Summary
Dynamic processes involving macromolecular complexes are essential to cell function. These processes take place over a wide variety of length scales from nanometers to micrometers, and over time scales from nanoseconds to many minutes. As a result, information from a variety of different experimental and computational approaches is required. We review the relevant sources of information and introduce a framework for integrating the data to produce representations of dynamic processes.
Keywords: biological complexes, assemblies, dynamics, process, structure, hybrid methods
Introduction
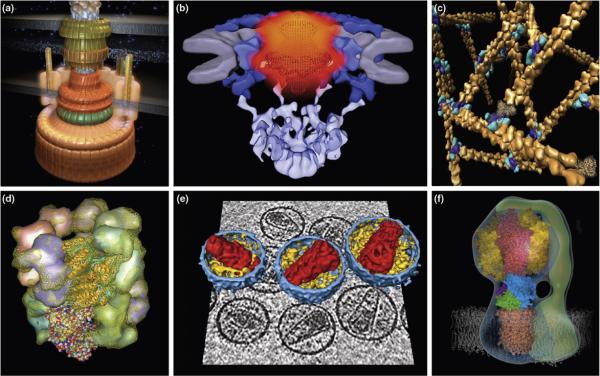
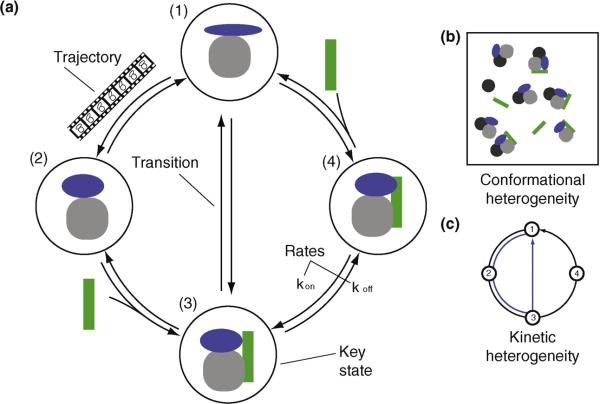
To understand the processes that maintain and replicate a living cell, we need to describe the structural dynamics of a few hundred core macromolecular processes [1•] (Figure 1), such as DNA replication by the replisome [2], transcription of DNA into RNA by RNA polymerase [3•], protein synthesis by the ribosome [4••,5], protein folding by chaperonins [6,7], nucleocytoplasmic transport through the nuclear pore complex [8], active transport with molecular motors [9–11], assembly pathways of large complexes [12–14], and protein degradation in the proteasome [15–17]. These processes take place over a wide variety of length scales from nanometers to micrometers, and over time scales from nanoseconds to many minutes. Moreover, the macromolecular systems can exist in different structural states (conformational heterogeneity) and follow different kinetic pathways during a single process (kinetic heterogeneity) (Figure 2, Table 1).
Figure 1. Examples of dynamic macromolecular processes.
[A] Locomotion of a cell is enabled by a reversible rotary propeller of the bacterial flagellum [119]. [B] Nucleocytoplasmic transport of macromolecules occurs in a regulated fashion through the nuclear pore complex [120•]. [C] A number of cellular functions, including muscle contraction, cell motility, cell division and cytokinesis, depend on the assembly and maintenance of branched actin filaments (http://www.cgl.ucsf.edu/chimera/ImageGallery/). [D] The folding of many proteins is catalyzed inside the chaperonin cavity [7] (http://www.cgl.ucsf.edu/chimera/ImageGallery/). [E] The HIV-1 core assembles inside the maturing virion [121]. [F] Synthesis of ATP in mitochondria and chloroplasts is catalyzed by ATP synthase (http://www.mrc-dunn.cam.ac.uk/research/atp_synthase).
Figure 2. A representation of a process.
[A] Several important terms used in the text are illustrated by a process of four key states (circled solid shapes) connected by transitions (arrows). [B] Illustration of conformational heterogeneity. [C] Illustration of kinetic heterogeneity, showing several paths through the graph. Definitions of the terms can be found in Table 1.
