 2. Differential phosphorylation kinetics for Akt, ERK1/2 and p38 after platelet activation with thrombin or Pam3CSK4.
Platelets were activated with 0.5 U/mL thrombin (closed circles) or 10 μg/mL Pam3CSK4 (open circles) for the indicated amount of time, lysates were blotted for (A) phospho-Akt(Ser473) and total Akt, (B) phospho-ERK1/2(Thr202/Tyr204) and total ERK1/2, and (C) phospho-p38(Thr180/Tyr182) and total p38. Protein bands were quantified by densitometry and normalized to the protein signal in resting platelets. Error bars indicate the standard error of the mean of 3 to 4 experiments.
Differential changes in protein-protein interactions after platelet activation with thrombin or Pam3CSK4
Identification of FXIIIa-associated proteins in platelets
Next, we investigated whether protein-protein interactions are also differentially affected by thrombin- vs. Pam3CSK4-stimulation of platelets. We selected FXIIIa to study such interactions because its intracellular function lies in regulating the cytoskeletal remodeling during platelet activation and aggregation, and in cross-linking cytoskeletal proteins such as actin, myosin and vinculin (18). In addition, FXIIIa is found abundantly in platelets and, therefore, is well-suited for a MS-based proteomics approach.
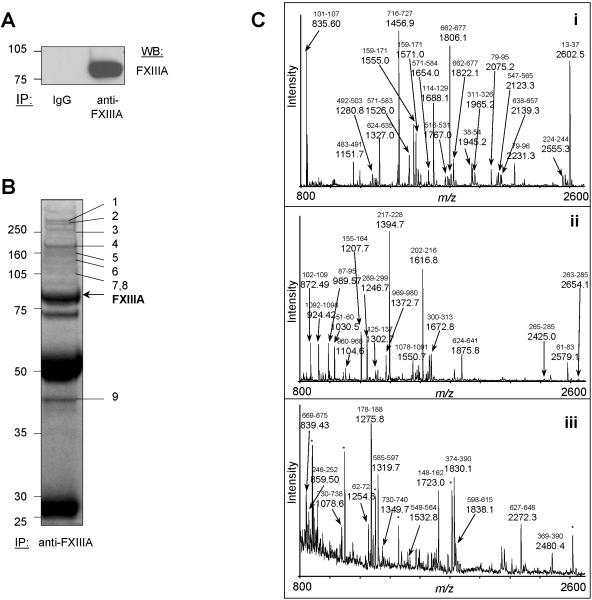
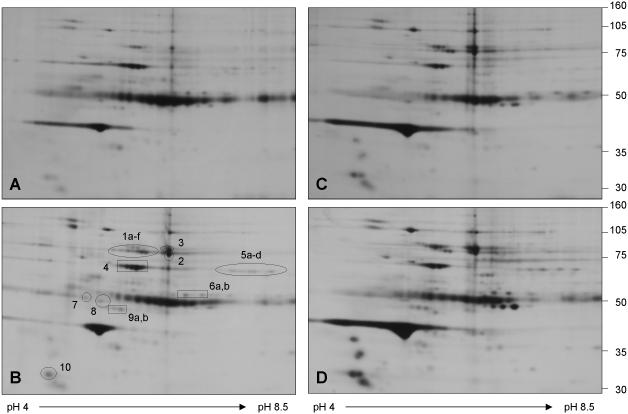
We had to first identify potential FXIIIa-binding partners by immunoprecipitating FXIIIa from platelets. We confirmed that the immunoprecipitation of FXIIIa was specific through immunoblotting (Figure 3A). Separation of protein mixtures by 1D- and 2D-GE often result in identification of complementary sets of proteins due to the differing nature of sample preparation and separation characteristics (19), therefore, the FXIIIa-immunoprecipitates were separated by both methods. In 1D-GE, we used only resting platelets and excised all distinguishable protein bands, then subjected them to tryptic in-gel digestion, MALDI-TOF MS and PMF analysis (Figures 3B, 3C). Details of the identification of all marked proteins are summarized in Table 1. In Figure 3C, we present examples of acquired MALDI-TOF mass spectra for three identified proteins. Major peptide ions of the proteins are labeled with their detected m/z values and corresponding amino acid intervals. In the 2D-GE experiment we also included samples of thrombin-and Pam3CSK4-activated platelets in order to establish whether differences in the FXIIIa-associated protein profiles exist. Gels were stained with either silver stain (spot detection) or Coomassie Blue (spot excision), and all excised protein spots were digested with trypsin and again analyzed by MALDI-TOF MS and PMF. In Figure 4, we show a representative set of silver stained 2D-gels. Control antibody and protein G+ beads bound many proteins non-specifically in resting platelets (Figure 4A). The FXIIIa-specific immunoprecipitate from resting platelets is shown in Figure 4B, where the positions of all excised and identified protein spots are indicated. The spots at position 1 and 2 correspond to FXIIIa and suggest the presence of various FXIIIa isoforms. The identity of the remaining spots are listed in Table 1. Figures 4C and 4D represent FXIIIa-immunoprecipitates from thrombin- and Pam3CSK4-activated platelets, respectively. All subunits of fibrinogen - fibrinogen α (Figure 4B, spots 5a-d), fibrinogen β (spots 6a,b), fibrinogen γ (spots 9a,b) - were absent in the immunoprecipitate from thrombin-activated platelets (Figure 4C) but remained present in the immunoprecipitate from Pam3CSK4-activated platelets (Figure 4D). The 2D patterns of immunoprecipitated proteins were reproducible, but spot intensities varied between experiments and, thus, necessitated thorough validation of the FXIIIa-protein interactions by auxiliary techniques. Several spots visible in silver stained gels were not detected in Coomassie Blue stained gels and, therefore, eluded identification.
Figure 3. Immunoprecipitation and 1D-gel separation of FXIIIa-associated proteins followed by tryptic digestion, MALDI-TOF characterization and PMF assignment of immunoprecipitated proteins.
(A) A specific signal was obtained for FXIIIa after immunoprecipitation from resting platelet lysates and immunoblotting (lane “anti-FXIIIa”) compared to the non-specific isotypic control (lane “IgG”). (B) FXIIIa was immunoprecipitated from resting platelet lysate at an antibody:protein ratio of 1:80. The immunoprecipitate was separated by 1D-GE and stained with Coomassie Blue. Shown are all proteins precipitated with the anti-FXIIIa sheep antibody. All distinguishable gel bands were excised, then subjected to tryptic in-gel digestion, and analyzed by MALDI-TOF MS and PMF. FXIIIa was identified as marked. All other proteins were assigned as indicated: 1-myosin, 2-talin, 3-filamin, 4-thrombospondin, 5-vinculin, 6-integrin αIIb, 7-α-actinin, 8-FAK, and 9-actin γ (for details, see Table 1). Experiments in (A) and (B) were repeated three times. IB indicates immunoblot. (C) MALDI-TOF mass spectra were obtained after tryptic in-gel digestion of excised protein bands/spots and were analyzed by PMF. (i) FXIIIa, (ii) thrombospondin, and (iii) gelsolin. Peaks of major peptide ions, which are derived from the identified protein, are labeled with their m/z values. The corresponding amino acid interval of the assigned peptide is indicated above the m/z value (small font). Abundant but unassignable ions are labeled with <*> symbols.
