Abstract
Androgen receptor (AR) signaling plays a critical role in the development and progression of prostate cancer. It has been reported previously that peroxiredoxin-1 (Prx1), a member of a novel family of peroxidases, interacts physically with AR to enhance AR transactivation of target genes. In the present study, we evaluated the biological significance of Prx1 in modulating dihydrotestosterone (DHT)-stimulated growth and AR target gene expression of prostate cancer cells. We also investigated the mechanism by which Prx1 might potentiate AR signaling. The contribution of Prx1 was assessed mainly by using the approach of stable Prx1 knockdown. The major observations are as follows. (i) A low level of Prx1 desensitizes cells to growth stimulation and AR target gene induction by DHT, such that exposure to a higher level of DHT is required to reach the same magnitude of response when Prx1 is depressed. (ii) Prx1 increases the affinity of AR to DHT and decreases the rate of DHT dissociation from the occupied receptor. (iii) Prx1 enhances the N-terminus and C-terminus interaction of AR. A stronger N-C interaction is consistent with a more robust AR activation signal by keeping DHT tight in the ligand binding pocket. (iv) The stimulatory effects of Prx1 on AR ligand binding affinity and AR N-C interaction are manifested regardless of a wild-type or mutant AR. The above findings lead us to believe that Prx1 may be a therapeutic target in blocking the transition of prostate cancer from an androgen-dependent to an androgen-refractory phenotype.
Keywords: Prx1, Androgen Receptor, Androgen, Prostate cancer
Introduction
Androgen receptor (AR) is a member of the nuclear receptor superfamily and serves as a ligand-dependent transcription factor. A functional AR signaling axis is integral to the development and progression of prostate cancer (1). For patients who are diagnosed with metastatic prostate cancer, androgen deprivation therapy (ADT) is a standard treatment of choice (2). ADT, however, is not curative; most tumors acquire resistance to this modality after a short period of improvement (3). There is a large body of evidence which suggests that AR still plays a key role in hormone-refractory prostate cancer (4,5). Several mechanisms have been proposed to account for the activation of AR signaling in a low or suboptimal androgen environment, including a mutated AR, a promiscuous AR, or an outlaw AR (4-6). Although many cellular and molecular events contributing to the development of hormone-refractory prostate cancer have been identified (7), the underlying factors responsible for the emergence of this phenotype remain to be elucidated.
Androgen deprivation has been shown to cause endothelial cell death, degeneration of capillaries, and vasoconstriction in the prostate (8). These findings suggest that a state of hypoxia is likely to occur in the malignant tissue after surgical or chemical androgen ablation. Previous studies reported that hypoxia increases AR activity in prostate cancer cells, and that peroxiredoxin-1, which is up-regulated by hypoxia, interacts with AR to enhance the expression of androgen-regulated genes (9, 10). Peroxiredoxin-1 (Prx1) belongs to a novel family of peroxidases which are usually involved in the destruction of peroxides (11). Elevated levels of Prx1 are found in several types of cancer (12-14), suggesting that Prx1 may confer a survival advantage to the cancer cells (15, 16). A recent paper showed that Prx1 is a target of Nrf2 (16), the latter is a redox-sensitive transcription factor which is generally up-regulated by hypoxia.
Despite the recognition of Prx1 as an antioxidant protein, the AR-stimulatory effect of Prx1 is not dependent on this function, since a mutant Prx1 lacking antioxidant activity behaves similarly as the wild-type towards AR (10). The physical interaction of Prx1 with AR is validated by reciprocal immunoprecipitation, chromatin immunoprecipitation, and in vitro pull-down assays (10). Thus Prx1, by acting as a “chaperone”, seems to sensitize AR to low levels of androgen (10). Upon binding to the ligand, the N-terminus of one AR molecule associates with the C-terminus of another AR molecule in a process called N-C dimerization (17). This is an important step in AR signaling because it keeps AR activated by holding the ligand in the ligand binding pocket. In the present study, we evaluated the ability of Prx1 to modulate AR ligand binding affinity as well as AR N-C dimerization. Additionally, we also determined the role of Prx1 in androgen-stimulated growth and gene expression of prostate cancer cells. The contribution of Prx1 was assessed mainly through the approach of stable Prx1 knockdown. Our interpretations are therefore based on comparing the response outcome in the presence of a high versus a low Prx1 expression status. Most of the experiments were carried out with the LNCaP cell model. Other prostate and non-prostate cancer cells were also used in some experiments in order to clarify certain issues.
Results
Prx1 Augments Androgen-Stimulated Growth of LNCaP Prostate Cancer Cells
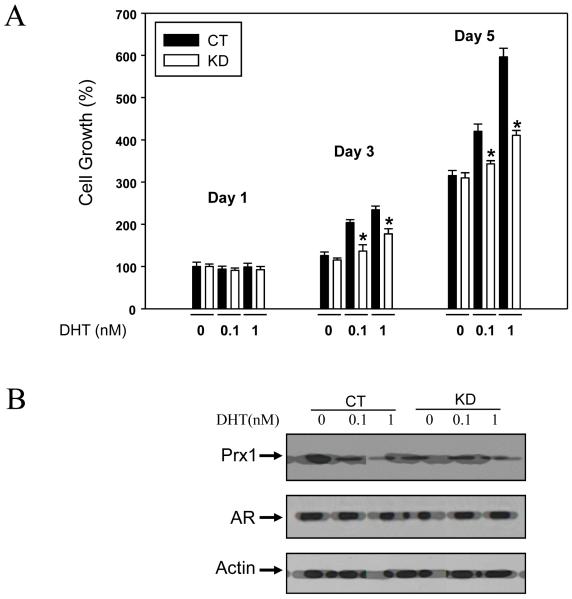
LNCaP cells express a mutated AR and are responsive to growth stimulation by androgens (18, 19). In the present experiment, LNCaP cells infected with either the Prx1-shRNA or scrambled-shRNA were cultured in a charcoal stripped-FBS medium and treated with 0, 0.1 or 1 nM DHT. The Prx1-shRNA and scrambled-shRNA subtypes are designated as KD (for Prx1 knockdown) and CT (for control), respectively. These abbreviations are used throughout the remaining text. Cell growth was monitored for 5 days using the MTT assay. The results are shown in Fig. 1A. The day 1 MTT data of the CT cells not exposed to DHT is set as 100%. All the other data are relative to this value. With the CT cells, growth was increased 3-fold on day 5 in the absence of DHT. These cells responded to DHT stimulation in a dose-dependent manner, such that at 1 nM DHT concentration, a 6-fold increase of cell growth was observed during the same period. In contrast, the KD cells were less responsive to DHT stimulation when compared to the CT cells, suggesting that decreased expression of Prx1 causes a desensitization to DHT signaling. Western blots of Prx1 in CT and KD cell extracts, with or without DHT treatment, are shown in Fig. 1B. The data confirm the successful knockdown of Prx1 by shRNA, and that DHT did not change the expression of Prx1. In order to ascertain that the differences in DHT-stimulated growth between the CT and KD cells were not due to changes in AR level, we also examined AR expression by Western blotting. It is evident from the data in Fig. 1B that the protein level of AR remained stable under these conditions. Both the Prx1 and AR Western blots were done using cell extracts under DHT treatment for 3 days.