Table 1.
Brief descriptions of important terms used in the review
| State | A state is described by a three dimensional structure of an assembly at some resolution. The structure may be flexible and its description may be incomplete. |
| Key state | The set of key states and transitions between them capture the essence of the process. Key states need not be stable and could correspond to transition states. |
| Transition | A transition occurs between a pair of key states that can interconvert directly without passing through other key states. |
| Trajectory | A trajectory is a detailed sequence of states describing a transition between two key states, like frames in a movie. |
| Rate of transition | Transition rates can be expressed in a variety of ways such as the probability of moving from one state to another in a given period of time or rate constants. |
| Conformational heterogeneity | Conformational heterogeneity implies that multiple states exist in a single sample of the system. Such heterogeneity complicates bulk experimental measurements, often requiring single-molecule experiments. |
| Kinetic heterogeneity | Kinetic heterogeneity results from different copies of the system following different transitions. For example, different parts of the secondary structure of a protein can form independently and asynchronously before the tertiary structure forms [110] and, during ribosome assembly, different interactions between proteins and RNA can stabilize independently of one another [13]. |
| Restraint | A restraint restricts geometric and/or temporal properties of an assembly, such as the distance between two components, the overall shape of the complex, or the time interval between two key states. A restraint is a scalar function that quantifies the agreement between a restrained feature and the data. |
| Process representation | A process is represented as a set of key states connected by transitions with associated trajectory and rate information. |
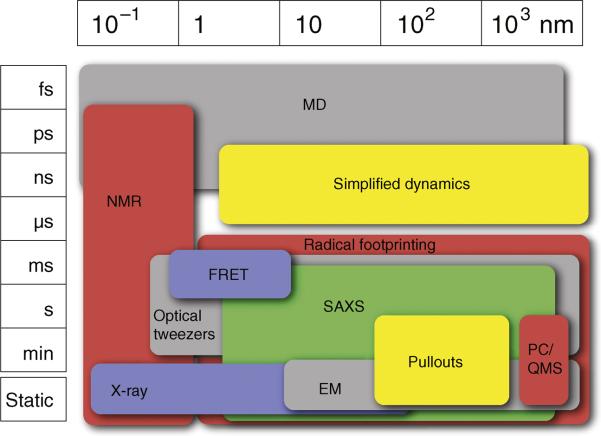
No single technique, computational or experimental, is able to span all relevant spatial and temporal scales (Figure 3). For static complexes, for example, X-ray crystallography can generate atomic structures of the components, while single particle cryo-electron microscopy (cryo-EM) can provide average mass density maps at nanometer resolution for the whole assembly; for processes, computer simulations are beginning to reach the microsecond time scale, while various single molecule and stopped-flow techniques come into play on the millisecond time scale. Thus, a key challenge is to integrate different kinds of static and dynamic characterizations at different resolutions to obtain a comprehensive description of a process. As when describing static structures [18••,19], such an integration of data will have to be achieved through computational approaches.
Figure 3. An overview of the spatial and temporal coverage of the various methods.
The x-axis represents the size of the systems that can be explored by each method in nanometers. The y-axis represents the time scales that can be reached. The methods and abbreviations are described in Table 2.
We expect that inspiration for the needed computational approaches will come from a wide range of fields that model dynamic systems. Examples include motion capture techniques for movies, where the motions of markers on an actor are tracked and used to restrain a general model of locomotion to make an animated character move like the actor [20]; kinematics in robotics, where motions are designed to connect a set of states subject to constraints, such as drive a vehicle from one point to another [21–23]; the master equation in chemical kinetics that captures rates of transitions between different states [24]; molecular dynamics simulations, where every attempt is made to make the trajectories correspond to reality [25,26]; simplified physical simulations, such as Brownian dynamics [25•,26] and modeling of transitions [27]; and diffusion-based models of biological processes [28]. However, none of these computational approaches is always accurate, applicable on all time and size scales of interest, capable of describing all properties of interest, and able to include all available experimental data and theoretical considerations. Such an integrative approach still needs to be developed.