Table 1.
Overview of all proteins identified after FXIIIA-immunoprecipitation from resting platelet lysates, followed by 1D- or 2D-GE, MALDI-TOF MS, and PMF analyses.
| Protein Name | Interaction with FXIIIa | Gel type & band/spot number◻ | Theoretical MW/pI | Experimental MW/pI | Mowse Score† | Number of matched peptides (Sequence coverage) | Accession number |
|---|---|---|---|---|---|---|---|
| Factor XIIIa | Reference | 1D-as indicated by arrow | 84 / 5.68 | 80/ND | 189 | 35 (46%) | P00488 |
| Factor XIIIa | Reference | 2D - #1 | 84 / 5.75 | a: 80 / 5.8 | 60 | a: 13 (17%) | P00488 |
| b: 80 / 5.9 | 112 | b: 19 (30%) | |||||
| c: 80 / 6.1 | 140 | c: 33 (42%) | |||||
| d: 80 / 6.2 | 217 | d: 39 (49%) | |||||
| e: 80 / 6.3 | 218 | e: 29 (33%) | |||||
| f: 80 / 6.4 | 127 | f: 18 (28%) | |||||
| Factor XIIIa | Reference | 2D - #2 | 84 / 5.75 | 80 / 6.8 | 193 | 35 (47%) | P00488 |
| Focal adhesion kinase | Specific | 1D - #8 | 100 / 5.97 | 110 / ND | 31 ‡ | 15 (21%) | Q05397 |
| Gelsolin | Specific | 2D - #3 | 86 / 5.90 | 85 / 6.8 | 85 | 16 (24%) | P06396 |
| Myosin IIA | Specific | 1D - #1 | 228 / 5.50 | >250 / ND | 194 | 73 (36%) | P35579 |
| Thrombospondin 1 | Specific | 1D - #4 | 133 / 4.71 | 180 / ND | 169 | 34 (28%) | P07996 |
| α-Actinin 1 | NSp | 1D - #7 | 104 / 5.25 | 110 / ND | 79 | 23 (24%) | P12814 |
| Actin γ | NSp | 1D - #9 | 42 / 5.29 | 42 / ND | 185 | 25 (62%) | P63261 |
| Albumin | NSp | 2D - #4 | 71 / 5.85 | 70 / 6.2 | 247 | 34 (64%) | P02768 |
| Fibrinogen α | NSp | 2D - #5 | a: 70 / 7.8 | 137 | a: 25 (42%) | p02671 | |
| a-c: 70 / 8.23 | b: 70 / 8.0 | 140 | b: 27 (46%) | ||||
| d: 70 / 7.98 | c: 70 / 8.2 | 186 | c: 20 (35%) | ||||
| d: 70 / 8.5 | 160 | d: 20 (40%) | |||||
| Fibrinogen β | NSp | 2D - #6 | a: 57 / 8.41 | a: 58 / 7.0 | 190 | a: 33 (56%) | P02675 |
| b: 57 / 8.54 | b: 58 / 7.3 | 185 | b: 30 (56%) | ||||
| Fibrinogen γ | NSp | 2D - #9 | 50 / 5.81 | a: 50 / 5.8 | 123 | a: 16 (41%) | P02679 |
| b: 50 / 5.9 | 193 | b: 22 (54%) | |||||
| Filamin A | NSp | 1D - #3 | 283 / 5.73 | 250 / ND | 302 | 62 (29%) | P21333 |
| Integrin αllb | NSp | 1D - #6 | 104 / 5.41 | 120 / ND | 102 | 20 (33%) | P08514 |
| Talin 1 | NSp | 1D - #2 | 272 / 5.72 | >250 / ND | 243 | 64 (34%) | Q9Y490 |
| Tropomyosin α4 | NSp | 2D - #10 | 28 / 4.67 | 33 / 4.7 | 173 | 19 (58%) | P67936 |
| Tubulin α3 | NSp | 2D - #7 | 51 / 4.94 | 54 / 5.3 | 60 | 10 (36%) | Q71U36 |
| Tubulin β1 | NSp | 2D - #8 | 51 / 5.05 | 52 / 5.6 | 164 | 28 (54%) | Q9H4B7 |
| Vinculin | NSp | 1D - #5 | 124 / 5.51 | 150 / ND | 140 | 32 (35%) | P1 8206 |
MW indicates molecular weight; pI, isoelectric point; 1D, one dimensional; 2D, two dimensional; ND, not determined; and NSp, non-specific.
1D-numbers correspond to gel bands indicated in Figure 3B; 2D-numbers correspond to gelspots indicated in Figure 4B.
the major protein species in each band or spot was identified by MascotTM PMF analysis with a high degree of confidence, in most cases with a p-value significantly less than 0.05.
although the assignment of FAK by PMF analysis was not statistically significant (p > 0.05), immunoblotting showed that FAK was present in the FXIIIa-immunoprecipitate and that its interaction with FXIIIa was specific (Figures 5A, 6A).
Figure 4. Immunoprecipitation and 2D-gel separation of FXIIIa-associated proteins followed by tryptic digestion and MALDI-TOF MS for protein identification.
FXIIIa was immunoprecipitated from lysates of resting and activated platelets at an 1:250 antibody:protein ratio and each immunoprecipitate was subjected to 2D-GE (pI 3-10). A representative set of silver-stained gels is shown. (A) A control immunoprecipitation was performed with a non-specific isotypic antibody for resting platelet lysate. FXIIIa-associated proteins were immunoprecipitated with an anti-FXIIIa antibody (B) from resting platelets, (C) from thrombin-activated platelets (0.5 U/mL, 15 min) and (D) from Pam3CSK4-activated platelets (10 μg/mL, 15 min), respectively. Proteins present in gel spots in panel (B) were identified by tryptic in-gel digestion, MALDI-TOF MS and PMF analysis. The proteins were assigned as: 1(a-f) and 2-FXIIIa; 3-gelsolin; 4-albumin; 5(a-d)-fibrinogen α; 6(a,b)-fibrinogen β; 7-tubulin α; 8-tubulin β; 9(a,b)-fibrinogen γ; and 10-tropomyosin α4. Details of the assignments are given in Table 1. The train of spots at 50 kDa in all four gels corresponds to the heavy chain of the precipitating antibody. This experiment was repeated three times.