Fig. 1. Effect of Prx1 knockdown on DHT-stimulated growth of LNCaP cells.
Panel A, MTT cell growth data, mean ± SD (n=3). CT, control cells; KD, Prx1 knockdown cells. The day 1 MTT data of CT cells not exposed to DHT is set as 100%. *P<0.05 compared to the corresponding CT value. Panel B, Western blots of Prx1 and AR from day 3 CT and KD samples of DHT treatment.
Prx1 Enhances Expression of Cyclin D1, PSA and KLK2 in Response to DHT
In order to verify that the reduced sensitivity of KD cells to DHT was also manifested at the molecular level, we examined selective genes which are known to be up-regulated by DHT in prostate cells. We chose cyclin D1, prostate specific antigen (PSA) and kallikrein 2 (KLK2) as markers of DHT signaling. DHT induces cyclin D1 via the mTOR-dependent protein translation mechanism, while PSA and KLK2 are direct transcriptional targets of AR. LNCaP CT or KD cells were treated with 0, 0.1 or 1 nM DHT for 24 hr, and then processed for Western blotting and quantitative PCR. The Western blot data are shown in Fig. 2A. DHT at 1 nM caused a 2.5-2.6 fold increase of all 3 molecular markers in the CT cells (i.e. with full expression of Prx1). In contrast, the same concentration of DHT produced only a 1.8-1.9 fold increase in the KD cells (i.e. with reduced expression of Prx1). Fig. 2B and 2C show the real-time PCR data of PSA and KLK2, respectively. As expected, the transcriptional control of these two genes in response to DHT was blunted in the KD cells. For PSA, the decrease was statistically significant at 0.1 and 1 nM DHT. For KLK2, the decrease was noticeable only at 1 nM DHT. Collectively, the results of Fig. 1 and 2 support the idea that Prx1 plays a key role in DHT signaling for stimulation of growth and AR target gene expression.
Fig. 2. Effect of Prx1 knockdown on expression of cyclin D1, PSA and KLK2 in response to DHT stimulation of LNCaP cells.
Panel A, Western blots of molecular markers of control (CT) and Prx1 knockdown (KD) cells. Panels B and C, real time RT-PCR analysis of PSA and KLK2, respectively. The results are expressed as fold of change relative to the value obtained without DHT treatment. *P<0.05 compared to the corresponding control value.
Decrease of AR Activity by Prx1 siRNA Mediated Knockdown is not Affected by DHT Level
The above experiments were done with cells which were stably infected with the Prx1 shRNA. In order to rule out any non-specific effect of the shRNA, we carried out another experiment with cells which were transiently transfected with one of two different Prx1 siRNAs (refer to Methods for detail). Instead of using AR target genes as markers, we used an androgen response element (ARE)-luciferase reporter assay as the readout to evaluate the effect of Prx1 knockdown in cells exposed to increasing levels of DHT at 0.1, 1 or 10 nM. Fig. 3A shows that both Prx1 siRNA 143701 and 11838 essentially abolished the expression of Prx1. Interestingly, there was a significant drop in the amount of α-tubulin in the siRNA-transfected cells. The same was true when we used actin or GAPDH as the loading control. This could be due to the fact that these transiently siRNA-transfected cells experienced ~30% decrease in growth (incidentally Prx1 is also a very important growth and survival factor). Aside from this complication, it can be seen from the results of Fig. 3B that both Prx1 siRNAs reduced the ARE-luciferase activity to the same level regardless of the concentration of DHT. The significance of the observation is discussed later. The ARE-luciferase activity in Fig. 3B was determined at 48 hr after DHT treatment. Similar results were obtained at 72 hr (data not shown). It should be noted that the experiment described in Fig. 3 was the only experiment in this paper using Prx1 siRNA-transfected cells. All subsequent experiments were again conducted with the stable Prx1 shRNA knockdown cells.
Fig. 3. Effect of transient Prx1 siRNA knockdown on DHT stimulation of AR activity in LNCaP cells.
Panel A, immunoblot of Prx1 in control and siRNA-transfected samples prepared for luciferase activity. Panel B, results of luciferase assay for AR transactivation in the presence of 0.1, 1 or 10 nM DHT. *P<0.05 compared to the corresponding control value.