In this review, we describe a process as a set of key states connected by transitions (Figure 2 and Table 1). Such a model is similar to the flowchart diagrams that are typically used to provide high-level views of processes. A representation of the process can vary in resolution, ranging from schemes involving two key states to high-resolution schemes with many key states and transitions between them. Some processes may be more concisely and efficiently modeled at a given resolution by a continuum diffusion model or molecular dynamics, particularly at high resolution where the number of states becomes large. As in the static structure case, we can use experimental and theoretical data to construct restraints that limit the set of possible process models [18••]. For example, a restraint can act to limit the distance between two components of the system as a function of time. More restraints are added as the evidence accumulates, reducing the number of acceptable models.
Important tasks required to build a structural dynamics model of a process are (i) discovering key states and determining their structures, (ii) finding which pairs of key states interconvert, (iii) determining rates of transitions between interconverting key states, and (iv) computing trajectories for the transitions between key states. The next four sections review how these four challenges can be addressed by different techniques (techniques referred to in italics are described in Table 2). These challenges are inter-related and progress toward resolving one may help resolve others.
Table 2.
A brief overview of the various experimental and computational methods mentioned in this review.
| Time resolved SAXS [46,111] | In time resolved small angle X-ray scattering (SAXS), a time course of the scattering profile is collected by repeatedly exposing a sample in solution. The scattering profile provides low-resolution information about the distribution of inter-atomic distances in the sample. |
| TROSY NMR [63,112] | Transverse Relaxation Optimized Spectroscopy (TROSY) is a variant of Nuclear Magnetic Resonance (NMR) spectroscopy that can be applied to large systems. This method isolates part of the system by replacing the remaining hydrogen atoms with deuterium atoms. The chemical shifts of the hydrogen atoms can then be monitored to measure local conformation and its changes. |
| PC/QMS [13] | In pulse-chase monitored by quantitative mass spectrometry (PC/QMS), a complex is allowed to assemble for some period, followed by a rapid dilution of non-bound proteins in solution with 14N labeled proteins. Quantitative mass spectrometry then measures the relative populations of the heavy and light molecules, producing an association rate estimate for accumulation in the complex. |
| Time resolved pullouts [113] | The cells are rapidly frozen and the media is ground. The ground substrate is thawed and the protein pulled out by affinity chromatography, bringing attached proteins along. These attached proteins can be identified with mass spectrometry or other methods. |
| Time resolved hydroxyl radical footprinting [31] | A brief pulse of synchrotron radiation is used to create radicals near the RNA that cleave the solvent exposed backbone. Sequencing of the resulting fragments allows the cleavage sites to be identified, and hence determines which parts of the backbone were exposed. Exposure information, coupled with secondary structure prediction, can be sufficient to reconstruct the shape of the RNA. |
| Flourescent tags [49] | Fluorescent tags are attached to particles and the system is observed through a microscope. When the marked particles are separated by tens of nanometers, the individual dyes can be located. Several different colors can be used at once to provide measurements of proximity. Fluorescent dyes suffer from photo bleaching that limits how long a single dye molecule can be used. |
| FRET [49,114]. | In Forster resonance energy transfer (FRET) spectroscopy, two particles are tagged with appropriate fluorophores. When the dyes are close to one another (tens of nanometers), they become coupled and excitation of one, the FRET donor, causes emission by the other, the FRET acceptor. The strength of this coupling depends on distance, allowing changes in distance to be detected. |
| Optical tweezers [115] | A micron sized polystyrene bead attached to part of the system is held in an optical trap. The trap can be used to either hold the bead at a specified force or, alternatively, to set the displacement over time. By restraining another part of the system (eg, by immobilizing a bead attached to another part of the system), a distance can be measured. Some recent setups allow a second bead to be trapped and manipulated independently. Optical tweezers can apply forces of up to hundreds of piconewtons. |
| Molecular dynamics [25,26,71•,74,75••] | The Newton's equations of motion are integrated for the atoms of the system, relying on a molecular mechanics force field. The result is a trajectory, sampled with time steps on the order of femtoseconds. The longest simulations approach microseconds in duration. Coarse-grained representations, combining multiple atoms into a single particle, can reach millisecond timescales. |