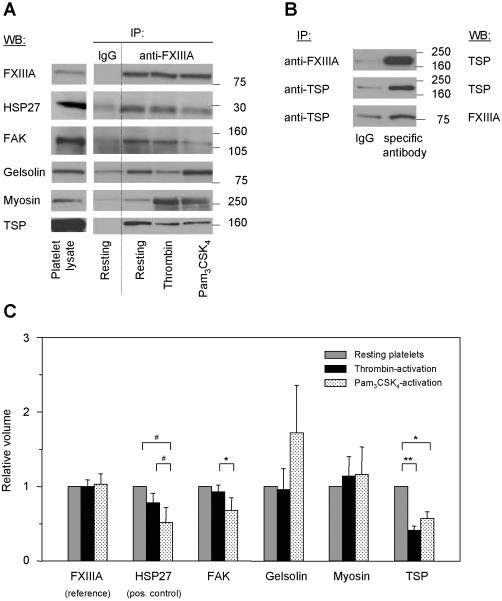
To confirm whether the identified proteins (Figures 3B, 4B, Table 1) were interacting specifically with FXIIIa, we performed immunoblot analysis for all 18 proteins and found, in resting platelets, FXIIIa binds specifically to four proteins: FAK, gelsolin, myosin IIA, and thrombospondin (Figure 5A, lanes “Resting”). All other identified proteins could not be confirmed to bind to FXIIIa in a specific manner as summarized in Table 1. While the identified FXIIIa-binding proteins myosin and thrombospondin are known substrates of FXIIIa (20), FAK and gelsolin have not previously been recognized. Since heat shock protein 27 (HSP27) had been reported to bind to FXIIIa in resting platelets (21), it was used as a positive control. However, it was not identified in our FXIIIa-immunoprecipitation experiments due to the low abundance of signaling molecules in platelets and the detection limit of the applied gel staining methods.
Figure 5. Validation of FXIIIa-binding proteins by immunoblotting and coimmunoprecipitation, and quantitative analysis of the FXIIIa-protein interactions.
(A) Proteins identified by 1D- and 2D-GE, MALDI-TOF MS and PMF analysis were tested by immunoblotting for the specificity and intensity of their FXIIIa-interaction. FXIIIa was immunoprecipitated at an 1:250 antibody:protein ratio from lysates of resting and activated platelets (all lanes “anti-FXIIIa”). As control, a non-specific antibody was used (lanes “IgG”, “Resting”). All immunoprecipitates were separated by 1D-GE, transferred and blotted for the identified proteins listed in Table 1. Here, representative immunoblots for the four specific FXIIIa-binding proteins only are shown. HSP27, as a known FXIIIa-interacting protein, was included as positive control. Lane “Platelet lysate” shows the position of the protein in the total lysate of resting platelets. (B) The interaction between FXIIIa and thrombospondin was tested by co-immunoprecipitation and immunoblotting in resting platelets. The FXIIIa-immunoprecipitate was separated by 1D-GE and blotted for thrombospondin (top image). Thrombospondin was immunoprecipitated with a specific antibody, separated by 1D-GE and blotted first for thrombospondin (middle image) and then for FXIIIa (bottom image). As control, corresponding non-specific antibodies (lanes “IgG”) were used. (C) To determine agonist-induced changes in FXIIIa-protein interactions, quantitative analysis of the immunoblot images in Figure 5A was performed by densitometry for all lanes “anti-FXIIIa”. Band intensities were quantified for each protein, then normalized to the band intensity of FXIIIa and to the protein's band intensity in resting platelets (see also Table 2). ** indicates p < 0.01; *, p < 0.05; #, p = 0.07; IP, immunoprecipitation; IB, immunoblot; TSP, thrombospondin; and FAK, focal adhesion kinase.
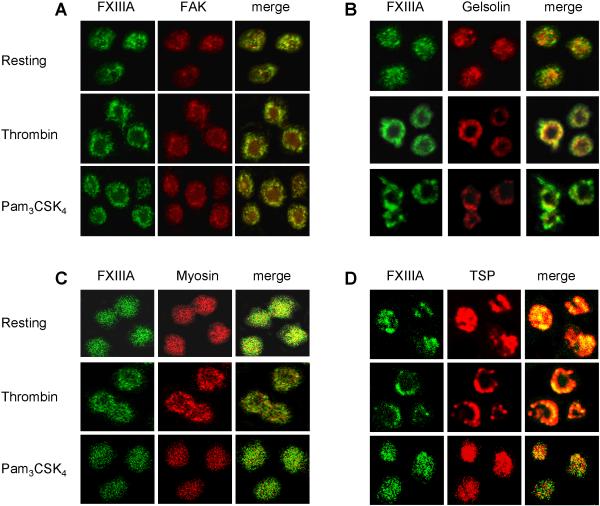
For further verification of the interaction between thrombospondin and FXIIIa, we performed co-immunoprecipitation experiments in resting platelets. Thrombospondin was immunoprecipitated and immunoblotted for FXIIIa (Figure 5B, bottom image) resulting in a much stronger signal for thrombospondin compared to the control, which demonstrates the specificity of the FXIIIa-thrombospondin interaction. Furthermore, we positively confirmed by confocal microscopy that the four FXIIIa-binding proteins co-localize with FXIIIa in resting as well as in activated platelets (Figure 6, last column of images in panels A-D).
Figure 6. Co-localization of FXIIIa with the four identified FXIIIa-binding proteins as determined by confocal microscopy.
The interaction between FXIIIa and the four specific FXIIIa-binding proteins was tested for co-localization by confocal microscopy in resting platelets (upper row of each panel), thrombin-activated platelets (middle row) and Pam3CSK4-activated platelets (lower row), respectively. The first column in each panel shows FXIIIa labeled with an FITC-conjugated secondary antibody, the middle row corresponds to the interacting protein labeled with a Texas Red-conjugated secondary antibody, and the last row shows co-localization of both proteins as indicated by the yellow color in the merged image. Platelets were activated for 10-20 min with either 0.5 U/mL thrombin or 10 μg/mL Pam3CSK4, respectively. (A) FAK, (B) gelsolin, (C) myosin, and (D) thrombospondin (TSP). All images were taken at 100X magnification.
Differential changes in FXIIIa-protein interactions after thrombin- or Pam3CSK4-activation
We determined whether the interaction between FXIIIa and the four identified binding proteins changed upon platelet activation with different agonists. We immunoprecipitated FXIIIa and then immunoblotted for FAK, gelsolin, myosin and thrombospondin (Figure 5A, lanes “anti-FXIIIa”). The signal of immunoprecipitated FXIIIa did not change upon platelet activation (Figure 5C). For HSP27, platelet activation with thrombin led to reduced FXIIIa-binding, compared to resting platelets. Activation with Pam3CSK4, however, showed an even stronger reduction in FXIIIa-association. FAK-binding to FXIIIa showed no significant difference for resting vs. thrombin-activated platelets, however, its binding to FXIIIa decreased significantly in Pam3CSK4-vs. thrombin-activated platelets. For gelsolin and myosin, band intensities varied greatly and the averaged densitometry values did not show significant differences before and after activation or between thrombin- and Pam3CSK4-activated platelets. Thrombospondin, however, showed a significant signal reduction after platelet activation which was independent of the agonist.