Prx1 Increases DHT Binding Affinity of AR in LNCaP Cells
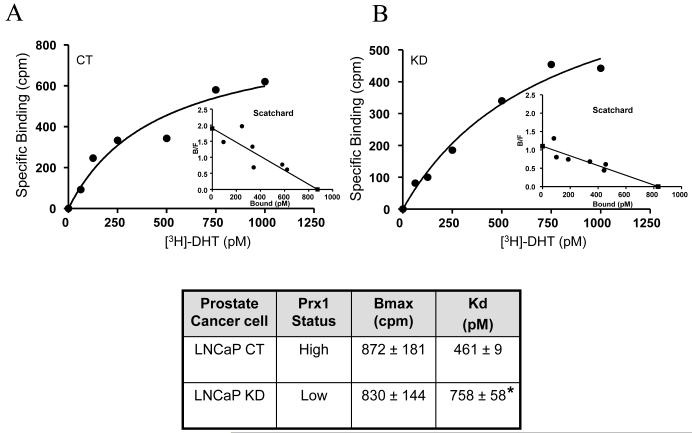
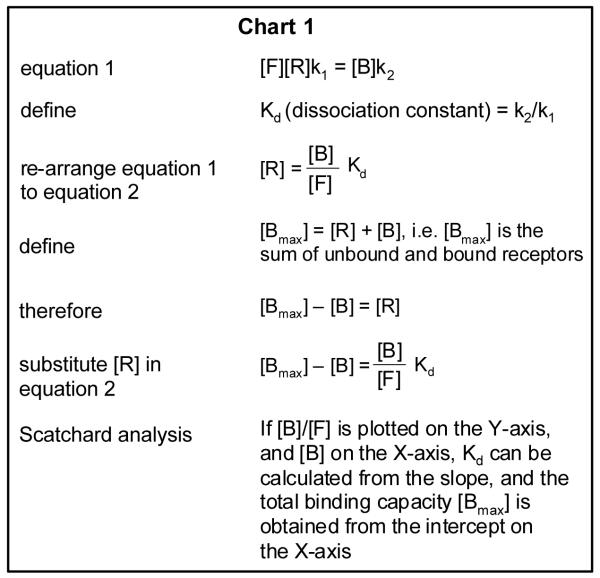
How might Prx1 facilitate AR transactivation? As noted in the Introduction, Prx1 interacts physically with AR. It is possible that this interaction may allow better access of DHT to the ligand binding pocket in AR. To test the hypothesis, we performed DHT binding assays in CT (high Prx1) or KD (low Prx1) cells. The specific binding data from representative experiments are shown in Fig. 4A and 4B. The raw data were transformed to a Scatchard plot, which provides two pieces of information: (i) the total binding capacity of AR in the cell (Bmax), as indicated by the intercept on the X-axis; and (ii) the dissociation constant (Kd) of DHT, which is derived from the slope of the plot and measures the affinity of AR to DHT. The details of the Scatchard analysis were described in the Methods. The chart in Fig. 4 summarizes the Bmax and Kd values from three separate experiments (mean ± SD). There was no change in total androgen binding capacity of AR (Bmax) between the CT and KD cells. The data are consistent with the AR Western blot results of Fig. 1B. On the other hand, the dissociation constant of DHT (Kd) was found to be significantly increased in the KD (low Prx1) cells. A greater Kd value means a lower affinity to DHT. In other words, the AR in the KD cells with a low level of Prx1 requires more DHT for activation than the AR in the CT cells with a high level of Prx1. The above conclusion is again consistent with the reduced sensitivity of the KD cells to DHT stimulation of growth and target gene expression, as demonstrated in Fig. 1 and 2.
Fig. 4. Effect of Prx1 knockdown on [3H]DHT specific binding in LNCaP cells.
Panel A, Control (CT) cells; Panel B, Prx1 knockdown (KD) cells. The accompanying table summarizes the Bmax and Kd values (derived from Scatchard analysis) of CT and KD cells (mean ± SD, n=3). *P<0.05 compared to the control.
Prx1 Increases DHT Binding Affinity of AR in PC3M Cells
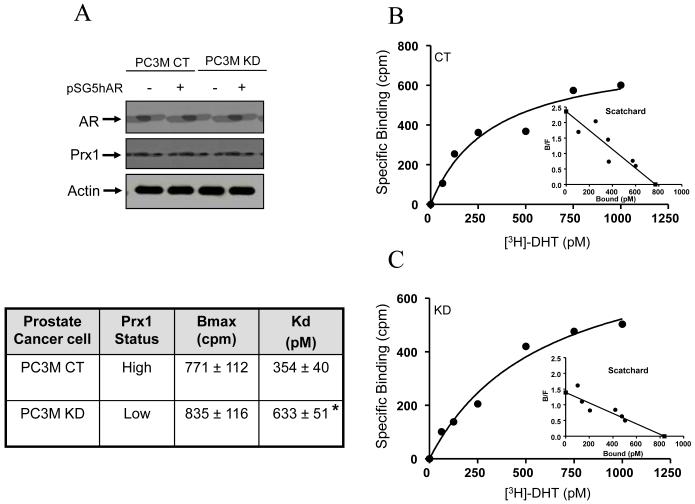
LNCaP cells contain a mutant AR. In order to verify that the effect of Prx1 on AR is not unique to LNCaP cells or the mutant AR, we carried out a parallel study with the PC3M prostate cancer cells. We developed stable Prx1 CT and KD subsets with the PC3M cells. Since there is no AR expression in the PC3M cells, we transfected them with the wild-type AR (pSG5hAR). The Western blots of AR and Prx1 expression of the four cell types (CT, CT/AR+, KD, KD/AR+) are shown in Fig. 5A. The data confirm the successful knock-in of AR. More importantly, the introduction of an exogenous AR did not affect Prx1 expression in either the CT or KD subset.
Fig. 5. Effect of Prx1 knockdown on [3H]DHT specific binding in PC3M cells transfected with the wild-type AR (pSG5hAR).
Panel A, Western blots of AR and Prx1 expression in the CT, CT/AR+, KD and KD/AR+ subtypes. Panels B and C, [3H]DHT specific binding in CT/AR+ and KD/AR+ cells, respectively. The accompanying table summarizes the Bmax and Kd values of CT and KD cells (mean ± SD, n=3). *P<0.05 compared to the control.
Once the models were validated, we used only the CT/AR+ and KD/AR+ cells for the DHT specific binding assays. The results from representative experiments are shown in Fig. 5B and 5C. The Bmax and Kd data (mean ± SD, n=3) are summarized in the accompanying chart. There was no change in the total binding capacity of AR. However, the dissociation constant (Kd) was increased from 354 pM in the CT cells (high Prx1) to 633 pM in the KD cells (low Prx1), suggesting that Prx1 plays a critical role in enhancing the affinity of AR to DHT. The results from the AR+/PC3M cells are in complete agreement with that from the LNCaP cells, thus supporting the idea that the effect of Prx1 is manifested with both the wild-type and mutant AR.