| Normal modes dynamics [116,117] | The assembly is represented as a collection of points connected by springs. The local dynamics of such an object is approximated by a linear combination of a small basis set of harmonic motions, each with a characteristic frequency. The trajectory is generated by an iterative extrapolation of local dynamics. |
| Motion planning [21,118] | Motion planning algorithms are a large family of techniques taken from robotics that search for non-colliding trajectories between two known states of the system. The most advanced techniques can handle hundreds of parameters. |
Discovering key states and determining their structures
Structural modeling of a dynamic process generally begins with the determination of key states and their structural characterization. If a conformationally homogenous sample of the key state can be purified, the whole arsenal of traditional structural biology techniques can be applied. These techniques are reviewed from a computational perspective in ref. [18••]. Key states that are not sufficiently stable can sometimes be stabilized by removing or modifying parts of the system (eg, ligands) to block the transition to another state. In studies of the ribosome-bound nascent chain by NMR spectroscopy, the RNA transcript was prepared without a stop codon. This modification led to translation arrest and allowed measurements to be taken on the partly produced protein [29]. In other work, the kiromycin antibiotic was used to stall translation in the Escherichia coli ribosome to take cryo-EM snapshots of the elongation factor Tu in complex with the ribosome [30]. Methods that can be used to characterize transient states and heterogeneous states directly are discussed next.
Characterizing transient states
Key states of the system often exist only for a brief time as the system changes from one stable state to another. To provide structural information about such a transient key state, the method must either quickly immobilize the key state or have high enough temporal resolution to take the measurement as the system changes. In the former category, hydroxyl radical footprinting breaks a structure into pieces over a few milliseconds, thus terminating the time evolution of the system. This approach has been used to monitor the early stages of ribosome assembly after rRNA-protein encounter, following changes in the structure of the 30S subunit [31••]. The results show that the initial RNA-protein complexes refold during the process. Fluorescent affinity tag purification allows visualization of the target protein in live cells, followed by extraction and detection of interacting macromolecular partners. It has been used to localize specific interactions of viral proteins with host-cell interaction partners at different stages during a viral infection [32].
NMR spectroscopy and fluorescence-based methods can monitor the assembly as it changes over time. Relaxation-dispersion NMR spectroscopy has been used to observe transient states of small complexes of proteins that only exist on the timescales of seconds [33–35••]. This new method has not yet been applied to macromolecular assemblies. Fluorescence tagging allows different types of measurements to be taken and has been used extensively in the determination of attributes of static structures. For example, the stoichiometry of a particular subunit in an assembly can be estimated by monitoring the fluctuations in intensity as complexes with tagged subunits move through the observed volume [36,37]. FRET can be used for structure determination by successively tagging pairs of proteins [38]. The detection of the addition or removal of a subunit from a single complex can be accomplished via fluorescence labeling of proteins [39] or polarization fluorescence spectroscopy [40].
Disentangling conformationally heterogeneous states
Methods that measure average properties of the system are typically comparatively easy to apply, but their precision is limited by conformational and kinetic heterogeneity in the sample. It is often difficult to create a sample containing only a single key state due to multiple kinetic pathways or inability to synchronize each step of a single pathway. To avoid averaging over various states in a sample, single molecule methods such as FRET and optical tweezers are required [41•,42]. Unfortunately, single molecule methods provide information about only a few variables at a time, thus making it difficult to reproduce the state of the whole assembly. Methods, such as cryo-electron tomography [43], not traditionally included in the category of single molecule techniques, also provide information about individual copies of the system. For example, the expectation-maximization algorithm, together with a maximum likelihood scoring function, is able to classify single particle cryo-EM images corresponding to different states of the complex. The classified images can then be used to produce structures of each of the well-populated states in the sample. This technique has been applied to the E. coli ribosome and the large T antigen of Simian Virus 40 [44].
