Abstract
Deciphering olfactory encoding requires a thorough description of the ligands that activate each odorant receptor (OR). In mammalian systems, however, ligands are known for fewer than 50 of over 1400 human and mouse ORs, greatly limiting our understanding of olfactory coding. We performed high-throughput screening of 93 odorants against 464 ORs expressed in heterologous cells and identified agonists for 52 mouse and 10 human ORs. We used the resulting interaction profiles to develop a predictive model relating physicochemical odorant properties, OR sequences, and their interactions. Our results provide a basis for translating odorants into receptor neuron responses and unraveling mammalian odor coding.
INTRODUCTION
Odorant receptors (ORs) in the olfactory sensory neurons of the nasal epithelium translate odorants into neural signals. Each OR is thought to be specialized to recognize physicochemical features, such as functional groups or molecular size, of odorant molecules; these features are then translated into a neural signal that in turn leads to an olfactory perception. The physicochemical features of an odorant, therefore, are a key determinant of the olfactory percept. The rules governing the translation of molecule into percept, however, remain largely unknown. To develop a theory that predicts olfactory percept from molecular structure, we must first be able to predict OR activation from molecular structure.
Mammalian ORs are heterotrimeric guanine nucleotide-binding (G-protein)-coupled receptors localized to the cell-surface membrane at the tips of olfactory sensory neuron dendrites (1). They comprise a multigene family; there are approximately 1035 and 387 putatively functional ORs in mice and humans, respectively (1-4), making it one of the largest gene families in the mammalian genome (5-7). The overall sequences of mammalian ORs are diverse, with amino acid sequence similarity between different ORs ranging from less than 40% to more than 90%. Based on phylogenetic analysis, OR family members can be divided into two subclasses, class I and class II (4,8). Most currently identified fish ORs belong to class I (but see 9), whereas amphibians and mammals express ORs of both classes (8). The dolphin Stenella Coeruleoalba has both class I and class II ORs, but the sequenced class II ORs are all pseudogenes. The preferential expression of class I ORs in marine mammals and fish suggests that water-soluble odorants may be the preferred agonists of class I but not class II ORs (10).
Elucidation of general properties of the olfactory system, such as determination of the general similarity between two odorants or two ORs, requires investigation of a wide panel of diverse ORs with a large number of chemically diverse odorants in a consistent assay. One of the roadblocks to studying receptor responses directly has been the inability to express ORs in heterologous systems suitable for high-throughput screening or mutational analysis. Agonists have been identified for fewer than 50 mammalian receptors in heterologous systems; moreover, these ORs were tested with various assay systems and odorant sets (11-24), greatly complicating any attempts to unify their analysis. Previously, we developed a system to determine odorant activation profiles for ORs using a set of molecules that induce the expression of mouse and human ORs in heterologous cells (17,21,25). Here, we leveraged this high-throughput system to identify agonists for a large number of mammalian ORs. We tested the responses of 219 mouse and 245 human receptors to a panel of 93 odorants, identifying 340 receptor-ligand interactions, with 62 ORs responding to at least one agonist. This study is the first to examine the interaction of a large number of mammalian ORs with a diverse set of odorants. We used these findings to address several distinct questions for which the lack of functional data has impeded progress, such as quantification of odorant similarity, quantification of receptor similarity, examination of the differences between class I and class II ORs, examination of the differences between human and mouse ORs, and development of a model to predict odorant-receptor activation.
RESULTS
High-throughput screening of mouse and human ORs
We generated libraries of mouse and human ORs that represent a large fraction of the total mouse and human OR families (Table S1). Our mouse OR library comprises 219 mouse ORs that represent over 21% of the total 1035 mouse OR genes and includes at least one member of 217 out of the 228 mouse OR subfamilies defined by Zhang and Firestein (4). Our human OR library comprises 245 ORs that represent 63% of the 387 human OR genes. Although many human OR genes are polymorphic (11,21,26), we used a single variant of each receptor in the screen (Table S1).
We stimulated the entire OR library with one of 8 odorant mixtures drawn from 93 odorants chosen to represent diverse functional groups, sizes, and structures (Table S2). We applied each mixture at five different concentrations. 121 human ORs (49.4%) and 169 mouse ORs (77.2%) showed a response to at least one mixture at one concentration. We then applied the 93 odorants individually at 100 μM to the 290 mixture-responsive ORs. 27 human ORs (11.0%) and 102 mouse ORs (46.6%) showed a significant response [p<0.05, uncorrected for multiple comparisons]to at least one of 67 odorants relative to a no-odor control. We then constructed dose-response curves for every combination of 129 receptors and 67 odorants that showed a response significantly above baseline in the previous step. In this more stringent test, we identified 52 mouse and 10 human ORs that responded to one or more of 63 odorants (Fig. 1, also see Fig. S1 to S3 and Table S3), representing 23.7% of the original mouse library and 4% of the original human library. The additional four odorants and 67 receptors did not elicit or show a significant response in this follow-up. Although our positive responses resulted in discovery of a large number of OR agonists, the failure of a specific OR to respond to any of the tested odorants may reflect a failure of the OR to function in our assay rather than a lack of sensitivity to the tested odorant. In further analyses we only looked at results for receptors that responded to at least one of the tested odorants, and were therefore unequivocally functional in our assay.
Figure 1.
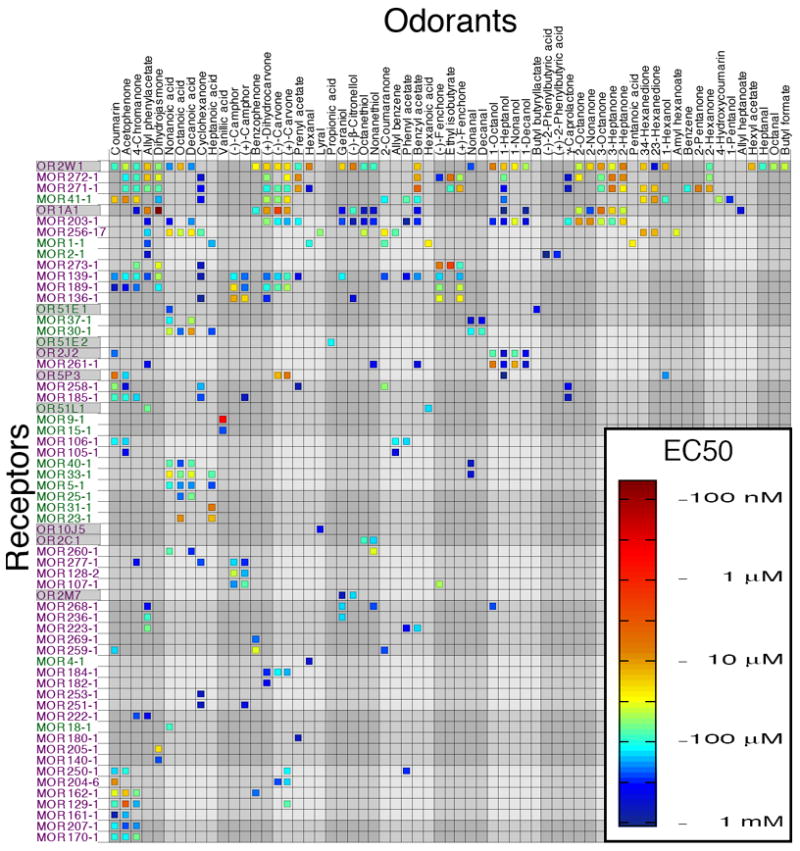
EC50 values for 62 odorant receptors and 63 odorants. Class I receptors are shown in green, class II receptors in purple. Human odorant receptors have a gray background. Odorant and receptor order were determined independently by cluster analysis using the receptor response data such that the most similar odorants (and receptors) are next to each other on their respective axes. A second version of this figure, with odorant order determined by functional group and receptor order determined by number of agonists is included for comparison (see Fig. S10).
Bias in odorant and receptor representation
One problem in the field of olfaction research is the lack of an agreed-upon metric to organize odorants; for example there is currently no agreed-upon metric to quantify odorant similarity (27). Without such a metric it is difficult to choose a random sample of odorants that fairly represents all odorants, as all odorants have discrete molecular structures that differ in such physicochemical properties as molecular weight, carbon number, functional groups, and hydrophobicity. A bias in odorant sampling may give a misleading estimate of a receptor’s preferred physicochemical features as well as lead to faulty extrapolation of odorant similarity. Due to the large number of physicochemical properties thought to determine odorant similarity, we use a broad descriptor set to show the bias in our chosen odorant set and then use principal component analysis (PCA) to simplify the organization of these odorants. PCA is a method for transforming a number of possibly correlated variables into a smaller number of uncorrelated variables.
To examine the bias in our odorant set, we constructed a 1664-dimensional space in which each dimension represents a physicochemical property. We plotted 2683 commercially available odorants (Table S4) in the resulting odorant space. A two-dimensional projection of this 1664-dimensional odorant space is shown in Fig. 2A. This is an incomplete representation of our bias in odor selection, because our choice of 2683 odorants is likely to itself be a biased representation of odorant space, but represents an intermediate between the limitations of our testing capability and the ideal case of sampling all odorous molecules.