PI3K-inhibition
Since thrombin and Pam3CSK4 activate PI3K, we investigated the effect of PI3K-inhibition by LY294002 on the FXIIIa-protein interactions. Although we showed in Figure 2A that the extent of Akt-phosphorylation for either agonist was comparable after 15 minutes of activation, the inhibition of PI3K had no effect on the interaction of FXIIIa and its binding proteins (data not shown) suggesting that the PI3K/Akt-signaling pathway is not involved in these protein-protein interactions.
Differential changes in the release of α-granule proteins after platelet activation with thrombin, Pam3CSK4, or ADP
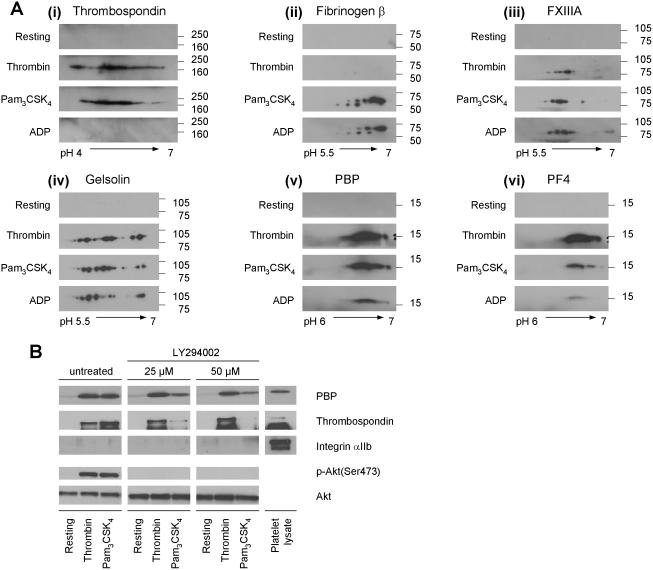
The observed differential changes in selected signaling pathways and protein-protein interactions that were induced by the thrombotic vs. immune stimulation of platelets, were further examined by studying the release of α-granule proteins after activation. Platelets were activated for 15 minutes with either agonist. Releasates were then separated by 2D-GE and immunoblotted for the following proteins: thrombospondin, fibrinogen β, FXIIIa and gelsolin (known α-granule proteins), as well as platelet basic protein (PBP) and platelet factor 4 (PF4) (platelet-specific chemokines) (Figure 7). Releasates tested negative for microvesicle contamination (data not shown). As control, immunoblots of releasates from unstimulated platelets are shown (Figure 7Ai-vi). Thrombospondin was present in the releasate of thrombin- and Pam3CSK4-activated platelets with an agonist-specific pattern, whereas ADP-stimulated platelets did not contain this protein in a detectable amount (Figure 7Ai). The presence of fibrinogen β in the releasate was similar for Pam3CSK4- and ADP-activated platelets, with small differences in isoform patterns and intensities (Figure 7Aii). The very weak signal after thrombin-stimulation might be attributed to the its conversion of fibrinogen into fibrin (22). FXIIIa displayed a similar pattern of isoforms in each releasate (Figure 7Aiii). Interestingly, the spot intensity of each isoform varied and appeared to be agonist-dependent. Particularly after Pam3CSK4-activation, an additional spot at pH 6.5 was observed. Gelsolin, known to be released from platelets after thrombin-stimulation (13), was also released after activation with Pam3CSK4 and ADP (Figures 7Aiv) with releasate patterns similar to each other. However, the relative abundance of several gelsolin isoforms varied between agonists. The chemokine PBP was released in a similar manner after platelet activation with thrombin and Pam3CSK4 (Figure 7Av), while ADP generated a smaller amount. The chemokine PF4 (Figure 7Avi) was highest in the thrombin stimulated platelets, followed by Pam3CSK4-activation. ADP-stimulation yielded the weakest signal for PF4.
Figure 7. Differential release of α-granule proteins after platelet activation with thrombin, Pam3CSK4 or ADP, and differential inhibition of α-granule release by LY294002 after platelet activation with thrombin or Pam3CSK4.
(A) Releasates from resting platelets and from platelets activated with 0.5 U/mL thrombin, 10 μg/mL Pam3CSK4, or 10 μM ADP, respectively, were tested by immunoblotting for selected released α-granule proteins. (i) Thrombospondin, (ii) fibrinogen β, (iii) FXIIIa, (iv) gelsolin, (v) platelet basic protein (PBP), and (vi) platelet factor 4 (PF4). Experiments were repeated up to three times for each protein. (B) Platelets were incubated with the PI3K-inhibitor LY294002 (0, 25 and 50 μM) for 30 min, then either left unstimulated (resting) or activated with thrombin (0.5 U/mL) or Pam3CSK4 (10 μg/mL) for 15 min. Immunoblotting for PBP and thrombospondin, as well as for integrin αIIb (to test for microvesicle contamination), phospho-Akt(Ser473) (to show PI3K inhibition) and total Akt (as lysate loading control) are shown.
In addition, the effect of PI3K-inhibition by LY294002 on platelet α-granule release was examined. The concentration of LY294002 used was based on previous work done in platelets (8). As shown in Figure 7B, the extent of Akt-phosphorylation for either agonist was comparable at 15 minutes of activation. Increasing amounts of LY294002 caused a decrease in the release of PBP and thrombospondin in Pam3CSK4 stimulated platelets, while, the release of these two proteins was not affected by LY294002 after thrombin-stimulation. This suggests that the PI3K/Akt-pathway is differentially involved in the thrombin- vs. Pam3CSK4-induced α-granule release.
Differential effects of thrombin and Pam3CSK4 on platelet aggregates
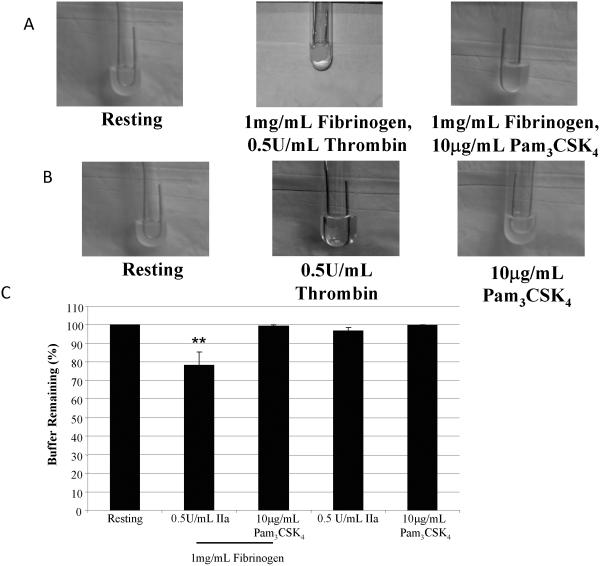
With the differences seen between thrombin and Pam3CSK4 in signaling, protein-protein interactions, αgranule release, and, specifically, FXIIIa, the question remains as to how these agonists affect platelet-dependent coagulation. Using the fibrin clot retraction assay, washed platelets were treated with fibrinogen and either 0.5 U/mL thrombin or 10 μg/mL Pam3CSK4, and incubated for 1 h. As seen in Figure 8A, the addition of thrombin resulted in a stable platelet aggregate, while Pam3CSK4 treatment did not cause any significant clot formation. Figure 8B shows quantitatively the amount of buffer remaining. Thrombin significantly reduces the volume to 82.6% ±5.4, while after Pam3CSK4 treatment the buffer volume remained the same (99.6% ±0.4), suggesting that FXIIIa released from platelets can crosslink fibrin monomers that were converted from fibrinogen by thrombin.