Finding which pairs of key states interconvert
Given the set of key states, the allowed transitions need to be determined. These transitions are between the pairs of key states that can interconvert directly (ie, without passing through other states). The effort to determine the connected key states involves using experimentally measured time series or computational searches.
Using experimental time series
When the process involves rapid transitions between a number of relatively stable key states, any temporal data that can distinguish between different mixtures of states can be used to determine connectivity. Such data fitting approaches enumerate sets of connections and determines if rates exist that reproduce the experimental data for each choice of connectivity. Successful applications include finding the topology of RNA folding using hydroxyl radical footprinting [45] and monitoring virus capsid assembly using time resolved SAXS [46••]. In the former work, a fivestate model of the folding process of a large RNA molecule was proposed. The number of key states was determined by clustering the rate at which various parts of the molecule were protected during folding. To find the best model, all possible graphs of five key states and assignments of protected regions to states were enumerated and scored. The assignment of backbone protection in each key state was sufficient to uniquely determine the RNA conformations.
Using simulations of system dynamics
Various computational methods can be used to find both key states and transitions between them. The most direct approach is to simply simulate the assembly with molecular dynamics, and connect key states based on the molecular dynamics trajectories. Such approaches require a high-resolution representation of the starting state. In addition, the methods suffer from an inability to connect states distant from each other, due to limitations in the accuracy and length of molecular dynamics trajectories. Nevertheless, this approach has been used extensively for building models of processes occurring on smaller spatial and temporal scales, such as protein folding [47] and lipid vesicle formation [48]. In both cases, many simulations were run and the frames from the resulting trajectories were clustered to give a small number of highly populated key states connected by less populous transitions.
Determining rates of transitions between interconverting states
Knowledge of how key states are connected captures much of what we want to know about dynamic processes, but a complete description requires determining the transition rates between directly connected states in the model. Different techniques can be applied to this problem, depending on the properties of the system.
Monitoring an order parameter
An order parameter is a simple structural feature of the system that changes during the transition of interest. Fluorescent tags are useful for following individual particles [49••], monitoring the accumulation in the target key state [50••], tracking the orientation of a molecule [51], and measuring relative distances [52•–54]. For example, FRET fluctuations have been used to determine transition rates between RNA folding states [55,56]. Dyes were attached to immobilized RNA molecules so that the two key states had different FRET efficiency [56]. Photons were then gathered from single molecules and averaged over thousands of transitions. By using the FRET efficiency measurements in narrow windows around transitions, it was possible to monitor transitions as short as the average photon emission interval as well as to measure the time spent in each key state. Atomic force microscopy can be used to monitor the height of an assembly attached to a support at millisecond time scale and nanometer resolution, as was done on GroEL [57]. Optical tweezers can be used to measure a single distance in a single copy of the complex; for example, monitoring the rate of RNA unfolding by a helicase [58] or translation by the ribosome [59••]. Such experiments involve holding part of the system under tension and monitoring how the distance changes as the process proceeds. When applied to translation, single translational steps could be observed to occur with a median interval of 2.8 seconds and to take less than a tenth of a second to complete. NMR spectroscopy can use changes in the local environment of certain atoms, such as their solvent accessibility, to measure rates of conformational transitions on the microsecond to millisecond time scale [60,61•]. NMR-based methods for monitoring enzyme kinetics on the picosecond to seconds timescale were reviewed in ref. [61•].
Determining rates of addition of components to a system
The process of assembling a complex is particularly amenable to rate measurements as the transitions between key states primarily involve the accumulation of new species. Pulse chase monitored by quantitative mass spectrometry can measure the rate of addition of new subunits to the assembly. The main application so far has been elucidation of the ribosome assembly process [13]. SAXS can monitor the size of an assembly as it is built, and was applied, for example, to measuring the rate of virus capsid assembly [46••].