Prx1 Decreases Rate of DHT Dissociation from AR
In addition to increasing AR affinity to DHT, Prx1 may also influence the rate of DHT dissociation from the receptor. The occupancy time of DHT in the DHT-AR complex is generally quantified by the half-life (t½) of the bound DHT in the dissociation curve. We carried out such a study with the CT and KD cells to see if Prx1 might modulate the dissociation rate of DHT. Both the LNCaP and AR+/PC3M models were used. Representative dissociation curves from each model are shown in Fig. 6. The mean t½ values from 3 independent experiments are indicated at the bottom of each panel. In the LNCaP model, the t½ was 225 min in the CT cells and 173 min in the KD cells. In the AR+/PC3M model, the t½ was 218 min in the CT cells and 168 min in the KD cells. The results thus support the conclusion that a low level of Prx1 hastens the rate of DHT dissociation from AR, and that this effect is evident regardless of whether the AR is wild-type (in AR+/PC3M cells) or a mutant (in LNCaP cells).
Fig. 6. Effect of Prx1 knockdown on dissociation rate of bound [3H]DHT.
Panel A, LNCaP cells. Panel B, AR+/PC3M cells. The accompanying tables summarize the half-life (t½) of the dissociation curve (mean ± SD, n=3) of CT and KD cells. *P<0.05 compared to the control.
Prx1 Enhances N-C Dimerization of AR
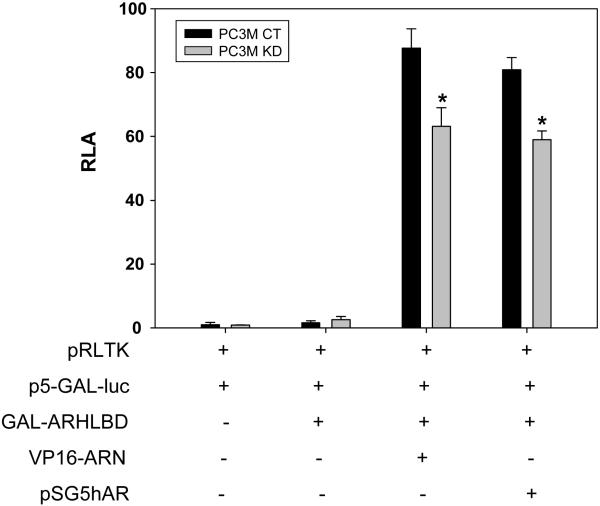
As noted in the Introduction, the N-terminus of one AR molecule associates with the C-terminus of another AR molecule after ligand binding. The N-C dimerization of AR holds the ligand in the ligand binding pocket (20, 21). A mammalian two-hybrid system was used to evaluate the intensity of N-C interaction in intact cells. The GAL DNA binding domain and VP16 transactivation domain fusion proteins were expressed as described in Methods. The p5-GAL-LUC reporter construct was co-transfected into the cells. The interaction of GAL-ARHLBD with VP16-ARN generates a luciferase signal which serves as a readout of N-C interaction between the AF-2 motif in the GAL-AR fragment and AF-1 in the N-terminal domain of VP16-ARN. We carried out the experiment with the PC3M CT and KD cells. The results are shown in Fig. 7. In the absence of VP16-ARN, GAL-ARHLBD alone did not generate any significant luciferase activity. The simultaneous presence of GAL-ARHLBD and VP16-ARN produced a very robust luciferase signal above the background in the CT cells. However, the knockdown of Prx1 reduced the intensity of the signal by about 50%. Introduction of the wild-type AR (pSG5hAR) in lieu of VP16-ARN gave essentially the same results. The data therefore suggest that decreasing the availability of Prx1 in the KD cells negatively regulates AR N-C interaction. The above conclusion is consistent with the increased rate of DHT dissociation in the KD cells as shown in Fig. 6.
Fig. 7. Effect of Prx1 knockdown on AR N-C interaction.
Increases of relative luciferase activity (shown on the Y-axis) were used as the readout of N-C interaction. pRLTK, Renilla luciferase activity for adjustment of transfection efficiency; p5-GAL-LUC, N-C interaction driven luciferase reporter; GAL-ARHLBD and VP16-ARN, components of the two-hybrid system for N-C interaction; pSG5hAR, wild-type AR plasmid. *P<0.05 compared to the control.
Prx1 Mediated Augmentation of AR N-C Interaction is not Unique to Prostate Cancer Cells and is Independent of the Antioxidant Activity of Prx1
In order to rule out that the N-C interaction promoting effect of Prx1 is unique only to prostate cancer cells, we conducted similar experiments with the A549 lung cancer cells and the MCF-7 breast cancer cells. These two cell models were chosen because they have a moderate level of endogenous Prx1. The abundance of Prx1 in each cell model was further boosted by transfection with either the wild-type Prx1 or the C52S mutant Prx1. The mutant Prx1 lacks antioxidant activity (10). The results of the N-C interaction driven luciferase reporter assay are shown in Fig. 8. Since A549 and MCF-7 cells naturally express Prx1, transfection of more wild-type Prx1 generated only a slight increase of the luciferase signal as compared to the empty vector transfection. Interestingly, transfection with the C52S mutant Prx1 produced essentially the same results as the wild-type Prx1 transfection, suggesting that the antioxidant activity of Prx1 is not important for Prx1 to promote AR N-C interaction.
Fig. 8. Effect of wild-type (wt) or mutant (mut) Prx1 transfection on AR N-C interaction in A549 or MCF-7 cells.
Increases of relative luciferase activity (shown on the Y-axis) were used as the readout of N-C interaction (refer to legend of Fig. 6). The mutant Prx1 lacks antioxidant activity.