Figure 2.
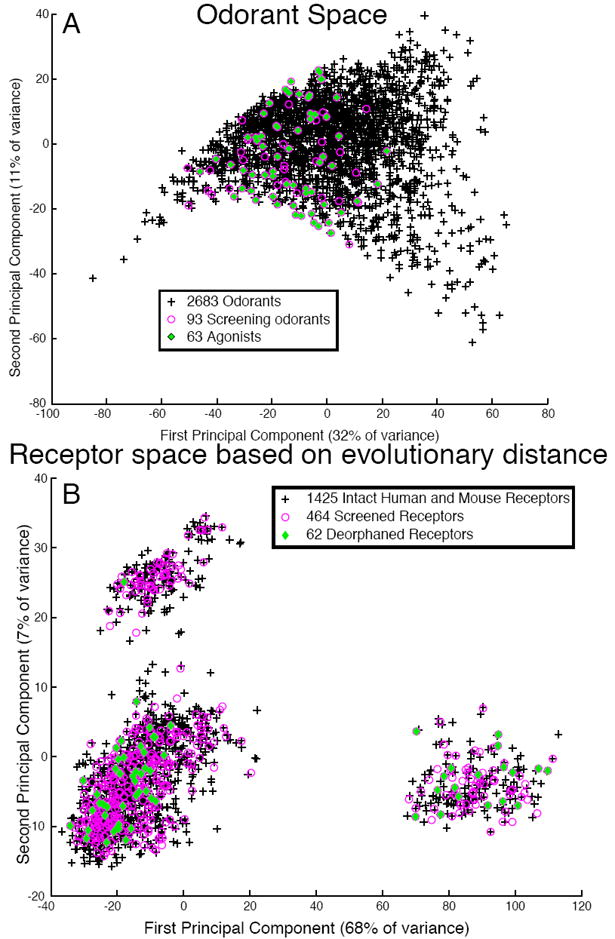
Bias in odorant and receptor sampling. (A) Odorant space. 1664 chemical descriptors were calculated for 2683 odorants. The odorants were projected onto a 2-dimensional space made of the first and second principal components. Odorants used in the mixture-screening phase are colored in magenta; odorants found to bind at least one receptor are shown in green. Black crosses represent untested odorants. (B) Receptor space. The Jukes-Cantor algorithm was used to calculate a distance matrix for 1425 intact odorant receptors. The matrix was visualized in two dimensions using principal component analysis. Receptors used in the mixture screening phase are colored in magenta, receptors deorphaned in this study are shown in green. Black crosses represent untested receptors.
To examine the bias in our resulting OR cohort, we mapped the distance between 1425 human and mouse ORs (2) (Table S1). We constructed a 1425 × 1425 distance matrix using the Jukes-Cantor method (28) and visualized the matrix in two-dimensional space using principal component analysis (Fig. 2B, Fig. S5).
The first principal component (PC1) of odorant space correlates with molecular size (29). Fig. 2A indicates that, although we have fairly dense coverage of the middle of PC1, some caution is warranted in regards to extrapolating to very large and very small odorants. Our criterion of including at least one member of 217 out of the 228 mouse receptor subfamilies insured a broad coverage in receptor space based on full-sequence similarity.
Physicochemical odorant properties predict functional data
Early studies focused on small sets of odorant features, such as carbon chain length and functional group, to explain response variability in the olfactory system. More recent studies have used more quantitative approaches using broader sets of physicochemical descriptors. In one such study, physicochemical descriptors of odorants predicted approximately 35% of the variation in perceived odorant pleasantness (29). This finding suggests that physicochemical descriptors might be useful for predicting earlier stages of olfactory perception. Indeed, two recent studies showed that physicochemical descriptors explain 43 to 72% of the variance in receptor neuron responses (30) and, in a meta-analysis, that a set of 32 physicochemical descriptors explained an average of 48% of the variance in neural responses for various olfactory data sets (31). Here we examined how well several proposed metrics predict our functional data (Fig. 3A).
Figure 3.
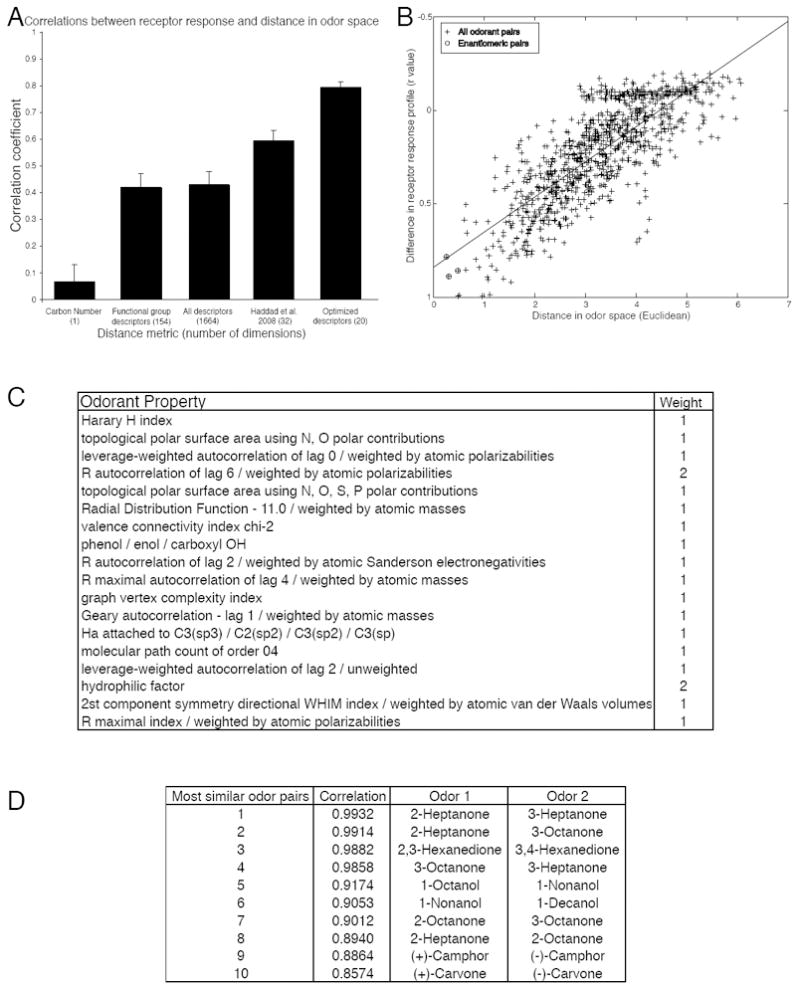
Distance in odorant space predicts similarity in receptor response. (A) Testing various odorant-similarity metrics against the functional data. (B) The difference in the receptor response profile is correlated with the distance between the two odorants calculated using 20 optimized descriptors (r = 0.79, p < 0.001). Enantiomeric pairs are circled. The absence of any completely non-correlated odorant pairs is due to the fact that all pairs of odorants have at least one receptor in common which was not activated by either odorant. (C) 18 physicochemical descriptors that explain over 62% of the variance in our dataset. Definitions of how the descriptors are calculated can be found in the Handbook of Molecular Descriptors (65). (D) The top ten most similar odorant pairs according to our assay. The second column is the Pearson correlation coefficient between the two odorant-response vectors represented in Fig. 1.
In our data set, carbon number alone described a very small portion of the variance in receptor response (r = 0.07, p < 0.04). Functional group descriptors (r = 0.42, p < 0.0001) explained nearly as much variance as all 1664 tested descriptors (r = 0.43, p < 0.0001). Haddad et al. (31) proposed a set of 32 descriptors, a subset of the 1664 used here, that described a large portion of the variance in a meta-analysis of 8 olfactory data sets. These descriptors outperformed carbon number, the set of functional group descriptors, and the full set of descriptors (r = 0.59, p < 0.0001). Haddad et al. optimized the set of 32 descriptors using a meta-analysis including data from different model organisms, different measurement techniques, and different levels of the olfactory system. We similarly optimized the descriptor set using a greedy optimization algorithm using a leave-10-out cross-validation scheme, but only on data from our assay system. On average, this technique explained 60% of the variance in the left-out dataset (r = 0.77). To verify that this result was not due to chance, we randomly shuffled each descriptor vector to create a new set of randomized descriptors from the same distribution. As expected, real descriptors significantly outperformed shuffled descriptors [shuffled r = 0.55, t(23) = 9.09, p < 0.0001]. Having validated this technique, we applied the algorithm to the entire dataset. To reduce overfitting, we chose descriptors that explained the most variance averaged over ten divisions of the dataset, resulting in a set of 18 descriptors that explain over 62% of the variance in our dataset (r = 0.79, p < 0.0001) (Fig. 3B-C). In other words, much of the variation in OR responses can be explained by a fairly small set of physicochemical descriptors.