Computing trajectories for the transitions between states
Trajectories connecting one key state to another can contain a great deal of information, especially for models with sparse key states. Generating a complete trajectory typically requires computation, because experimental methods generally cannot monitor all structural coordinates for each molecule in an ensemble. The computational methods range from highly physically accurate, but expensive methods (eg, molecular dynamics simulation) to less physically realistic but cheaper approaches that allow us to compute trajectories between key states more separated in time and space (eg, normal modes dynamics and motion planning).
Experimental restraints on trajectories
Hydroxyl radical footprinting has been used to monitor the assembly process of the ribosome where the solvent accessibility of the RNA backbone could be determined with 10 millisecond precision in vivo [31••]. The solvent accessibility information it provided was sufficient to determine the folding nucleation sites and rates. While time resolved SAXS can generally monitor only coarse shape of the assembly, under certain circumstances with regular and well defined structures, the time-resolved data can be used to reconstruct the whole trajectory. Examples include formation of insulin fibrils [62] and virus capsids [46••]. Recently introduced TROSY NMR spectroscopy allows detecting changes in the local environment of a small part of the assembly, typically methyl groups, during a dynamic process [63]. Applications include monitoring conformational changes of a protein in the GroEL-GroES chaperonin [64] and following the transition between two conformations of the ClpP protease [65••].
Computing trajectories
Computational approaches are generally needed to obtain trajectories between key states. The gold standard is to perform all atom molecular dynamics simulations of the system in solvent. Such simulations require detailed structural information about the components of the system and their starting state, and can only simulate processes lasting less than microseconds. Fortunately, molecular dynamics software parallelizes efficiently and so can handle large systems consisting of millions of atoms [66••–68]. Coarse graining and multiscale methods can extend the reachable time scales to fractions of a millisecond by representing many atoms with a single particle and using force fields derived from more detailed all-atom computations [69•–73•]. Adding intermediate key states along the transition can make it easier to explore longer time scales [74]. An orthogonal approach to speeding simulations, at the risk of loosing the physical accuracy, is to a add forces that guide the evolution of the system. These forces can minimize the time the simulation spends stuck in dead ends and local minima. For example, each atom can be constrained inside a ball centered at the final position of the atom, as given by an X-ray crystallographic structure [75•]. The radius of the ball is initially large enough to include the initial position of the atom, and gradually shrinks to zero during the simulation, forcing the atom to converge on its final location.
Techniques such as normal modes dynamics and motion planning further sacrifice physical realism to handle long time scales. Normal modes dynamics has been applied to ribosomes [76,77], viruses [78–80], myosin [81,82], and chaperonins [83] at time scales up to 10–8 second [84]. Motion planning approaches have been applied to connecting the open and closed conformations of the K-channel [85], studying the folding pathways of proteins [86] and RNA [87], and computing large-amplitude motions [88].
Example process: the ubiquitin-proteasome protein degradation pathway
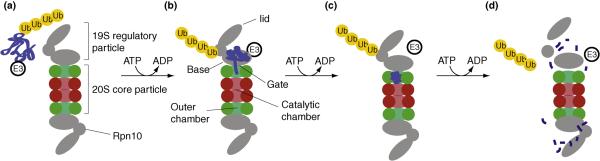
To illustrate our perspective on dynamic processes outlined above, we review recent studies toward elucidating the dynamics of the 26S proteasome as part of the ubiquitin-proteasome pathway (Figure 4). This pathway plays a key role in regulating protein levels in eukaryotes [17,89]. The ubiquitinproteasome pathway involves tagging the substrate protein by covalently attaching multiple ubiquitin molecules, followed by degradation of the tagged protein inside the 26S proteasome and the release of the ubiquitin molecules. A number of key biological questions remain unanswered, such as how tagged substrates are recognized by the proteasome for degradation, whether the proteasome disassembles during the catalytic cycle, and how the substrate is degraded within the proteasome. Addressing these questions is challenging, as some key states are transient and it is difficult to prepare sufficient quantities of tagged substrates.