Figure 8. Fibrin clot formation using washed platelets stimulated with thrombin or Pam3CSK4.
Representative photographs (A) were taken of clots that formed in the presence or absence of 1 mg/mL fibrinogen upon stimulation with either 0.5 U/mL thrombin or 10 μg/mL Pam3CSK4. Bar graphs (B) show the percent buffer remaining that was calculated, as described in the Methods, in the presence or absence of fibrinogen. The average and standard error from 6 experiments are graphed. ** p<0.01 compared to resting platelets using an ANOVA analysis, followed by a Dunnett's post test.
The releasate of thrombin-stimulated platelets contains a small amount of fibrinogen β, unlike Pam3CSK4-stimulation (Figure 7Aii). It is hypothesized that this discrepancy is due to the presence of thrombin converting fibrinogen into fibrin. If this is true, then any fibrinogen released from the platelets would be involved in aggregate formation, albeit small, in the presence of thrombin. Using the fibrin clot retraction assay (Figure 8A,B) without the addition of fibrinogen, thrombin stimulation of platelets resulted in a small, stable clot, which was absent after Pam3CSK4-stimulation. Therefore, fibrinogen released from the platelets was converted into fibrin monomers by thrombin, which binds αIIbβ3. FXIIIa released by the platelet would crosslink the fibrin to form a network onto which platelets adhere and form a stable aggregate (23).
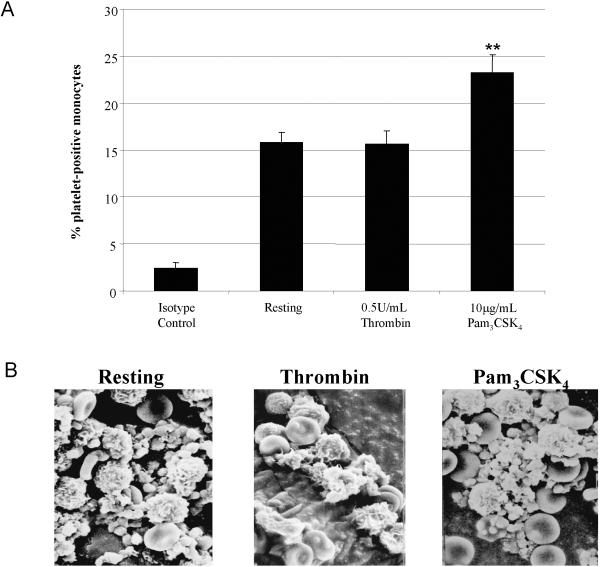
Since Pam3CSK4-stimulation does not affect platelet-platelet interactions (homotypic aggreagates), it is possible that Pam3CSK4 could affect the interaction of platelets with other cells, such as monocytes (heterotypic aggregates). Whole blood left untreated or stimulated with 0.5 U/mL thrombin or 10 μg/mL Pam3CSK4 for 10 minutes was analyzed by flow cytometry to detect and quantitate the percent of platelet-positive monocytes. As seen in Figure 9A, thrombin stimulation did not significantly increase the number of platelet-positive monocytes over resting (15.7±1.4 vs. 15.8±1.1, respectively). Interestingly, Pam3CSK4-stimulation did significantly increase the percent of platelet-positive monocytes (23.2±1.9) over resting or thrombin. Using SEM, we were able to visualize the differences between thrombin- and Pam3CSK4-stimulation on platelet interactions with other blood cells. In Figure 9B, untreated samples contain platelets with a round, spherical appearance. Upon activation with thrombin, platelets underwent a shape change as seen with the formation of pseudopodia, which interact with one another and, to a lesser extent, with circulating white cells. Pam3CSK4-stimulation also resulted in a change of platelet shape, however, with a larger number of platelets binding to white cells. More than one platelet appeared to bind onto each white cell. Pam3CSK4-activated platelets bound to white cells were also binding one another. This interaction is causing white cells to localize and interact with themselves. Therefore, Pam3CSK4 bound to TLR2/1 to activates platelets during inflammation in order to localize and form interactions between white cells. Platelets stimulated with Pam3CSK4 utilize white cells to form a stable clot, unlike thrombin-stimulated platelets, which form a clot via crosslinked fibrin monomers converted by the actions of thrombin and FXIIIa.
Figure 9. Differential interactions of platelets with monocytes upon stimulation with thrombin or Pam3CSK4 using SEM.
Bar graph (A) shows the percent of platelet-positive monocytes determined through flow cytometry of whole blood samples untreated (Resting) or treated with 0.5 U/mL thrombin or 10 μg/mL Pam3CSK4 for 10 minutes at room temperature. The average percent and standard error from 7 experiments are graphed. ** p<0.01 compared to Resting and Thrombin treatments using an ANOVA analysis, followed by a Bonferroni post test. Scanning electron micrographs (B) of control (no agonist) and samples treated with 0.5 U/mL thrombin or with 10μg/mL Pam3CSK4. Magnification at 15,000X.
Discussion
We activated human platelets with thrombin or Pam3CSK4, and examined agonist-dependent effects on the phosphorylation kinetics of signaling proteins, on FXIIIa-protein interactions, and on α-granule release. Three major signaling proteins - Akt, ERK1/2, and p38 - were examined as to whether they are differentially affected by thrombin- vs. Pam3CSK4-stimulation. Thrombin-induced signaling cascades in platelets have been intensively studied (9), and PI3K is a known key player in these events (24). However, the details of signaling pathways involving ERK1/2 or p38 remain controversial (16, 17). TLR2 signaling, on the other hand, culminates in NF-κB activation in various cell types (2, 25), and PI3K has been implicated as a regulator of TLR signaling (26). Although anuclear platelets express NF-κB (27), no reports describing mechanistic details of the TLR2 signaling pathway within platelets have been published. Recent data from our laboratory have shown that PI3K/Akt play a major role in Pam3CSK4-induced platelet responses (8). Here, we further demonstrate that Pam3CSK4 also induces phosphorylation of ERK1/2 and p38 in a time-dependent and agonist-dependent manner. This clearly suggests that other pathways beside PI3K/Akt are involved in TLR2 signaling and Pam3CSK4-induced platelet responses.