Discussion
A functional AR continues to play a critical role in the transcriptional control and proliferative growth of prostate cancer after androgen deprivation therapy. Past research was focused mainly on AR amplification or mutation as an adaptive mechanism in sensitizing cells to a low androgen environment (22). Recent interest, however, has shifted to distinct molecular factors which are capable of modulating AR activity (23). These factors include chaperones, co-activators and non-steroidal ligands. Prx1 is fast emerging as a viable candidate. The increasing attention on Prx1 is justified by several lines of supportive evidence. First, Prx1 interacts physically with AR and co-localizes with AR to the nucleus in the transcriptional control of AR target genes (10). Second, the expression of Prx1 is up-regulated by hypoxia (16), the latter is a common occurrence in androgen-sensitive tissues when the supply of androgen is interrupted. This context-dependent phenomenon thus takes into consideration the microenvironmental changes which are taking place in the tissue. Third, AR-positive prostate cancer cells which are able to sustain growth in a very low androgen condition generally show an elevated level of Prx1 (10), suggesting that Prx1 may help boost AR signaling. To put things in perspective, Prx1 is not the only chaperone protein known to increase AR function. A recent study showed that heat shock protein 27 (hsp27) also interacts with AR to enhance AR stability, nuclear shuttling and transcriptional activity (24). However, it is unclear whether hsp27 can sensitize AR to low levels of DHT, a characteristic which seems to be unique to Prx1. Another example is the role of a chaperone/co-chaperone complex, hsp70-Bag-IL, in regulating the transcriptional activity of AR by enhancing its binding to the androgen response element (25).
How might Prx1 affect AR activation, especially when the availability of DHT is limiting? Our study showed that Prx1 increases AR affinity to DHT and AR N-C interaction. The purpose of N-C interaction presumably is to retain DHT in the ligand binding pocket (21, 26). A stronger N-C interaction means a more stable DHT-AR complex, and this is precisely what we found from the bound DHT dissociation curve when Prx1 is present in abundance. It is noteworthy to point out that the enhancement of AR activation by Prx1, as measured by a number of biochemical parameters, is reflected in DHT stimulation of cell growth and transcription of AR targets. The stimulatory effect of Prx1 on AR is manifested regardless of a wild-type or mutant AR. The observation has significant implications because a substantial proportion of prostate cancer carries mutations on the AR gene (27). Incidentally, the AR T877A mutation in LNCaP cells is not acquired due to cell passaging, and is found sporadically in human prostate cancer specimens (28).
The availability of DHT and the time of occupancy of DHT on the receptor are important determinants of AR activity. If the affinity of AR to DHT remains unchanged, decreases in binding would be expected with exposure to low levels of DHT, since the binding equilibrium is governed by the following equation: free bound DHT-AR complex, where Kd (dissociation constant) = k2/k1. Studies with AR deletion mutants showed that the interaction between the N-terminus and C-terminus of AR stabilizes the DHT-AR complex and slows down the dissociation rate of bound DHT (26). By associating physically with AR, Prx1 is likely to cause a conformational change in AR, and in doing so, increases the affinity of AR to DHT, and simultaneously stabilizes the DHT-AR complex so that AR stays activated longer despite operating in a low androgen environment. The ability of Prx1 to modulate the activity of other cellular proteins through a physical interaction mechanism has been described in a number of publications (29-31).
In our DHT dose response experiment shown in Fig. 1, we treated cells with up to 1 nM DHT. This concentration of DHT is similar to the level found in recurrent prostate cancer after castration (32). Some of the DHT may be the product of de novo synthesis by prostate cancer cells, as described in several recent publications (33, 34). Regardless of the availability of Prx1, 1 nM DHT is clearly adequate for AR activation. It is interesting to note that based on our results shown in Fig. 1, 0.1 nM DHT in a high Prx1 environment (i.e. CT cells) produced the same magnitude of growth stimulation as 1 nM DHT in a low Prx1 environment (i.e. KD cells). We do not wish to belabor the issue of the threshold level of DHT needed to fully activate AR when there is an abundance of Prx1. The evidence is sufficiently compelling that Prx1 enables AR to function in a normal or near normal capacity even if the availability of DHT is suboptimal.
Based on the experiment described in Fig. 3B, it can be seen that Prx1 knockdown decreases AR activity to the same extent in the presence of either 1 or 10 nM DHT. If DHT binding affinity to AR is the only thing modulated by Prx1, one would expect that the decrease of AR activity by Prx1 knockdown would be more pronounced at 1 nM DHT than at 10 nM DHT. This is clearly not the case. Prx1 may have multiple roles in activating AR. Previous reports have shown that the steroid receptor coactivator-1, or SRC-1, interacts with the AF-1 (N-terminus) and AF-2 (C-terminus) domains of AR (35, 36), and that SRC-1 potentiates the activity of AR at suboptimal levels of androgen (37). It is therefore tempting to hypothesize that Prx1 may also stimulate the recruitment of SRC-1 to the ligand-bound AR complex. Androgen deprivation therapy is often accompanied by hypoxic conditions in the prostate tumor. Prx1 is up-regulated rapidly by hypoxia (16). Consequently, Prx1 may allow the cancer cells to continue to rely on AR signaling despite the stress of androgen ablation, without having to undergo selection pressure for other adaptive changes. As such, Prx1 may represent a legitimate therapeutic target in controlling androgen-refractory prostate cancer.
Materials and Methods
Cell Cultures
Prostate LNCaP, prostate PC3M, breast MCF-7 and lung A549 cancer cell lines were obtained from the American Type Culture Collection. All cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mmol/L of glutamine, 100 units/mL of penicillin, and 100 μg/mL of streptomycin at 37°C in an atmosphere of 5% CO2 and 95% air.
Cell Growth Assay
LNCaP Cells were seeded at 2 × 103 per well in 96-well plates and allowed to grow for 24 h. Cells were washed with PBS, then switched to a phenol red-free medium containing 10% charcoal-stripped FBS (Hyclone) and supplemented with 0, 0.1 or 1 nM dihydrotestosterone (DHT; Sigma). Growth rate was measured every other day for 5 days by the MTT assay as described in our previous publication (38).