Receptor sequence predicts functional data
OR genes are classified into families and subfamilies based on sequence alignment of the full-length proteins. Because of the paucity of functional data regarding these receptors, it is unclear if these divisions correspond to functional variability (but see (19)). In other words, the assumption that full-length protein sequence variability predicts functional variability has not been tested on a comprehensive set of receptors. Here we test this assumption by examining how well the properties of amino acids residues predict our functional data (Fig. 4A).
Figure 4.
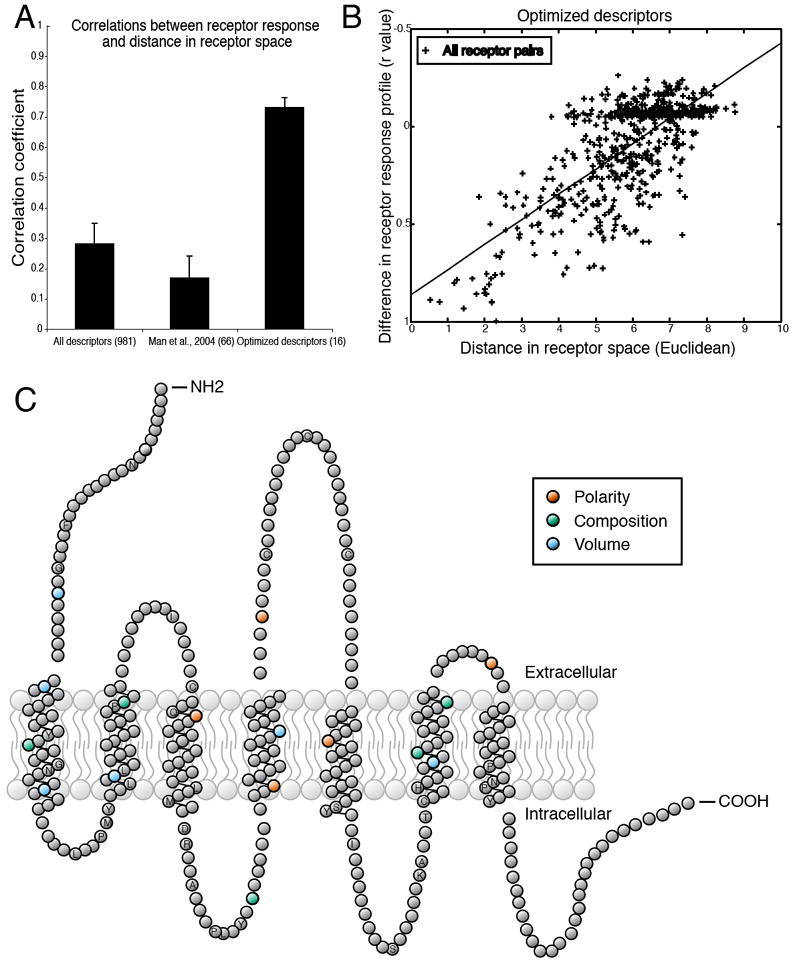
Distance in receptor space predicts similarity in responses to odorants. (A) Testing various receptor-similarity metrics against the functional data shows that our optimized descriptors predict functional responses better than full-sequence similarity or similarity at previously suggested residues (34). (B) Differences in the odorant response profiles of two receptors are correlated with distances between the same receptors, calculated using 16 optimized descriptors (r = 0.73, p < 0.001). Each point represents one pair of receptors. The absence of any completely non-correlated receptor pairs is due to the fact that all pairs of receptors have at least one odorant in common which fails to activate both receptors. (C) Snake plot of a typical OR in which amino acid residues with ligand-specificity-determining properties are highlighted. Residue properties selected by the greedy optimization algorithm are indicated by color. Amino acid positions conserved in at least 90% of the 1425 receptors are labeled with their single-letter amino acid code. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; and Y, Tyr.
As with odorants, we constructed a set of descriptors for ORs. Using a multiple alignment of 1425 intact mouse and human ORs (Table S1), we calculated amino acid properties (polarity, composition, and volume) as defined by Grantham (32) for 327 amino acid residues common to at least 10% of the ORs. The entire set of descriptors explained only 7% of the variance (r = 0.28, p < 0.0001). Recently, Man et al. (33) proposed that comparison of predicted ligand-binding residues might predict functional variation more accurately than full-length comparisons. Restricting the set of descriptors to the 66 properties of 22 predicted binding site residues (34), however, did not improve prediction of functional variation (r = 0.17, p < 0.0001). We optimized the descriptor set using a greedy optimization algorithm using a leave-10-out cross-validation scheme. On average, this technique explained 40% of the variance in the left-out dataset (r = 0.63). To verify that this result was not due to chance, we randomly shuffled each descriptor vector to create a new set of randomized descriptors from the same distribution. As expected, real descriptors significantly outperformed shuffled descriptors [r = 0.18, t(12) = 8.53, p < 0.0001]. Having validated this technique, we applied the algorithm to the entire dataset. To reduce overfitting, we chose descriptors that explained the most variance averaged over ten divisions of the dataset, resulting in a set of 16 descriptors that explain over 53% of the variance in our dataset (r = 0.73, p < 0.0001) (Fig. 4B and C, also see Table S4).
Breadth of tuning
In color vision, three broadly-tuned receptors sense the entire visible range of wavelengths (35). In audition, approximately 3,500 narrowly-tuned cochlear hair cells sense the audible spectrum of frequencies (36). In olfaction, it is not clear if receptors are broadly or narrowly tuned. Given the aforementioned lack of an agreed-upon metric for measuring odorant similarity this question has traditionally been answered in terms of a receptor’s number of agonists. These values (number of agonists) are listed for five receptors in Fig. 5A. On the second line of Fig. 5A we incorporated the sensitivity of the receptor to each odorant to create a tuning curve (for sensitivity-ordered tuning curves for all receptors see Fig. S6). Note that the x-axis is ordered to place the most sensitive odorant in the center and is different for each receptor. A metric used to define an odorant receptor as “broadly” or “narrowly” tuned should, however, take into account not only the number of agonists to which it responds, but also the similarity of those agonists to each other. On the third line of Fig. 5A we ordered the x-axis to reflect odorant similarity, in other words the x-axis represents the odorant’s position along the first principal component of Haddad et al’s 32-dimensional odorant space (31) (for one-dimensional tuning curves based on the first principal component for all receptors see Fig. S7). This first principal component only describes 19.4% of the variance in odorant space. Using more principal components, as in the fourth line of Fig. 5A, describes more of the variance in odorant space, but beyond three principal components is difficult to display in a figure (for two-dimensional tuning curves based on the first two principal components for all receptors see Fig. S8). The radius of a circle enclosing all of the agonists, as in the fourth line of Fig. 5A, gives us a measure of tuning breadth that, unlike the figure, can be scaled to high-dimensional space. The radius of a hypersphere enclosing all five receptors’ agonists in Haddad et al’s 32-dimensional odorant space is listed on the fifth line of Fig. 5A, and for all receptors in Fig. 5B. The results reveal that the mammalian ORs vary along a continuum of tuning breadths. In other words, some receptors are broadly tuned, responding to a large number of odorants that occupy a large area of odorant space (i.e. are structurally dissimilar), others are more narrowly tuned, and some respond to only a small number of closely related odorants.
Figure 5.