The increased phosphorylation of Akt, ERK1/2, and p38 after activation with thrombin results in the formation of a more stable clot, as seen in the platelet aggregation and fibrin clot retraction assays, compared to Pam3CSK4-stimulation. Furthermore, the phosphorylation kinetics of Akt is faster with thrombin treatment compared to Pam3CSK4. PI3K/Akt pathways have been implicated in the outside-in signaling that occurs when fibrinogen binds to αIIbβ3 and activates signaling pathways that stabilize platelet aggregates (28). Important to clot stabilization is the greater extent of phosphorylation of both ERK1/2 and p38 with thrombin compared to Pam3CSK4. Both kinases are phosphorylated in more rapid manner, suggestive of their role in inside-out signaling, when signaling events activated by an agonist lead to conformational changes in αIIbβ3 (29). Therefore, the more stable clot formation occurring with thrombin is a result of the rapid phosphorylation of ERK1/2 and p38 causing conformational change in αIIbβ3 to increase fibrinogen binding, which results in an increased phosphorylation of Akt to further stabilize the aggregate.
The second part of our study focused on protein-protein interactions with particular emphasis on FXIIIa. For anuclear platelets, MS-based proteomic methods have proven to be an important tool for characterizing protein expression, post-translational modifications, and protein release (13, 30-35). Eighteen FXIIIa-binding proteins were identified and validated in resting and activated platelets. Only four of these proteins were found to bind to FXIIIa specifically. Known FXIIIa-substrates have been classified into three groups: coagulatory/fibrinolytic proteins, adhesive proteins, and cytoskeletal proteins (20). The four FXIIIa-binding proteins we identified in this study were: FAK, gelsolin, myosin IIA and thrombospondin. FAK and gelsolin are novel FXIIIa-interacting partners, while myosin IIA and thrombospondin had been reported to function as substrates for FXIIIa. FAK is a tyrosine kinase that mediates signaling through integrins and translocates to the cytoskeleton in platelets in the second wave of cytoskeletal rearrangements (36-38). The FAK-FXIIIa interaction could lead to crosslinking of FAK to other proteins, or phosphorylation of FXIIIa by FAK. Gelsolin, a cytoskeletal protein that regulates actin assembly (39), has also been found in α-granules (35), where its interaction with FXIIIa might.
It was examined whether these FXIIIa-protein interactions are differentially altered upon thrombotic vs. immune stimulation in comparison to resting platelets. We found differences in FXIIIa-binding to FAK, thrombospondin, and HSP27. The FXIIIa-FAK interaction changed in a statistically significant, agonist-dependent manner. The intensity of the interaction was not altered significantly after thrombin-stimulation, indicative of their translocation to the cytoskeleton where both proteins are thought to function after activation. In Pam3CSK4-stimulated platelets, the FAK-FXIIIa interaction decreased significantly compared to thrombin-activated platelets, suggesting a decrease in binding due to differences in signaling. The FXIIIathrombospondin interaction under resting conditions stems from α-granules, since thrombospondin is located only in these granules. After thrombin- or Pam3CSK4-activation, the intensity of the FXIIIa-thrombospondin interaction decreased significantly indicating that less thrombospondin was bound to FXIIIa after activation with either agonist. The FXIIIa-HSP27 binding for thrombin-activated platelets decreased compared to resting but not to the extent as that seen with Pam3CSK4. HSP27, like FAK, is thought to translocate to the cytoskeleton upon platelet stimulation (40). The observed differences in signaling might have led to the differences in FXIIIa binding. Additional experiments showed the inhibition of PI3K did not affect the observed differences in these FXIIIa-interactions, suggesting that the PI3K/Akt-pathway is not involved in regulating FXIIIa-protein interactions.
Platelet activation with thrombin, Pam3CSK4 or ADP was examined whether it causes a differential release of various α-granule proteins. Platelet α-granules are the major storage compartment for adhesive proteins, chemokines, receptors, coagulatory and fibrinolytic proteins (35, 41). Granules are distributed randomly within platelets, but, upon stimulation, fuse with the plasma membrane or open canalicular systems to release their contents. Our data suggest a differential, agonist-dependent release for selected proteins upon platelet activation with thrombin, Pam3CSK4, or ADP. The releasate of ADP-stimulated platelets lacked thrombospondin, while the releasate of thrombin-activated platelets lacked fibrinogen β, likely because it had been converted into fibrin (22). For FXIIIa and gelsolin, differences occurred in protein spots representing multiple isoforms and in spot intensity. For PBP and PF4, the most abundant chemokines released from platelets (44) were differently abundant in the respective releasates. One might expect that Pam3CSK4-stimulation could lead to an excessive release of these chemokines, however, we did not observe this. Pam3CSK4-stimulation of platelets that had been treated with increasing amounts of PI3K-inhibitor, caused a decrease in the release of PBP and thrombospondin. The thrombin-stimulated release of these two proteins was not affected by the inhibitor, suggesting that the PI3K/Akt signaling pathway is significantly involved in Pam3CSK4-induced, but not in thrombin-induced, α-granule release.
Finally, these data demonstrate the functional differences that occur when platelets are stimulated in the context of inflammation and thrombosis. Using thrombin as the agonist, platelets release fibrinogen, which is converted into fibrin by thrombin. FXIIIa crosslinks fibrin to form a stable, platelet clot. In normal hemostasis, this stable clot prevents hemorrhage and allows for proper wound healing. However, in a pathologic setting, the same clot can also prevent blood flow and cause an ischemic event to occur, such as in myocardial infarction. Stimulation with Pam3CSK4, which binds to TLR2/1, leads to the release of fibrinogen, which not only binds to platelets, but also to immune cells such as neutrophils and monocytes. Platelets will bind these cells and each other to localize the immune response to the site of injury.
In conclusion, our data demonstrate for the first time that platelet stimulation with thrombin or Pam3CSK4 leads to differential activation of signaling proteins, changes in protein-protein interactions, as well as α-granule release. These results highlight the differences in the platelet's immune vs. thrombotic responses and form the basis for further functional and mechanistic studies to better understand the platelet's role in innate immunity and inflammation.
Table 2.
Quantitative changes in specific FXIIIa-protein interactions after plateletactivation with thrombin vs. Pam3CSK4.
| FXIIIa-binding protein | Relative volume◻: Thrombin | Relative volume◻: Pam3CSK4 | Protein location in resting platelets |
|---|---|---|---|
| Factor XIIIa (reference) | 1.00 ± 0.09 | 1.03 ± 0.14 | α-granules (35, 45) cytosol (46) |
| Heat shock protein 27 (positive control) | 0.78 ± 0.13 | 0.52 ± 0.20 | cytosol (21) |
| Focal adhesion kinase | 0.93 ± 0.09 | 0.68 ± 0.17 | cytosol (38) membrane skeleton (38) |
| Gelsolin | 0.96 ± 0.28 | 1.72 ± 0.64 | α-granules (35) cytosol (47) cytoskeleton (39, 48) |
| Myosin IIA | 1.14 ± 0.26 | 1.16 ± 0.37 | cytosol (49) cytoskeleton (48) |
| Thrombospondin 1 | 0.41 ± 0.06 | 0.57 ± 0.09 | α-granules (35, 50) |
presented are data of 5 to 6 experiments for each protein. Protein bands from immunoblot images were quantified by densitometry, normalized to the corresponding band intensity of FXIIIa for each experimental condition and then further normalized to the protein's band intensity in resting platelets which was arbitrary set to 1.