Figure 4. A model of the substrate degradation by the proteasome.
The degradation process is part of a larger pathway, consisting of activation of ubiquitin, conjugation of the protein substrate and ubiquitins, degradation of the tagged protein, and deubiquitination to recycle the ubiquitins (bottom). The degradation process (top) is modeled by four key states and transitions between them, discussed in more detail in the text. The modeled system involves the 26S proteasome, the E3 ligase enzyme and the ubiquinated substrate protein. The four key states of the model are: [A] stable holo 26S, [B] recruitment of polyubiquinated substrate, [C] storage of substrate inside the proteasome and [D] disassociated and disassembled regulator particle. Arrows show transitions between states. As more information is obtained, the model can become more detailed by adding key states, increasing the spatial resolution of each key state, and mapping trajectories between key states.
The 26S proteasome consists of a 28-protein 20S core particle chamber that is capped on both sides by a ~13-protein 19S regulatory particle. We chose to describe the degradation process using four key states, as suggested by the “chew and spew” model [90], although alternative models have also been proposed [91]. The “chew and spew” model proposes that ATP hydrolysis in the presence of product peptides triggers controlled disassociation and disassembly of the 19S regulatory particle from the 26S proteasome (Figure 4). Next, we describe each of the key states and the experimental and computational techniques used for their characterization.
The first key state of the model consists of the ubiquitinated substrate bound to the E3 ligase enzyme and a holo 26S proteasome. The structure of the 20S core particle was solved by X-ray crystallography [92]. The identities and interactions of proteins in the 19S regulatory particle were revealed by affinity purification studies followed by mass spectrometry [93–97]. A low-resolution structure of the entire 26S holo molecule was determined by cryo-EM [98].
In the second key state, the ubiquinated-substrate/E3 complex is bound to the holo 26S proteasome via one of two known ubiquitin receptors, Rpn10 or Rpn13 subunits of the 19S regulatory particle [99–102]. Biochemical studies have revealed that Rpn10 is bound to the polyubiquitin chain [103] and recognizes targets via its ubiquitin-binding motif [104]. The amino-terminal domain of Rpn13 shows no similarity to Rpn10 ubiquitin-binding motif, as revealed by NMR spectroscopy and X-ray crystallography.
In the third key state of our model, the protein substrate is located within the 20S cavities, prior to degradation. The existence of this key state is suggested by cryo-EM and tandem mass spectrometry that determined the stoichiometry and location of substrates within the 26S proteasome [16]. This study also observed structural differences between the free and substrate-bound 26S structure.
In the fourth key state, the 19S particle is disassembled and disassociated from the holo 26S and the peptides have been released. The disassembled complex has been mapped by negative stain EM [90].
The transitions between the key states are less well characterized. Between key states two and three, substrates enter the 20S core particle through a gated channel after being unfolded by energydependent translocation through the ATPase ring [102,105]. The residues involved in this gate as well as its open and closed structure have been localized in the 20S core particle of an archaeal proteasome using cryo-EM [106,107]. The mechanism appears to be conserved in mammals [108].
The transition from the third to fourth key state involves degradation of the substrate and disassociation of the complex. NMR spectroscopy has been used to monitor the 20S core particle during the degradation process. The study revealed motions occurring on the tens of nanoseconds timescale in the outer chamber walls that are correlated with much slower motions on the millisecond time scale in the catalytic chamber [109••]. The coupling between substrate degradation and disassociation of 19S regulatory particle is likely to be linked to conformational changes in its ATPasering. These changes have been observed using biochemical studies [90].
Conclusions
Understanding macromolecular processes requires a wide range of experimental and computational techniques. As a result, we expect that integrative approaches will be critical, even more so than in the static structure case. Most existing models of dynamic processes have been the result of ad hoc integration of experimental results via simple models or mental constructions. But moving to higher accuracy, precision, coverage, and efficiency through incorporating all the available information will require novel computational approaches.