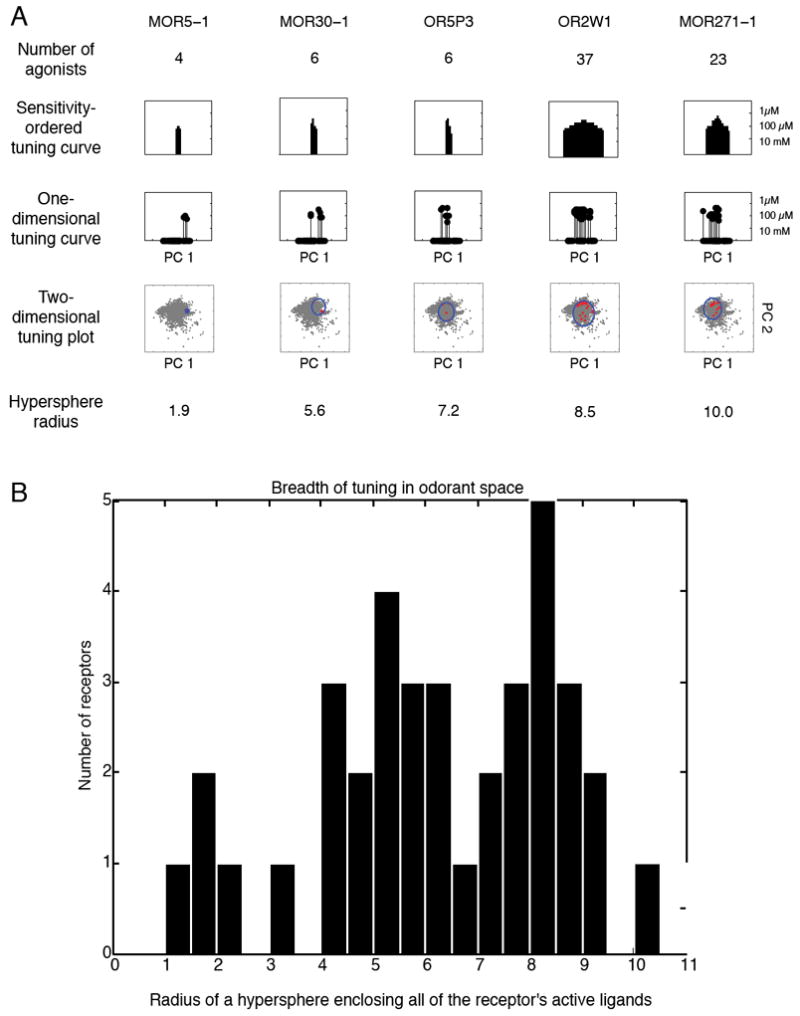
Breadth of tuning in odorant space. A) Table of assorted breadth of tuning representations. The sensitivity-ordered tuning curve displays the 63 tested odorants on the x-axis ordered according to their EC50 for the given receptor. Odorants that activated the receptor at the lowest concentrations are placed near the center of the distribution, whereas those that did not elicit a response are placed at the edges of the distribution. The order of odorants is thus different for different receptors. The one-dimensional tuning curves are stem-plot versions of the sensitivity-ordered tuning curves with the x-axis representing the value of the odorant along the first principal component of Haddad et al.’s odorant space (31). The 2D odorant space figure retains the x-axis of the 1D odorant space figure, but plots the value of the odorant along the second principal component of Haddad et al.’s odorant space on the y-axis, thus forming a 2D projection of odorant space. 2683 odorants are plotted in grey. The odorants activating the receptor are plotted in red and a circle circumscribing the odorants is plotted in blue. The final row is the radius of a hypersphere that encloses all of the receptor’s agonists in the 32-dimensional odorant space. B) A histogram of the hypersphere radius measure for all 62 receptors. For comparison, a hypersphere enclosing 2,683 odorants (listed in Supplementary Table 5) has a radius of 26; a hypersphere enclosing the 93 odorants in our test set has a radius of 14; a hypersphere enclosing the 63 odorants that activated at least one receptor has a radius of 12.
Receptor-response to enantiomers
Enantiomers—stereoisomers that are nonsuperimposable mirror images of each other—impose an interesting constraint on olfactory theories because, even though they have many identical properties, mammals can discriminate between the members of some pairs (37-40). We examined four pairs of enantiomers: phenylbutyric acid, carvone, fenchone, and camphor. The (+) and (-) enantiomers of carvone activate overlapping but distinct sets of olfactory neurons in mice (41). Consistent with these previous reports, we found that the response of an OR to one enantiomer is highly correlated with its response to the other enantiomer (r=0.85, p < 0.0001). We did, however, find a receptor basis for the ability to perceptually distinguish between three of the four enantiomers: for example MOR107-1 is activated only by the (-) enantiomer of fenchone and MOR271-1 is only activated by the (+) enantiomer of fenchone. Although at some doses, MOR2-1 responded more strongly to the (+) enantiomer of 2-phenylbutyric acid, no receptors responded to only one enantiomer of 2-phenylbutyric acid (Fig. 6). Enantiomers were the most similar odorant pairs in terms of physicochemical descriptors, and among the hardest to discriminate in our dataset (Fig. 3B,D).
Figure 6.
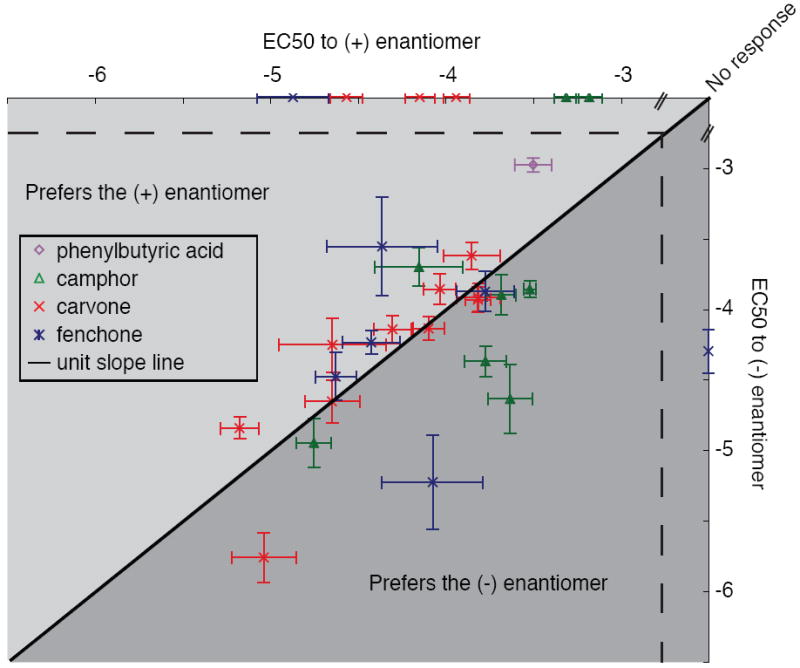
Responses of olfactory receptors to enantiomeric pairs. All receptors that responded to at least one member of the 4 tested enantiomeric pairs are plotted. The black line represents the unit slope line. Points that fall above the line represent receptors more sensitive to the (+) enantiomer, points below the line represent receptors more sensitive to the (-) enantiomer. Error bars represent standard error.
Functional comparison of class I and class II odorant receptors
Class I and class II ORs did not differ in number of agonists (p = 0.11, Mann-Witney U-test) (Fig. 7A), breadth of odor tuning in odorant space (p = 0.21, Mann-Witney U-test) (Fig. 7B), or sensitivity to odorant concentration (p = 0.99, Mann-Whitney U-test) (Fig. 7C). However, we were able to differentiate class I OR agonists from class II OR agonists using a machine-learning algorithm trained on the physicochemical descriptors of the agonists (d’ = 1.98, p < 0.0001).
Figure 7.
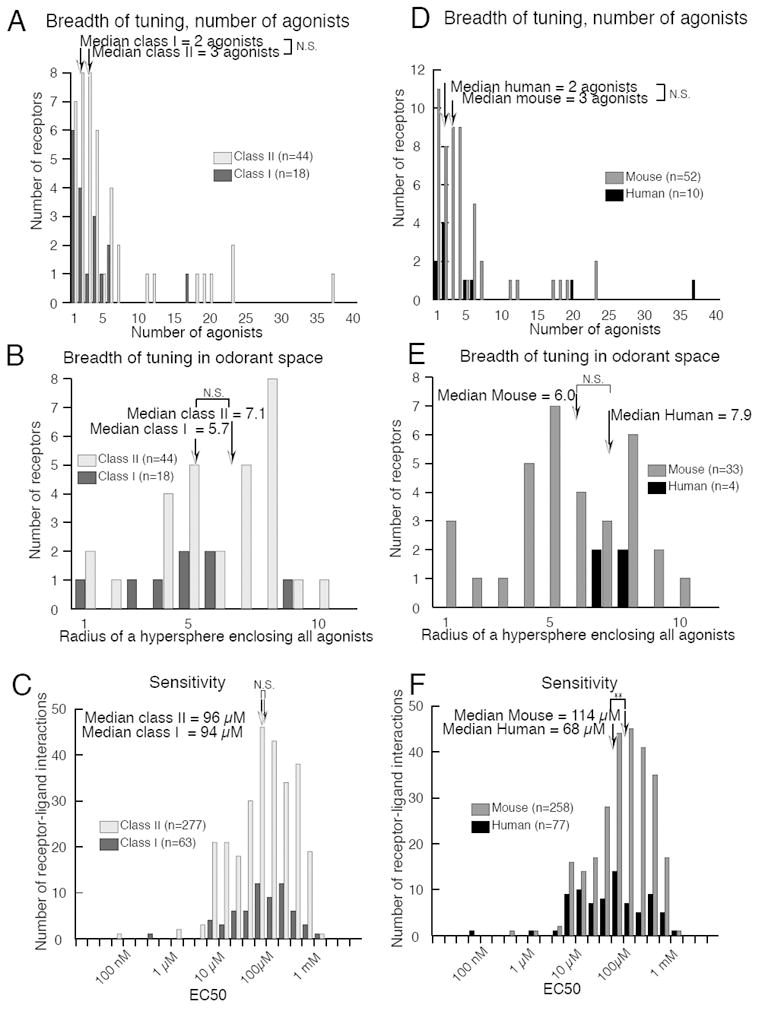
Receptor comparisons by classification. Breadth of tuning did not differ between Class I and Class II receptors by either (A) number of agonists or (B) coverage of odorant space. (C) Sensitivity did not differ between Class I and Class II receptors. Breadth of tuning did not differ between human and mouse receptors by either (D) number of agonists or (E) distance in odorant space. (F) Human receptors were significantly more sensitive to odorants than mouse receptors (p < 0.008).