Acknowledgments
We thank Dr. Kahraman Tanriverdi for his assistance with gel imaging.
This work was supported by NIH-NHLBI grant P01 HL083801 (J.E.F.), NIH-NHLBI contract N01 HV28178 (C.E.C.), NIH-NCRR grants P41 RR10888 and S10 RR15942 (C.E.C.), NIH NHLBI grant T32 HL07224 (L.M.B.).
References
- 1.Beutler B. Innate immunity: an overview. Mol Immunol. 2004;40(12):845–59. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Akira S. Toll-like receptor signaling. J Biol Chem. 2003;278(40):38105–8. doi: 10.1074/jbc.R300028200. [DOI] [PubMed] [Google Scholar]
- 3.Shiraki R, Inoue N, Kawasaki S, Takei A, Kadotani M, Ohnishi Y, et al. Expression of Toll-like receptors on human platelets. Thromb Res. 2004;113(6):379–85. doi: 10.1016/j.thromres.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Cognasse F, Hamzeh H, Chavarin P, Acquart S, Genin C, Garraud O. Evidence of Toll-like receptor molecules on human platelets. Immunol Cell Biol. 2005;83(2):196–8. doi: 10.1111/j.1440-1711.2005.01314.x. [DOI] [PubMed] [Google Scholar]
- 5.Weyrich AS, Zimmerman GA. Platelets: signaling cells in the immune continuum. Trends Immunol. 2004;25(9):489–95. doi: 10.1016/j.it.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Wetzler LM. The role of Toll-like receptor 2 in microbial disease and immunity. Vaccine. 2003;21:S2/55–S2/60. doi: 10.1016/s0264-410x(03)00201-9. [DOI] [PubMed] [Google Scholar]
- 7.Berg M, Offermanns S, Seifert R, Schultz G. Synthetic lipopeptide Pam3CysSer(Lys)4 is an effective activator of human platelets. Am J Physiol. 1994;266(6 Pt 1):C1684–91. doi: 10.1152/ajpcell.1994.266.6.C1684. [DOI] [PubMed] [Google Scholar]
- 8.Blair P, Rex S, Vitseva O, Beaulieu L, Tanriverdi K, Chakrabarti S, et al. Stimulation of Toll-like Receptor 2 in Human Platelets Induces a Thrombo-Inflammatory Response Through Activation of Phosphoinositide 3-Kinase. Circ Res. doi: 10.1161/CIRCRESAHA.108.185785. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brass LF, Stalker TJ, Zhu L, Woulfe DS. Signal Transduction During Platelet Plug Formation. In: Michelson AD, editor. Platelets. 2nd ed. Elsevier Science; New York: 2007. pp. 319–346. [Google Scholar]
- 10.Freedman JE, Loscalzo J, Barnard MR, Alpert C, Keaney JF, Michelson AD. Nitric oxide released from activated platelets inhibits platelet recruitment. J Clin Invest. 1997;100(2):350–6. doi: 10.1172/JCI119540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achyuthan KE, Dobson JV, Greenberg CS. Gly-Pro-Arg-Pro modifies the glutamine residues in the alpha- and gamma-chains of fibrinogen: inhibition of transglutaminase cross-linking. Biochim Biophys Acta. 1986;872(3):261–8. doi: 10.1016/0167-4838(86)90279-7. [DOI] [PubMed] [Google Scholar]
- 12.Garcia BA, Smalley DM, Cho H, Shabanowitz J, Ley K, Hunt DF. The platelet microparticle proteome. J Proteome Res. 2005;4(5):1516–21. doi: 10.1021/pr0500760. [DOI] [PubMed] [Google Scholar]
- 13.Coppinger JA, Cagney G, Toomey S, Kislinger T, Belton O, McRedmond JP, et al. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood. 2004;103(6):2096–104. doi: 10.1182/blood-2003-08-2804. [DOI] [PubMed] [Google Scholar]
- 14.Bizios R, Lai L, Fenton JW, 2nd, Malik AB. Thrombin-induced thromboxane generation by neutrophils and lymphocytes: dependence on enzymic site. J Cell Physiol. 1987;132(2):359–62. doi: 10.1002/jcp.1041320224. [DOI] [PubMed] [Google Scholar]
- 15.Yoon HY, Hwang SH, Lee EY, Kim TU, Cho EH, Cho SW. Effects of ADP on different inhibitory properties of brain glutamate dehydrogenase isoproteins by perphenazine. Biochimie. 2001;83(9):907–13. doi: 10.1016/s0300-9084(01)01325-6. [DOI] [PubMed] [Google Scholar]
- 16.Nadal-Wollbold F, Pawlowski M, Levy-Toledano S, Berrou E, Rosa JP, Bryckaert M. Platelet ERK2 activation by thrombin is dependent on calcium and conventional protein kinases C but not Raf-1 or B-Raf. FEBS Lett. 2002;531(3):475–82. doi: 10.1016/s0014-5793(02)03587-1. [DOI] [PubMed] [Google Scholar]
- 17.Begonja AJ, Geiger J, Rukoyatkina N, Rauchfuss S, Gambaryan S, Walter U. Thrombin stimulation of p38 MAP kinase in human platelets is mediated by ADP and thromboxane A2 and inhibited by cGMP/cGMP-dependent protein kinase. Blood. 2007;109(2):616–8. doi: 10.1182/blood-2006-07-038158. [DOI] [PubMed] [Google Scholar]
- 18.Adany R, Bardos H. Factor XIII subunit A as an intracellular transglutaminase. Cell Mol Life Sci. 2003;60(6):1049–60. doi: 10.1007/s00018-003-2178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson SP, Bahou WF, Fitzgerald D, Ouwehand W, Rao AK, Leavitt AD. Mapping the platelet proteome: a report of the ISTH Platelet Physiology Subcommittee. J Thromb Haemost. 2005;3(9):2098–101. doi: 10.1111/j.1538-7836.2005.01550.x. [DOI] [PubMed] [Google Scholar]
- 20.Muszbek L, Yee VC, Hevessy Z. Blood coagulation factor XIII: structure and function. Thromb Res. 1999;94(5):271–305. doi: 10.1016/s0049-3848(99)00023-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, Tassi L, Lane W, Mendelsohn ME. Specific binding of the transglutaminase, platelet factor XIII, to HSP27. J Biol Chem. 1994;269(35):22379–84. [PubMed] [Google Scholar]
- 22.Siebenlist KR, Meh DA, Mosesson MW. Protransglutaminase (factor XIII) mediated crosslinking of fibrinogen and fibrin. Thromb Haemost. 2001;86(5):1221–8. [PubMed] [Google Scholar]
- 23.Shen L, Lorand L. Contribution of fibrin stabilization to clot strength. Supplementation of factor XIII-deficient plasma with the purified zymogen. J Clin Invest. 1983;71(5):1336–41. doi: 10.1172/JCI110885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson SP, Yap CL, Anderson KE. Phosphoinositide 3-kinases and the regulation of platelet function. Biochem Soc Trans. 2004;32(Pt 2):387–92. doi: 10.1042/bst0320387. [DOI] [PubMed] [Google Scholar]