Western Blot Analysis
Equal amounts of protein were analyzed in duplicate by SDS-PAGE. Protein concentrations were measured by the BCA protein assay kit as per manufacturer’s protocol (Pierce). The following monoclonal antibodies were used: anti-Prx1 (Lab Frontier), anti-AR (BD Pharmingen), anti-PSA (Neomarker), anti-KLK2 (Abcam), cyclin D1 (Santa Cruz), anti-β-actin (Sigma), and anti-α-tubulin (Upstate). Immunoreactive proteins were detected with a HRP-conjugated secondary antibody (Biorad) and visualized by using an enhanced chemiluminescence detection system (Amersham Bioscience).
Real-Time Reverse Transcription-PCR
First-strand cDNA was synthesized from 100 ng of total RNA by SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. The PCR primers for GAPDH, PSA, and KLK2 were Assays-on-Demand products from Invitrogen. An aliquot of 2 μl of first-strand cDNA was added to 25 μl of SYBR GreenER™ qPCR Supermix for ABI PRISM (Invitrogen) and 2.5 μl of 100 pM primer mixture in a final 50μl volume. Temperature cycling and real-time fluorescence measurement were performed with an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The PCR conditions were as follows: an initial incubation at 50°C for 2 min, then a denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The relative quantitation of gene expression was computed by using the comparative Ct (ΔΔCt) method (39). Briefly, the threshold cycle number (Ct) was obtained as the first cycle at which a statistically significant increase in fluorescence signal was detected. Data normalization was carried out by subtracting the Ct value of GAPDH from that of the target gene. The ΔΔCt was calculated as the difference between the normalized Ct values (ΔCt) of the treatment and control samples. Lastly, ΔΔCt was converted to fold of change by the following formula: Fold of change = 2-ΔΔCt.
Prx1 Knockdown by shRNA
LNCaP and PC3M cells were subjected to Prx1 knockdown. The pSilencer 5.1 system (Ambion) was used for the expression of short hairpin RNA (shRNA). The sense and antisense sequences of oligonucleotides targeting human Prx1 are: 5′-GATCCGTTCTCACTTCTGTCATCTATTCAAGAGATAGATGACAGAAGTGATAATTTT TTGGAAA-3′, and 5′-AGCTTTTCCAAAAAATTCTCACTTCTGTCATCTATCTCTTGAATAGATGACAGAAGT GAGAACG-3′, respectively. The sense and antisense scrambled sequences are: 5′-GATCCCGTTCTCCGAACGGTGCACGTTTCAAGAGAACGTGCACCGTTCGGAGAAT TTTTTGGAAA-3′, and 5′-AGCTTTTCCAAAAAATTCTCCGAACGGTGCACGTTCTCTTGAAACGTGCACCGTTC GGAGAACGG-3′, respectively. Both the shRNA and scrambled sequences were checked by searching the genome database (BLAST) to ensure that they do not share any significant sequence homology with other human genes. Oligonucleotides were annealed, and cloned into the pSilencer vector. The sequence accuracy of the constructs was confirmed with an ABI 3700 capillary sequencer (Applied Biosystems). Phoenix packaging cells were transfected with either the Prx1-shRNA or scrambled-shRNA expression vector by using LipofectAMINE 2000 reagents (Invitrogen). The culture supernatants were collected at 48 h after transfection and filtered. Prostate cancer cells were infected with either the Prx1-shRNA or scrambled-shRNA viral preparation in the presence of 4 μg/mL of polybrene (Sigma). Fresh viral suspensions were added to the cells every 8 h during the next 48 h. Infected cells were selected in a growth medium containing 250 μg/mL of neomycin. Cells expressing the scrambled-shRNA or Prx1-shRNA are designated as CT (for control) or KD (for knockdown), respectively.
Prx1 Knockdown by siRNA
All materials for siRNA transfection were purchased from Ambion. Transient transfection of siRNA was done using a protocol recommended by the manufacturer. The two siRNA sequences, which were obtained online from the Ambion siRNA library (ID number 143701 and 11838), were directed against exon 6 of Prx1. The information f o r t h e t w o s i R N A s i s a s f o l l o w s: s i R N A 1 4 3 7 0 1, sense: 5′GCCGAAUUGUGGUGUCUUATT-3′, antisense: 5′-UAAGACACCACAAUUCGGCTG-3′; siRNA 11838, sense: 5′-GGCUACUGGUUUGUAUGAUTT-3′, antisense: 5′-AUCAUACAAACCAGUAGCCTG-3′. Non-silencing siRNA sequence was used as the negative control. All siRNAs, obtained in lyophilized and annealed form, were resuspended in diethylpyrocarbonate-treated distilled water to achieve a stock concentration of 15 μM, and stored at -20°C in 50-μl aliquots. Approximately 5 × 105 LNCaP cells were plated onto 6-well plates to achieve 40% to 50% confluency. For each well, 5 μl of Lipofectamin 2000 transfection Reagent (Invitrogen) was incubated with 100 μl of fresh medium. The ARE-luciferase reporter plasmid, which contains three repeats of the ARE region ligated in tandem to the luciferase reporter (40), along with pRLTK plasmid (for Renilla luciferase activity), were incubated with 100 μl of fresh medium with or without 75 nM of siRNA for 5 min. The transfection reagent and the plasmid-siRNA mixture were then combined and incubated for 25 min at room temperature to allow complex formation. After adding 2 ml of fresh media to the cells, the siRNA-transfection reagent complexes were added drop-wise. Transfected cells were cultured with various DHT concentrations for 48 h at 37°C in 5% CO2 and harvested for luciferase assay as described in the following section.
Plasmids and Expression Vectors
The pSG5hAR expression vector was originally described by Yeh and Chang (41). We have used this vector in our previous publications (9,10). The pSG5hAR vector was used to express the wild-type AR in PC3M cells, which are AR-null. A mammalian two-hybrid system consisting of the N-terminus of AR fused with VP16 (VP16-ARN), and the C-terminal hinge and ligand binding domain of AR fused with GAL-4-DBD (GAL-ARHLBD), was described previously by Onate (35). Included in the system is the p5-GAL-LUC reporter construct, which generates a luciferase signal as the readout for AR N-C interaction. For the over-expression of Prx1 in MCF-7 and A549 cells, pCR3.1 plasmids containing either the wild-type Prx1 or mutant Prx1 (pCR C52S, lacking antioxidant activity) were described previously (42).