We then asked what molecular features best differentiated class I from class II agonists. The top 10 descriptors are shown in Table 1A. As predicted (10), agonists for class I ORs are significantly more hydrophilic than agonists for class II ORs (median class I hydrophilic factor = -0.2440, mean class II hydrophilic factor = -0.8020, p<0.001, Mann-Whitney U-test after Bonferroni correction for 1664 descriptors), and have a higher topological polar surface area (median class I TPSA = 37.3, mean class II TPSA = 17.1, p<0.001, Mann-Whitney U-test after Bonferroni correction for 1664 descriptors).
Table 1.
The top 10 physicochemical descriptors for distinguishing between agonists for (A) class I and class II receptors or (B) human and mouse receptors. Abrreviations are as follows: 3D-MoRSE (3D-MOlecule Representation of Structures based on Electron diffraction), WHIM (Weighted Holistic Invariant Molecular descriptors). Definitions of how the descriptors are calculated can be found in the Handbook of Molecular Descriptors (65).
| A. Top ten descriptors that differentiate class I from class II ligands | Class I average value | SE | Class II average value | SE |
|---|---|---|---|---|
| Geary autocorrelation - lag 2 / weighted by atomic masses | 0.643 | 0.0021 | 0.733 | 0.0007 |
| Eigenvalue sum from van der Waals weighted distance matrix | -1.908 | 0.0103 | -0.954 | 0.002 |
| Eigenvalue sum from polarizability weighted distance matrix | -2.4 | 0.0129 | -1.2 | 0.0026 |
| Radial Distribution Function - 4.5 / weighted by atomic polarizabilities | 0.748 | 0.0213 | 1.759 | 0.0058 |
| 3D-MoRSE - signal 25 / unweighted | 0.376 | 0.0036 | 0.534 | 0.0007 |
| 3D-MoRSE - signal 23 / weighted by atomic Sanderson electronegativities | -0.873 | 0.0067 | -0.445 | 0.0015 |
| 2nd component symmetry directional WHIM index / unweighted | 0.193 | 0.0008 | 0.23 | 0.0003 |
| 3rd component accessibility directional WHIM index / weighted by atomic electrotopological states | 0 | 0.0017 | 0.007 | 0.0004 |
| H autocorrelation of lag 2 / unweighted | 0.989 | 0.0064 | 1.405 | 0.0017 |
| R maximal autocorrelation of lag 2 / weighted by atomic polarizabilities | 0.024 | 0.0001 | 0.035 | 0 |
| B. Top ten descriptors that differentiate human from mouse ligands | Human average value | SE | Mouse average value | SE |
| molecular electrotopological variation | 7.347 | 0.0423 | 10.151 | 0.0105 |
| self-returning walk count of order 10 | 2528 | 40.9437 | 5378 | 21.7641 |
| Eigenvalue 01 from edge adjacency matrix weighted by dipole moments | 3.562 | 0.0082 | 3.6245 | 0.0019 |
| Spectral moment 09 from edge adjacency matrix weighted by dipole moments | 11.4595 | 0.0306 | 11.6935 | 0.0068 |
| 1st component symmetry directional WHIM index / unweighted | 0.186 | 0.0017 | 0.197 | 0.0007 |
| 1st component symmetry directional WHIM index / weighted by atomic van der Waals volumes | 0.177 | 0.0017 | 0.191 | 0.0004 |
| 1st component symmetry directional WHIM index / weighted by atomic Sanderson electronegativities | 0.177 | 0.0017 | 0.191 | 0.0004 |
| leverage-weighted autocorrelation of lag 2 / weighted by atomic masses | 0.055 | 0.001 | 0.076 | 0.0003 |
| first eigenvalue of the R matrix | 0.8295 | 0.0008 | 0.86 | 0.0003 |
| R autocorrelation of lag 2 / weighted by atomic masses | 0.3085 | 0.0022 | 0.415 | 0.0007 |
Functional comparison of human and mouse receptors
Humans have fewer than 400 ORs with an intact open reading frame whereas mice have more than 1000 (4,8,42-44). One hypothesis to account for this difference in OR number is that humans may have preferentially retained broadly-tuned receptors to retain the ability to detect most odorants at the expense of sensitivity to low-concentration odorants (45). We did not find a significant difference between mouse and human receptors in the number of agonists (p = 0.25, Mann-Whitney U-test) (Fig. 7D) or breadth of tuning in odorant space (p = 0.14, Mann-Whitney U-test) (Fig. 7E). On average, however, human ORs were significantly more sensitive than mouse receptors (p < 0.008, Mann-Whitney U-test) (Fig. 7F). Although this finding may not generalize to the overall OR population due to the large discrepancy in the relative percentages of human and mouse ORs uncovered by our screening process, it provides preliminary evidence contradicting the hypothesis that mouse ORs are more sensitive to low-concentration odorants than human ORs.
Using a machine-learning algorithm trained on the physicochemical descriptors of the odorants as above, we were able to differentiate odorants that activated human ORs from odorants that activated mouse ORs (d’ = 0.62, p < 0.013). The 10 molecular features that best differentiate odorants activating human receptors from odorants activating mouse receptors are shown in Table 1B.
Predicting odorant-receptor interactions
Except for a few examples (12,13,19,46-48), our knowledge of what makes an OR-ligand interaction effective is very limited. Here, we have greatly increased the number of ORs with known agonists. Moreover, all 62 receptors were tested with the same 63 odorants in a consistent assay, allowing us to look for general rules that govern the interaction between odorant and OR.
We used logistic regression to differentiate odorant-OR combinations that result in a response from odorant-OR combinations that fail to elicit a response. Our initial work suggested that a subset of physicochemical odorant descriptors and amino acid properties predict variation in odorant-receptor activation. Here we combined both physicochemical descriptors and amino acid properties in an attempt to predict the interaction of novel odorants with novel ORs.
We used a leave-10-out cross validation procedure to validate our model, in each round selecting a set of descriptors that best predicted the training data and applying those descriptors to the test data. We selected the values to leave out using two different methods. First, we reserved 10% of the receptors, simulating a situation in which an investigator is searching for the response of novel ORs to odorants that activate a known OR. Our model predicted the response of the ‘novel’ ORs to the 63 odorants [area under the receiver operating characteristic curve (AUC) = 0.59, p < 0.0001, Mann-Whitney U-test] (Fig. 8A). Second, we reserved 10% of the odorants, simulating a situation in which an investigator is searching for the response of ORs with known agonists to untested odorants. Our model predicted the response of all 62 ORs to the ‘novel’ odorants (AUC = 0.64, p < 0.0001, Mann-Whitney U-test) (Fig. 8B).
Figure 8.
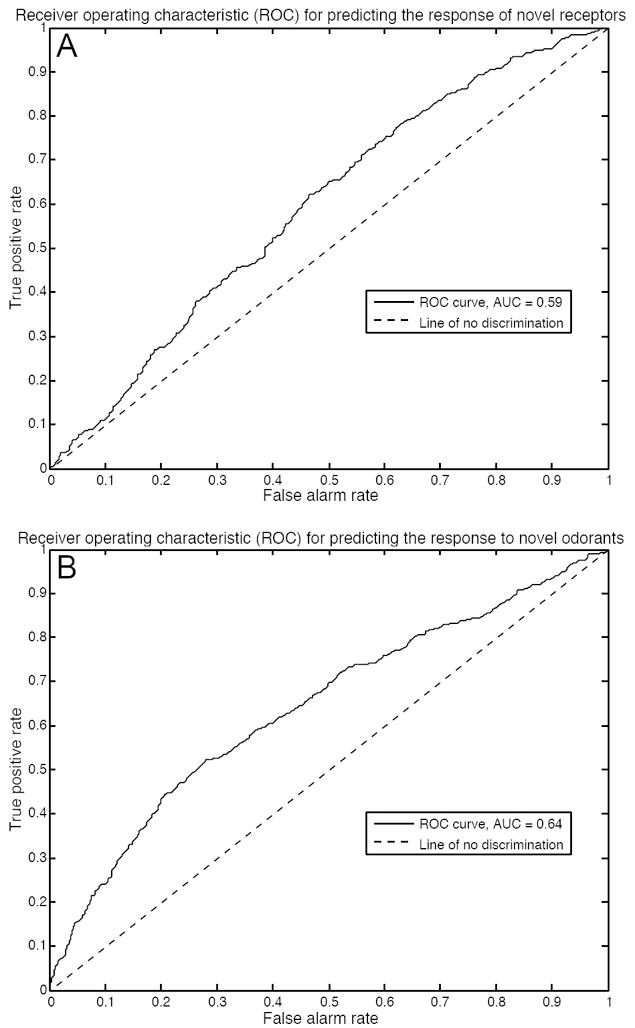
Receiver operating characteristic curves for our ligand-receptor interaction classifier. (A) Validation of our model when predicting the response of novel receptors to tested odorants. The area under the curve indicates that our model successfully predicts if a tested odorant is an agonist of a novel receptor 59% of the time. (B) Validation of our model when predicting the response of tested receptors to novel odorants. The area under the curve indicates that our model successfully predicts if a novel odorant is an agonist of a tested receptor 64% of the time.