- 25.Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285(5428):736–9. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 26.Hazeki K, Nigorikawa K, Hazeki O. Role of phosphoinositide 3-kinase in innate immunity. Biol Pharm Bull. 2007;30(9):1617–23. doi: 10.1248/bpb.30.1617. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Morris S, Epps J, Carroll R. Demonstration of an activation regulated NF-kappaB/I-kappaBalpha complex in human platelets. Thromb Res. 2002;106(45):199–203. doi: 10.1016/s0049-3848(02)00130-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Banfic H, Straforini F, Tosi L, Volinia S, Rittenhouse SE. A type II phosphoinositide 3-kinase is stimulated via activated integrin in platelets. A source of phosphatidylinositol 3-phosphate. J Biol Chem. 1998;273(23):14081–4. doi: 10.1074/jbc.273.23.14081. [DOI] [PubMed] [Google Scholar]
- 29.Flevaris P, Li Z, Zhang G, Zheng Y, Liu J, Du X. Two distinct roles of mitogen-activated protein kinases in platelets and a novel Rac1-MAPK-dependent integrin outside-in retractile signaling pathway. Blood. 2008 doi: 10.1182/blood-2008-05-155978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maguire PB, Wynne KJ, Harney DF, O'Donoghue NM, Stephens G, Fitzgerald DJ. Identification of the phosphotyrosine proteome from thrombin activated platelets. Proteomics. 2002;2(6):642–8. doi: 10.1002/1615-9861(200206)2:6<642::AID-PROT642>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Garcia A, Prabhakar S, Brock CJ, Pearce AC, Dwek RA, Watson SP, et al. Extensive analysis of the human platelet proteome by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2004;4(3):656–68. doi: 10.1002/pmic.200300665. [DOI] [PubMed] [Google Scholar]
- 32.Garcia A, Prabhakar S, Hughan S, Anderson TW, Brock CJ, Pearce AC, et al. Differential proteome analysis of TRAP-activated platelets: involvement of DOK-2 and phosphorylation of RGS proteins. Blood. 2004;103(6):2088–95. doi: 10.1182/blood-2003-07-2392. [DOI] [PubMed] [Google Scholar]
- 33.Zahedi RP, Begonja AJ, Gambaryan S, Sickmann A. Phosphoproteomics of human platelets: A quest for novel activation pathways. Biochim Biophys Acta. 2006;1764(12):1963–76. doi: 10.1016/j.bbapap.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Garcia A. Proteome analysis of signaling cascades in human platelets. Blood Cells Mol Dis. 2006;36(2):152–6. doi: 10.1016/j.bcmd.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Maynard DM, Heijnen HF, Horne MK, White JG, Gahl WA. Proteomic analysis of platelet alpha-granules using mass spectrometry. J Thromb Haemost. 2007;5(9):1945–55. doi: 10.1111/j.1538-7836.2007.02690.x. [DOI] [PubMed] [Google Scholar]
- 36.Clark EA, Shattil SJ, Brugge JS. Regulation of protein tyrosine kinases in platelets. Trends Biochem Sci. 1994;19(11):464–9. doi: 10.1016/0968-0004(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 37.Shattil SJ, Haimovich B, Cunningham M, Lipfert L, Parsons JT, Ginsberg MH, et al. Tyrosine phosphorylation of pp125FAK in platelets requires coordinated signaling through integrin and agonist receptors. J Biol Chem. 1994;269(20):14738–45. [PubMed] [Google Scholar]
- 38.Ohmori T, Yatomi Y, Asazuma N, Satoh K, Ozaki Y. Involvement of proline-rich tyrosine kinase 2 in platelet activation: tyrosine phosphorylation mostly dependent on alphaIIbbeta3 integrin and protein kinase C, translocation to the cytoskeleton and association with Shc through Grb2. Biochem J. 2000;347(Pt 2):561–9. doi: 10.1042/0264-6021:3470561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fox JE. Cytoskeletal proteins and platelet signaling. Thromb Haemost. 2001;86(1):198–213. [PubMed] [Google Scholar]
- 40.Zhu Y, O'Neill S, Saklatvala J, Tassi L, Mendelsohn ME. Phosphorylated HSP27 associates with the activation-dependent cytoskeleton in human platelets. Blood. 1994;84(11):3715–23. [PubMed] [Google Scholar]
- 41.Reed GL. Platelet Secretion. In: Michelson AD, editor. Platelets. 2nd ed. Elsevier Science; New York: 2007. pp. 309–318. [Google Scholar]
- 42.Flaumenhaft R. Molecular basis of platelet granule secretion. Arterioscler Thromb Vasc Biol. 2003;23(7):1152–60. doi: 10.1161/01.ATV.0000075965.88456.48. [DOI] [PubMed] [Google Scholar]
- 43.Sehgal S, Storrie B. Evidence that differential packaging of the major platelet granule proteins von Willebrand factor and fibrinogen can support their differential release. J Thromb Haemost. 2007;5(10):2009–16. doi: 10.1111/j.1538-7836.2007.02698.x. [DOI] [PubMed] [Google Scholar]
- 44.von Hundelshausen P, Petersen F, Brandt E. Platelet-derived chemokines in vascular biology. Thromb Haemost. 2007;97(5):704–13. doi: 10.1160/th07-01-0066. [DOI] [PubMed] [Google Scholar]
- 45.Marx G, Korner G, Mou X, Gorodetsky R. Packaging zinc, fibrinogen, and factor XIII in platelet alpha-granules. J Cell Physiol. 1993;156(3):437–42. doi: 10.1002/jcp.1041560302. [DOI] [PubMed] [Google Scholar]
- 46.Lopaciuk S, Lovette KM, McDonagh J, Chuang HY, McDonagh Subcellular distribution of fibrinogen and factor XIII in human blood platelets. Thromb Res. 1976;8(4):453–65. doi: 10.1016/0049-3848(76)90223-1. [DOI] [PubMed] [Google Scholar]
- 47.Yin HL, Kwiatkowski DJ, Mole JE, Cole FS. Structure and biosynthesis of cytoplasmic and secreted variants of gelsolin. J Biol Chem. 1984;259(8):5271–6. [PubMed] [Google Scholar]
- 48.Hartwig JH. The Platelet Cytoskeleton. In: Michelson AD, editor. Platelets. 2nd ed. Elsevier Science; New York: 2007. pp. 75–97. [Google Scholar]
- 49.Sixma JJ, van den Berg A, Jockusch BM, Hartwig J. Immunoelectron microscopic localization of actin, alpha-actinin, actin-binding protein and myosin in resting and activated human blood platelets. Eur J Cell Biol. 1989;48(2):271–81. [PubMed] [Google Scholar]
- 50.Lawler J, Derick LH, Connolly JE, Chen JH, Chao FC. The structure of human platelet thrombospondin. J Biol Chem. 1985;260(6):3762–72. [PubMed] [Google Scholar]