Transfection for AR N-C Interaction and Luciferase Assay
An aliquot of 5 × 104 cells was placed in a 24-well plate and transfected with DNA (described in Plasmids and Expression Vectors) using Fugene 6 reagent (Roche). The total amount of plasmid DNA was normalized to 200 ng/well by the addition of empty plasmid. Luciferase activities were measured after 24 h of transfection by using the Luciferase Assay System (Promega). Relative luciferace activities (RLA) were calculated by normalizing against the Renilla luciferase activity from the pRLTK plasmid. All transfection experiments were repeated at least three times.
Scatchard Analysis and Androgen Dissociation Assays
LNCaP or PC3M (transfected with the pSG5hAR expression vector) clones were cultured in a medium containing charcoal stripped-FBS for 24 h prior to the start of the experiment. For the determination of apparent equilibrium binding affinity, whole cell binding assays were performed as previously described (43). Briefly, cells were cultured to confluency, and then incubated for 2 h at 37°C with increasing concentrations (0.06-1.0 nM) of 3H-DHT (Amersham Biosciences), either in the absence or presence of 200-fold excess of nonradioactive DHT. Cells were then placed on ice, washed four times with ice-cold PBS, and subsequently lysed in 0.5 M NaOH. Radioactivity in the lysate was measured in a scintillation counter. Scatchard analysis was carried out to determine the dissociation constant (Kd) by using the Graph Pad Prism5 software. The dissociation constant measures the affinity of AR to DHT.
The derivation of the Scatchard plot is described below. It starts with the concept that free DHT, the unbound AR, and the bound DHT-AR complex are in a state of equilibrium as represented by the following: , where F = free DHT, R = unbound receptor, B = bound receptor complex, k1 = rate constant of association, and k2 = rate constant of dissociation. The equilibrium can be expressed in an equation form as shown by equation 1 in Chart 1. The subsequent steps detailed in Chart 1 describe how the various binding parameters are obtained from the Scatchard plot analysis.
The stability of the AR-DHT complex was quantified by a different assay as follows. DHT dissociation rates were determined by incubating cells with 3 nM 3H-DHT for 2 h at 37°C, followed by a chase with 6 μM nonradioactive DHT for different time periods up to 240 min. Cells were then washed and lysed, and radioactivity was determined as described above. The data are presented as the natural logarithmic ratio (ln) of bound radioactivity at a given time (B) to that at time zero (B0) (44). Nonspecific binding was determined in parallel experiments in which the cells were exposed to 200-fold excess of cold DHT during the entire incubation period.
Statistical Analysis
Statistical significance was evaluated by using Student’s t tests. Values were reported as means ± SD. P<0.05 is considered significant.
Acknowledgments
Grant Support: NIH/NCI P01 CA126804 (C. Ip, P.I.) and partially supported by shared resources of NIH/NCI P30 CA16056 (Roswell Park Cancer Center Support Grant).
Abbreviations
- AR
Androgen Receptor
- DHT
Dihydrotestosterone
- Prx 1
Peroxoredoxin 1
- ADT
Androgen Deprivation Therapy
- PSA
Prostate Specific Antigen
- KLK2
Kallikrein 2
- shRNA
short hairpin RNA
- siRNA
small interference RNA
- CT
Cells expressing scrambled-shRNA
- KD
Cells expressing Prx1-shRNA for its KnockDown
- Ct
Threshold Cycle
- RLA
Relative Luciferase Activity
- Kd
Dissociation Constant
- t½
Half-life of receptor bound DHT
- ARE
Androgen Responsive Element
- SD
Standard Deviation
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Brinkmann AO, Trapman J. Genetic analysis of androgen receptors in development and disease. Adv Pharmacol. 2000;47:317–41. doi: 10.1016/s1054-3589(08)60115-5. [DOI] [PubMed] [Google Scholar]
- 2.Santen RJ. Clinical review 37: Endocrine treatment of prostate cancer. J Clin Endocrinol Metab. 1992;75:685–9. doi: 10.1210/jcem.75.3.1517354. [DOI] [PubMed] [Google Scholar]
- 3.Di Lorenzo G, De Placido S. Hormone refractory prostate cancer (HRPC): present and future approaches of therapy. Int J Immunopathol Pharmacol. 2006;19:11–34. [PubMed] [Google Scholar]
- 4.Buchanan G, Irvine RA, Coetzee GA, Tilley WD. Contribution of the androgen receptor to prostate cancer predisposition and progression. Cancer Metastasis Rev. 2001;20:207–23. doi: 10.1023/a:1015531326689. [DOI] [PubMed] [Google Scholar]
- 5.Grossmann ME, Huang H, Tindall DJ. Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst. 2001;93:1687–97. doi: 10.1093/jnci/93.22.1687. [DOI] [PubMed] [Google Scholar]
- 6.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 7.Pienta KJ, Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res. 2006;12:1665–71. doi: 10.1158/1078-0432.CCR-06-0067. [DOI] [PubMed] [Google Scholar]
- 8.Jain RK, Safabakhsh N, Sckell A, et al. Endothelial cell death, angiogenesis, and microvascular function after castration in an androgen-dependent tumor: role of vascular endothelial growth factor. Proc Natl Acad Sci U S A. 1998;95:10820–5. doi: 10.1073/pnas.95.18.10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park SY, Kim YJ, Gao AC, et al. Hypoxia increases androgen receptor activity in prostate cancer cells. Cancer Res. 2006;66:5121–9. doi: 10.1158/0008-5472.CAN-05-1341. [DOI] [PubMed] [Google Scholar]
- 10.Park SY, Yu X, Ip C, Mohler JL, Bogner PN, Park YM. Peroxiredoxin 1 interacts with androgen receptor and enhances its transactivation. Cancer Res. 2007;67:9294–303. doi: 10.1158/0008-5472.CAN-07-0651. [DOI] [PubMed] [Google Scholar]
- 11.Fujii J, Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002;7:123–30. doi: 10.1179/135100002125000352. [DOI] [PubMed] [Google Scholar]
- 12.Yanagawa T, Ishikawa T, Ishii T, et al. Peroxiredoxin I expression in human thyroid tumors. Cancer Lett. 1999;145:127–32. doi: 10.1016/s0304-3835(99)00243-8. [DOI] [PubMed] [Google Scholar]