DISCUSSION
Starting with over 450 mouse and human ORs, we identified agonists for 52 mouse and 10 human ORs. These data were used to confirm the utility of a multidimensional metric for odorant similarity (31), develop a multidimensional metric for receptor similarity, quantify the breadth of receptor tuning in odorant space, identify ORs capable of discriminating three enantiomeric odorant pairs, distinguish activation profiles of class I and class II ORs, distinguish activation profiles of human and mouse ORs, and develop a model to predict odorant-receptor activation.
Our in vitro assay lacks many components of an in vivo olfactory system, including odorant binding proteins, a mucosal layer, intracellular molecules, and sniffing behaviors. In addition, although some odorants inhibit ORs (14,22,47,49-51), our assay is not designed to detect inhibitory responses. In spite of these shortcomings, previous results from in vitro systems predict human olfactory perception (11,21), suggesting that in vitro assays are relevant to the intact olfactory system.
Despite starting with similar numbers of human and mouse ORs, we identified agonists for over five times more mouse ORs than human ORs. This may indicate, as previously suggested (52,53), that mice have even more functional ORs relative to humans than the genome sequences predict. This could reflect a large fraction of nonfunctional genes among the intact human ORs (54) or a large fraction of nonfunctional variants among the human OR clones we used for screening. Alternatively, this difference in mouse and human OR response to agonist may be due to bias in odorant choice or technical problems in the heterologous system specific to the human ORs.
We identified 18 physicochemical odorant descriptors that predict our functional data. Odorant similarity calculated using these descriptors is highly correlated with similarity using Haddad et al.’s previously identified 32 descriptors (r = 0.77, p < 0.0001) across a set of over 2500 odorants (data not shown). Our descriptors outperform Haddad et al.’s descriptors for our data, but it is unlikely that they are universally better. Although there is some universality in odorant similarity measures across various studies (31), there are also large differences between studies, which would suggest that optimization to a specific organism and technique would outperform a general set of descriptors.
We identified properties of 16 OR amino acid residues that predict our functional data. Of these 16 residues, 12 occur in predicted transmembrane domains, three occur in predicted extracellular domains and one occurs in a predicted intracellular domain. This is consistent with functional evidence and computational predictions suggesting that the binding pocket of olfactory receptors is formed by the transmembrane domains (34,48,55,56), however these residues do not necessarily correspond to the binding site of the ORs, because amino acid residues far from the binding pocket may affect ligand specificity by changing the global conformation of the folded protein. The properties of the residues predicted by Man et al. to play a role in ligand-binding (34) did not predict our data (see the comparison in Fig. S10), perhaps because several of Man et al.’s putative orthologs do not share similar odorant specificities or because the amino acid sites predicted to play a role in ligand-binding by Man et al. (34) may be more important for a set of odorants that was not well represented in our data set. With more functional data, we expect the set of ligand-specificity-determining residues to change somewhat and improve.
Enantiomers have very similar physicochemical properties, but mammals can discriminate between members of some enantiomeric pairs (37-40). The (+) and (-) enantiomers of carvone activate overlapping but distinct sets of olfactory neurons in mice (41). Consistent with these previous reports, we find that some ORs can distinguish between enantiomers, although many cannot. We have identified candidate ORs capable of supplying sufficient information for the discrimination of three enantiomeric pairs. These ORs are ideal for future structure-function analysis to understand how ORs discriminate between physicochemically-similar compounds.
With two exceptions, class I ORs are expressed only in the anterior-dorsal-most zone of the olfactory epithelium in mice (57). In agreement with the theory that the olfactory mucosa serves as a chromatographic separator of odorants, with ORs responding to fast-sorbing hydrophilic compounds expressed early in the airstream and ORs responding to slow-sorbing hydrophobic compounds expressed at locations further along in the airstream (58,59), we found that class I ORs prefer polar (and therefore hydrophilic) compounds. This suggests that the location of receptor expression along the mucosa may be related to the ligands that bind the receptor.
Humans have approximately 387 potentially functional ORs (in other words, ORs with an intact open reading frame); mice have approximately 1035 (2). Despite this huge difference in OR number, behavioral studies have failed to show a clear distinction between rodents and primates in odorant detection threshold (60). This may be due to a lack of data, or it may be due to compensatory mechanisms such as a shortened nose or more computation at later neural stages (61). In our data set we found, unexpectedly, that human ORs are more sensitive than mouse ORs. Although our results may fail to generalize to the entire OR repertoire due to both the small absolute number of human ORs in our data set and the bias in relative number of human and mouse ORs that were activated in our screen, our predictions provide a tentative framework for identifying odorants that are more likely to activate either human or mouse ORs. This narrows the field of compounds to test in behavioral studies searching for differences between mouse and human perception. Understanding perceptual differences between species, in turn, will give insight into the evolutionary pressures driving changes in olfaction and aid the translation of research from one species to another.
In addition to identifying active agonists for 62 ORs, we developed a model to predict the interaction of ORs and their ligands. Why is our model better at predicting a tested OR’s response to a novel odorant than a novel OR’s response to a tested odorant? This is likely because of the relative quality of information we have about odorant similarity compared to receptor similarity. More detailed modeling of the OR that includes 3D information about the protein structure and binding pocket will undoubtedly improve the ability to predict the activity of novel ORs and improve our initial model relating molecular structure to OR response.
MATERIALS AND METHODS
Cloning mouse and human Ors
219 mouse and 245 human odorant receptors were cloned with sequence information from The Olfactory Receptor Database (http://senselab.med.yale.edu/senselab/ORDB/default.asp)(3). We adopted the nomenclature proposed by the D. Lancet group for the human ORs (8) and by the S. Firestein group for the mouse ORs (4). OR open reading frames were amplified from genomic DNA using proofreading KOD DNA polymerase (Toyobo/Novagen) and subcloned into pCI expression vectors (Promega) containing the first 20 residues of human rhodopsin (Rho tag). The sequences of the cloned receptors were verified by sequencing (3100 Genetic Analyzer, ABI Biosystems).
Immunocytochemistry and FACS analysis
For live cell-surface staining, the mouse monoclonal anti-rhodopsin antibody, 4D2 (gift from Dr. R. Molday) (62) and Cy3-conjugated donkey anti-mouse IgG (Jackson Immunologicals) were used. For FACS analysis we used phycoerythrin (PE)-conjugated anti-mouse IgG (Jackson Immunologicals). In order to monitor the transfection efficiency, we co-transfected Green Fluorescent Protein (GFP) with the OR. We quantified the intensity of receptor cell surface expression as the ratio of PE and GFP expression. We added 7-amino-actinomycin D (Calbiochem) before flow cytometry to mark dead cells which were excluded from the analysis. We normalized the gate and mean of PE positive cells by GFP values.
Luciferase assay
We used Dual-Glo™ Luciferase Assay System (Promega) for the luciferase assay as previously described (17). Odorant receptor activation leads to an increase in intracellular cAMP; we used CRE-luciferase (Stratagene) to measure this change. Renilla luciferase driven by a constitutively active SV40 promoter (pRL-SV40; Promega) served as an internal control for cell viability and transfection efficiency. We plated Hana3A cells on poly-D-lysine-coated 96-well plates (BioCoat; Becton Dickinson). We transfected ORs with Rho tag in a Hana3A cell line along with RTP1S(25), CRE-luciferase, and pRL-SV40 using Lipofectamine2000 (Invitrogen). For each 96-well plate we transfected, 1 μg of CRE-Luc, 1μg of pRL-SV40, 5 μg of odorant receptor plasmid, and 1 μg of RTP1S. Approximately 24 hours post-transfection, we replaced the medium with CD293 chemically defined medium (Gibco) and then incubated the plate for 30 min at 37°C. We then replaced the medium with 25uL of odorant solution diluted in CD293 and incubated the plate for 4 hrs at 37°C and 5% CO2. We followed the manufacturer’s protocols for measuring luciferase and Renilla luciferase activities. We measured luminescence using a Wallac Victor 1420 plate reader (Perkin-Elmer). First, we divided all luminescence values by the Renilla Luciferase activity to control for transfection efficiency in a given well. We calculated normalized luciferase activity using the formula (LN-Lmin)/(Lmax-Lmin), where LN is the luminescence of firefly luciferase in response to the odorant, Lmin is the minimum luciferase value on a plate or set of plates, and Lmax is the maximum luciferase value on a plate or set of plates. We analyzed the data using Microsoft Excel and GraphPad Prism 4.
Screening procedure
We stimulated the entire OR library with eight separate odorant mixtures formed from 93 odorants (Fig. S4). We applied the mixtures at 100 μM and eliminated all ORs that did not show activity (the ratio of CRE-Luciferase to Renilla luciferase was less than 0.1 above baseline). We then applied the mixtures at five different doses (1 μM, 10 μM, 100 μM, 300 μM and 1 mM) and eliminated all ORs that did not show dose-dependent activity to any of the eight mixtures (statistical significance was not assessed for this screening stage), leaving 121 human receptors (49.4%) and 169 mouse receptors (77.2%). We then took the 290 receptors and performed a comparison between the 93 individual odorants at a 100 μM dose and a no-odor control. Each comparison was performed in triplicate; statistical significance was assessed by t-test (uncorrected for multiple comparisons). In addition, we confirmed the consistency of our experimental conditions with two positive controls: MOR203-1 with nonanoic acid, and MOR32-1 with nonanoic acid. 27 human ORs (11.0%) and 102 mouse ORs (46.6%) showed a significant response to at least one of 67 odorants relative to a no-odor control. We then constructed dose-response curves ranging from 10 nM to 3 mM for each combination of 129 receptors and 67 odorants. On each plate we used a single odorant to avoid cross-contamination, and each OR-odorant dose was tested at least three times. We fit the data to a sigmoidal curve. We counted an odorant/receptor pair as a significant activation if both the normalized activity at 100 uM was significantly different than the baseline activity (using a t-test), and the standard deviation of the fitted log EC50 was less than 0.5 log units. We confirmed that the raw CRE-Luc curve and the normalized (Luc/RL) curve EC50 values did not differ by more than 1 log step. As a result, we identified 52 mouse (23.7%) and 10 human (0.04%) ORs that showed a significant dose-dependent response to one or more of 63 odorants (Fig. 1). Four odorants failed to activate any OR in this final stage.
Physicochemical descriptors
We obtained molecular structure files for each odorant, from PubChem (http://pubchem.ncbi.nlm.nih.gov/search/), and input these structures into the Virtual Computational Chemistry Laboratory (http://www.vcclab.org) (63). There we used CORINA (64) to obtain 3D coordinates and Dragon (Talete) to compute 1664 physicochemical descriptors (65).
Receptor descriptors
We aligned 1425 OR amino acid sequences from Niimura and Nei (2) and 464 OR amino acid sequences from our OR library using the MUSCLE algorithm (66) in Seaview (67) with manual adjustment for conserved domains (Table S1). We eliminated all sites that were gaps in over 90% of the 1425 ORs, leaving 327 amino acids. Our set of 981 descriptors consisted of the polarity, composition and volume of these 327 residues, as defined by Grantham (32). Differences between disease alleles and wild-type alleles computed using these properties are on average greater than those observed between putatively neutral polymorphic alleles, suggesting that these properties are functionally relevant (68).
Correlations between descriptors and responses
Each descriptor was z-scored across 2683 odorants (Table S4) or all 1425 receptors (Table S1). We eliminated all odorants for which we had fewer than 3 responsive receptors. For all remaining odorant pairs we calculated the Pearson correlation between EC50 vectors and the Euclidean distance between descriptor vectors. We then measured the Pearson correlation between these two sets of distances.
Optimizing descriptors
Testing all possible combinations of the descriptors is an intractable task, so we used a greedy optimization algorithm, as in (31), to determine the best set. In this method we begin with an empty set of descriptors. We then combine each descriptor with the previous set of descriptors and compute correlation values for all of these candidate sets of descriptors. To reduce overfitting, we divide the data randomly into ten subsets and compute correlation values for each leave-one-out subset. The final correlation coefficient for each set of descriptors is the average of the correlation coefficients for each subset. The set with the best correlation coefficient then becomes the new set of descriptors and the process is repeated until the correlation coefficient increases by less than 0.004 in three consecutive iterations.
We validated this method on each dataset using a leave-10-out cross-validation scheme. In other words, we optimized the descriptors using 90% of the data, and then tested the descriptors on the remaining 10% of the data. We repeated this division ten times such that all subdivisions were test sets and reported the average performance over all test sets. We then verified that this optimization does not work as well for randomly shuffled vectors. We used the same descriptor values as in the actual vectors, but shuffled each descriptor independently so that any given object had a random set of descriptor values. We created 30 sets of these objects with shuffled descriptor values. For each of these 30 sets we optimized descriptors for 90% of the data and tested the descriptors on the remaining 10% of the data. We reported the average of all 30 values. We compared the performance of the real optimized descriptors to the shuffled optimized descriptors using a two-sample t-test with unequal variance.
Breadth of tuning
We define a receptor’s breadth of tuning as the radius of a hypersphere, centered on the center of mass of all of the receptor’s agonists and enclosing all of the receptor’s agonists in Haddad et al’s (31) odorant space. For reference, a hypersphere enclosing 2,683 odorants (Table S5) has a radius of 26; a hypersphere enclosing the 93 odorants in our test set has a radius of 14; a hypersphere enclosing the 63 odorants that activated at least one receptor has a radius of 12.
Machine-learning algorithm
We used the support vector machines (SVM) functions in the Bioinformatics Toolbox of Matlab (Mathworks) to classify the odorants in our data set. To estimate the discrimination ability of our classifier for class I/class II discriminations and human/mouse discriminations, we jackknifed the data set. In other words, we trained the classifier on all but one instance and then tested the classifier on that instance. We repeated this n times, where n was the total number of instances. We used signal detection theory (69) to compute the sensitivity index (d’), which is the separation of the means of the two distributions (class I and class II agonists, or mouse and human agonists) in units of standard deviation. We confirmed that the d’ was significantly different from zero according to Marascuilo’s test (70). We used the rankfeatures function in the MATLAB Bioinformatics Toolbox to determine the physicochemical properties that best predict membership in a class. We applied a cross-correlation weighting value of 0.7 to reduce the number of highly correlated properties.
Predicting odorant-OR interactions
Each of the 3,906 tested odorant-OR interactions were represented by a vector of 1664 physicochemical descriptors and 981 receptor descriptors. We then used a cross-validation procedure to determine if a subset of these vectors could predict if the odorant-OR combination resulted in activation (i.e. all colored squares in Fig. 1). We used a ten-fold validation procedure in two different ways. In the first method, we divided all 62 rows of odorant-OR interactions into 10 sets (leave-receptor-out). In the second method, we divided all 63 columns of odorant interactions into ten sets (leave-odorant-out). After choosing the test set for a round we used the greedy optimization method described above to calculate optimized descriptor sets for the other nine sets (training data). After this selection of attributes we then performed logistic regression in JMP v6 (SAS Institute) on the training set and used the resulting coefficients to predict the probability of activation for the test set. We repeated this method ten times for each of the three selection processes, rotating the test set each iteration, such that we evaluated the entire data set using a model that was not trained on its respective test set. We generated receiver operating characteristic curves according to (71). We determined statistical significance using a Mann-Whitney U-test comparing the distribution of predicted odds for interactions resulting in activation to the distribution of predicted odds for interactions resulting in no activation.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is the author’s version of the work. It is posted here by permission of the AAAS for personal use, not for redistribution. The definitive version was published in Science Signaling Volume 2 Issue 60, 3 March 2009, [doi:10.1126/scisignal.2000016] http://stke.sciencemag.org/cgi/content/full/sigtrans;2/60/ra9
REFERENCES AND NOTES
- 1.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Niimura Y, Nei M. Evolutionary changes of the number of olfactory receptor genes in the human and mouse lineages. Gene. 2005;346:23–8. doi: 10.1016/j.gene.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Healy MD, Smith JE, Singer MS, Nadkarni PM, Skoufos E, Miller PL, Shepherd GM. Olfactory receptor database (ORDB): a resource for sharing and analyzing published and unpublished data. Chemical Senses. 1997;22:321–6. doi: 10.1093/chemse/22.3.321. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–33. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian AD, Wyman A, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 7.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 8.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 9.Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freitag J, Ludwig G, Andreini I, Rossler P, Breer H. Olfactory receptors in aquatic and terrestrial vertebrates. J Comp Physiol [A] 1998;183:635–50. doi: 10.1007/s003590050287. [DOI] [PubMed] [Google Scholar]