- 13.Yanagawa T, Iwasa S, Ishii T, et al. Peroxiredoxin I expression in oral cancer: a potential new tumor marker. Cancer Lett. 2000;156:27–35. doi: 10.1016/s0304-3835(00)00434-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Bogner PN, Baek SH, et al. Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clin Cancer Res. 2008;14:2326–33. doi: 10.1158/1078-0432.CCR-07-4457. [DOI] [PubMed] [Google Scholar]
- 15.Chen MF, Keng PC, Shau H, et al. Inhibition of lung tumor growth and augmentation of radiosensitivity by decreasing peroxiredoxin I expression. Int J Radiat Oncol Biol Phys. 2006;64:581–91. doi: 10.1016/j.ijrobp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–54. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 17.Doesburg P, Kuil CW, Berrevoets CA, et al. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–64. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 18.Horoszewicz JS, Leong SS, Kawinski E, et al. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–18. [PubMed] [Google Scholar]
- 19.Veldscholte J, Ris-Stalpers C, Kuiper GG, et al. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem Biophys Res Commun. 1990;173:534–40. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 20.Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–8. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 21.He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J Biol Chem. 1999;274:37219–25. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- 22.Li TH, Zhao H, Peng Y, Beliakoff J, Brooks JD, Sun Z. A promoting role of androgen receptor in androgen-sensitive and -insensitive prostate cancer cells. Nucleic Acids Res. 2007;35:2767–76. doi: 10.1093/nar/gkm198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards J, Bartlett JM. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 1: Modifications to the androgen receptor. BJU Int. 2005;95:1320–6. doi: 10.1111/j.1464-410X.2005.05526.x. [DOI] [PubMed] [Google Scholar]
- 24.Zoubeidi A, Zardan A, Beraldi E, et al. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res. 2007;67:10455–65. doi: 10.1158/0008-5472.CAN-07-2057. [DOI] [PubMed] [Google Scholar]
- 25.Shatkina L, Mink S, Rogatsch H, et al. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol Cell Biol. 2003;23:7189–97. doi: 10.1128/MCB.23.20.7189-7197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou ZX, Lane MV, Kemppainen JA, French FS, Wilson EM. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol Endocrinol. 1995;9:208–18. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
- 27.Brooke GN, Parker MG, Bevan CL. Mechanisms of androgen receptor activation in advanced prostate cancer: differential co-activator recruitment and gene expression. Oncogene. 2008;27:2941–50. doi: 10.1038/sj.onc.1210955. [DOI] [PubMed] [Google Scholar]
- 28.Shi XB, Ma AH, Xia L, Kung HJ, de Vere White RW. Functional analysis of 44 mutant androgen receptors from human prostate cancer. Cancer Res. 2002;62:1496–502. [PubMed] [Google Scholar]
- 29.Jung H, Kim T, Chae HZ, Kim KT, Ha H. Regulation of macrophage migration inhibitory factor and thiol-specific antioxidant protein PAG by direct interaction. J Biol Chem. 2001;276:15504–10. doi: 10.1074/jbc.M009620200. [DOI] [PubMed] [Google Scholar]
- 30.Mu ZM, Yin XY, Prochownik EV. Pag, a putative tumor suppressor, interacts with the Myc Box II domain of c-Myc and selectively alters its biological function and target gene expression. J Biol Chem. 2002;277:43175–84. doi: 10.1074/jbc.M206066200. [DOI] [PubMed] [Google Scholar]
- 31.Wen ST, Van Etten RA. The PAG gene product, a stress-induced protein with antioxidant properties, is an Abl SH3-binding protein and a physiological inhibitor of c-Abl tyrosine kinase activity. Genes Dev. 1997;11:2456–67. doi: 10.1101/gad.11.19.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohler JL, Gregory CW, Ford OH, 3rd, et al. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10:440–8. doi: 10.1158/1078-0432.ccr-1146-03. [DOI] [PubMed] [Google Scholar]
- 33.Locke JA, Guns ES, Lubik AA, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 2008;68:6407–15. doi: 10.1158/0008-5472.CAN-07-5997. [DOI] [PubMed] [Google Scholar]
- 34.Montgomery RB, Mostaghel EA, Vessella R, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–54. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onate SA, Boonyaratanakornkit V, Spencer TE, et al. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–8. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 36.Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19:8383–92. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agoulnik IU, Vaid A, Bingman WE, 3rd, et al. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–67. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 38.Chhipa RR, Kumari R, Upadhyay AK, Bhat MK. Abrogation of p53 by its antisense in MCF-7 breast carcinoma cells increases cyclin D1 via activation of Akt and promotion of cell proliferation. Exp Cell Res. 2007;313:3945–58. doi: 10.1016/j.yexcr.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Dong Y, Zhang H, Gao AC, Marshall JR, Ip C. Androgen receptor signaling intensity is a key factor in determining the sensitivity of prostate cancer cells to selenium inhibition of growth and cancer-specific biomarkers. Mol Cancer Ther. 2005;4:1047–55. doi: 10.1158/1535-7163.MCT-05-0124. [DOI] [PubMed] [Google Scholar]
- 41.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YJ, Lee WS, Ip C, Chae HZ, Park EM, Park YM. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66:7136–42. doi: 10.1158/0008-5472.CAN-05-4446. [DOI] [PubMed] [Google Scholar]
- 43.Umar A, Berrevoets CA, Van NM, et al. Functional analysis of a novel androgen receptor mutation, Q902K, in an individual with partial androgen insensitivity. J Clin Endocrinol Metab. 2005;90:507–15. doi: 10.1210/jc.2004-0057. [DOI] [PubMed] [Google Scholar]
- 44.Dubbink HJ, Hersmus R, Verma CS, et al. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol Endocrinol. 2004;18:2132–50. doi: 10.1210/me.2003-0375. [DOI] [PubMed] [Google Scholar]