- 11.Menashe I, Abaffy T, Hasin Y, Goshen S, Yahalom V, Luetje CW, Lancet D. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:e284. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquier V, Pick H, Vogel H. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. J Neurochem. 2006;97:537–44. doi: 10.1111/j.1471-4159.2006.03771.x. [DOI] [PubMed] [Google Scholar]
- 13.Schmiedeberg K, Shirokova E, Weber HP, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–12. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Shirokova E, Schmiedeberg K, Bedner P, Niessen H, Willecke K, Raguse JD, Meyerhof W, Krautwurst D. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J Biol Chem. 2005;280:11807–15. doi: 10.1074/jbc.M411508200. [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa K, Touhara K. Myr-Ric-8A Enhances G{alpha}15-Mediated Ca2+ Response of Vertebrate Olfactory Receptors. Chemical Senses. 2008 doi: 10.1093/chemse/bjn047. [DOI] [PubMed] [Google Scholar]
- 16.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–78. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–91. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 18.Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, Touhara K. Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 2006;52:857–69. doi: 10.1016/j.neuron.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Abaffy T, Matsunami H, Luetje CW. Functional analysis of a mammalian odorant receptor subfamily. J Neurochem. 2006;97:1506–18. doi: 10.1111/j.1471-4159.2006.03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuhaus EM, Mashukova A, Zhang W, Barbour J, Hatt H. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chemical Senses. 2006;31:445–52. doi: 10.1093/chemse/bjj049. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–72. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 22.Sanz G, Schlegel C, Pernollet JC, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chemical Senses. 2005;30:69–80. doi: 10.1093/chemse/bji002. [DOI] [PubMed] [Google Scholar]
- 23.Bush CF, Jones SV, Lyle AN, Minneman KP, Ressler KJ, Hall RA. Specificity of olfactory receptor interactions with other G protein-coupled receptors. J Biol Chem. 2007;282:19042–51. doi: 10.1074/jbc.M610781200. [DOI] [PubMed] [Google Scholar]
- 24.Matarazzo V, Clot-Faybesse O, Marcet B, Guiraudie-Capraz G, Atanasova B, Devauchelle G, Cerutti M, Etievant P, Ronin C. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chemical Senses. 2005;30:195–207. doi: 10.1093/chemse/bji015. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–93. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 26.Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34:143–4. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- 27.Haddad R, Lapid H, Harel D, Sobel N. Measuring Smells. Current Opinion in Neurobiology. 2008;18:1–7. doi: 10.1016/j.conb.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- 29.Khan RM, Luk CH, Flinker A, Aggarwal A, Lapid H, Haddad R, Sobel N. Predicting odor pleasantness from odorant structure: pleasantness as a reflection of the physical world. J Neurosci. 2007;27:10015–23. doi: 10.1523/JNEUROSCI.1158-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmuker M, de Bruyne M, Hahnel M, Schneider G. Predicting olfactory receptor neuron responses from odorant structure. Chem Cent J. 2007;1:11. doi: 10.1186/1752-153X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haddad R, Khan R, Takahashi YK, Mori K, Harel D, Sobel N. A metric for odorant comparison. Nature Methods. 2008;5:425–429. doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- 32.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 33.Man O, Willhite DC, Crasto CJ, Shepherd GM, Gilad Y. A framework for exploring functional variability in olfactory receptor genes. PLoS ONE. 2007;2:e682. doi: 10.1371/journal.pone.0000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004;13:240–54. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Valois RL, De Valois KK. A multi-stage color model. Vision Res. 1993;33:1053–65. doi: 10.1016/0042-6989(93)90240-w. [DOI] [PubMed] [Google Scholar]
- 36.Pickles JO. An Introduction to the Physiology of Hearing. Academic; New York: 1988. [Google Scholar]
- 37.Laska M, Shepherd GM. Olfactory discrimination ability of CD-1 mice for a large array of enantiomers. Neuroscience. 2007;144:295–301. doi: 10.1016/j.neuroscience.2006.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubin BD, Katz LC. Spatial coding of enantiomers in the rat olfactory bulb. Nat Neurosci. 2001;4:355–6. doi: 10.1038/85997. [DOI] [PubMed] [Google Scholar]
- 39.Joshi D, Volkl M, Shepherd GM, Laska M. Olfactory sensitivity for enantiomers and their racemic mixtures--a comparative study in CD-1 mice and spider monkeys. Chemical Senses. 2006;31:655–64. doi: 10.1093/chemse/bjl006. [DOI] [PubMed] [Google Scholar]
- 40.Linster C, Johnson BA, Morse A, Yue E, Leon M. Spontaneous versus reinforced olfactory discriminations. J Neurosci. 2002;22:6842–5. doi: 10.1523/JNEUROSCI.22-16-06842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma M, Shepherd GM. Functional mosaic organization of mouse olfactory receptor neurons. Proc Natl Acad Sci U S A. 2000;97:12869–74. doi: 10.1073/pnas.220301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci U S A. 2003;100:12235–40. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2584–9. doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2156–61. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lapidot M, Pilpel Y, Gilad Y, Falcovitz A, Sharon D, Haaf T, Lancet D. Mouse-human orthology relationships in an olfactory receptor gene cluster. Genomics. 2001;71:296–306. doi: 10.1006/geno.2000.6431. [DOI] [PubMed] [Google Scholar]
- 46.Sanz G, Thomas-Danguin T, Hamdani el H, Le Poupon C, Briand L, Pernollet JC, Guichard E, Tromelin A. Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. Chemical Senses. 2008;33:639–53. doi: 10.1093/chemse/bjn032. [DOI] [PubMed] [Google Scholar]
- 47.Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–55. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 48.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–15. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. Embo J. 2004;23:120–6. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–60. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 51.Kurahashi T, Lowe G, Gold GH. Suppression of odorant responses by odorants in olfactory receptor cells. Science. 1994;265:118–20. doi: 10.1126/science.8016645. [DOI] [PubMed] [Google Scholar]
- 52.Menashe I, Aloni R, Lancet D. A probabilistic classifier for olfactory receptor pseudogenes. BMC Bioinformatics. 2006;7:393. doi: 10.1186/1471-2105-7-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–46. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- 54.Mainland RC. Absence of olfactory sensation. J Heredity. 1945;36:143–144. [Google Scholar]
- 55.Zhang X, Rodriguez I, Mombaerts P, Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004;83:802–11. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Baldwin JM. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994;6:180–90. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 57.Tsuboi A, Miyazaki T, Imai T, Sakano H. Olfactory sensory neurons expressing class I odorant receptors converge their axons on an antero-dorsal domain of the olfactory bulb in the mouse. Eur J Neurosci. 2006;23:1436–44. doi: 10.1111/j.1460-9568.2006.04675.x. [DOI] [PubMed] [Google Scholar]
- 58.Mozell EJ, Sabanathan S, Mearns AJ, Bickford-Smith PJ, Majid MR, Zografos G. Continuous extrapleural intercostal nerve block after pleurectomy. Thorax. 1991;46:21–4. doi: 10.1136/thx.46.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoenfeld TA, Cleland TA. Anatomical contributions to odorant sampling and representation in rodents: zoning in on sniffing behavior. Chemical Senses. 2006;31:131–44. doi: 10.1093/chemse/bjj015. [DOI] [PubMed] [Google Scholar]
- 60.Laska M, Seibt A, Weber A. ‘Microsmatic’ primates revisited: Olfactory sensitivity in the squirrel monkey. Chemical Senses. 2000;25:47–53. doi: 10.1093/chemse/25.1.47. [DOI] [PubMed] [Google Scholar]
- 61.Shepherd GM. The human sense of smell: are we better than we think? PLoS Biol. 2004;2:E146. doi: 10.1371/journal.pbio.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laird DW, Molday RS. Evidence against the role of rhodopsin in rod outer segment binding to RPE cells. Invest Ophthalmol Vis Sci. 1988;29:419–28. [PubMed] [Google Scholar]
- 63.Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. Virtual computational chemistry laboratory--design and description. J Comput Aid Mol Des. 2005;19:453–63. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 64.Gasteiger J, Rudolph C, Sadowski J. Automatic Generation of 3D Atomic Coordinates for Organic Molecules. Tetrahedron Comput Method. 1992;3:537–547. [Google Scholar]
- 65.Todeschini R, Consonni V. Handbook of Molecular Descriptors. Wiley-VCH; Weinheim: 2000. [Google Scholar]
- 66.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–8. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 68.Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10:2319–28. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- 69.Green DM, Swets JA. Signal Detection Theory and Psychophysics. Wiley; New York: 1966. [Google Scholar]
- 70.Marascuilo LA. Extensions of the significance test for one-parameter signal detection hypotheses. Psychometrika. 1970;35:237–243. [Google Scholar]
- 71.Fawcett T. An introduction to ROC analysis. Pattern Recgonition Letters. 2006;27:861–874. [Google Scholar]
- 72.This work was supported by grants from the NIH, HFSP and by an NRSA postdoctoral fellowship to J.D.M. The authors thank I. Davison, R. Haddad, N. Sobel, H. Lapid, and M. Caron as well as N. Pillai from the Duke SCC for helpful discussions. We thank D. Marchuk and R. Valdivia for sharing equipment. We thank A. Toyama and M. Kubota for expert technical assistance. J.-T. Chi for initial analysis, M. Cook and O. Awonuga for FACS analysis. H.S. performed functional assays and preliminary analysis. Q.C. created the OR libraries. H.Z. optimized the assay conditions. H.M. supervised the project, interpreted data and wrote the paper. J.D.M. analyzed and interpreted data and wrote the paper. All authors commented on the manuscript.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