Acknowledgements
We thank Yifan Cheng, Friederich Förster, Joerg Gsponer, Avner Schlessinger, and Elizabeth Villa for helpful discussions. KL, JVM, and DSD have been funded by the Clore Foundation Predoctoral Fellowship, Spanish Ministry of Education Postdoctoral Fellowship, and Weizmann Institute Advancing Women in Science Postdoctoral Fellowship, respectively. We also acknowledge support from the Sandler Family Supporting Foundation, NIH/NCRR U54 RR022220, NIH R01 GM083960, NIH PN2 EY016525, NSF IIS 0705196, and Pfizer Inc. And we are grateful for computer hardware gifts from Ron Conway, Mike Homer, Intel, Hewlett-Packard, IBM, and Netapp.
Bibiography
- • 1.Sali A, Chiu W. Macromolecular Assemblies Highlighted. Structure. 2005;13:339–341. [Google Scholar]
- 2.van Oijen AM. Single-molecule studies of complex systems: the replisome. Mol Biosyst. 2007;3:117–125. doi: 10.1039/b612545j. [DOI] [PubMed] [Google Scholar]
- • 3.Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu Rev Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 4.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]; A review of single molecule experiments used to study translation in the ribosome.
- 5.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn AY, Melville MW, Frydman J. Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol. 2001;135:176–184. doi: 10.1006/jsbi.2001.4380. [DOI] [PubMed] [Google Scholar]
- 7.Clare DK, Stagg S, Quispe J, Farr GW, Horwich AL, Saibil HR. Multiple states of a nucleotidebound group 2 chaperonin. Structure. 2008;16:528–534. doi: 10.1016/j.str.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 9.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 11.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 12.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008 doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talkington M, Siuzdak G, Williamson J. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misra N, Lees D, Zhang T, Schwartz R. Pathway complexity of model virus capsid assembly systems. Computational and mathematic models in medicine. 2008;9:277–293. [Google Scholar]
- 15.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 16.Sharon M, Witt S, Felderer K, Rockel B, Baumeister W, Robinson CV. 20S proteasomes have the potential to keep substrates in store for continual degradation. J Biol Chem. 2006;281:9569–9575. doi: 10.1074/jbc.M511951200. [DOI] [PubMed] [Google Scholar]
- 17.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- •• 18.Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]; The paper describes an approach for integrating experimental and computational methods toward determination of static macromolecular assemblies.
- 19.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 20.Moeslund T, Granum E. A Survey of Computer Vision-Based Human Motion Capture. Computer Vision and Image Understanding. 2001;81:231–268. [Google Scholar]
- 21.Latombe J-C. Motion Planning: A Journey of Robots, Molecules, Digital Actors, and Other Artifacts. 1999 Edited by: unknown. [Google Scholar]
- 22.Agarwal PK, Guibas LJ, Edelsbrunner H, Erickson J, Isard M, Har-Peled S, Hershberger J, Jensen C, Kavraki L, Koehl P, et al. Algorithmic issues in modeling motion. ACM Comput. Surv. 2002;34:550–572. [Google Scholar]
- 23.Thrun S, Montemerlo M, Dahlkamp H, Stavens D, Aron A, Diebel J, Fong P, Gale J, Halpenny M, Hoffmann G, et al. Stanley, The robot that won the DARPA Grand Challenge. Journal of Robotic Systems. 2006;23:661–692. [Google Scholar]
- 24.MacNamara S, Burrage K, Sidje RB. Multiscale Modeling of Chemical Kinetics via the Master Equation. Multiscale Modeling & Simulation. 2008;6:1146–1168. [Google Scholar]
- • 25.McGuffee SR, Elcock AH. Atomically detailed simulations of concentrated protein solutions: the effects of salt, pH, point mutations, and protein concentration in simulations of 1000-molecule systems. J Am Chem Soc. 2006;128:12098–12110. doi: 10.1021/ja0614058. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Chang CE, Yu Z, Zhang Y, Sun M, Leyh TS, Holst MJ, McCammon JA. Diffusional channeling in the sulfate-activating complex: combined continuum modeling and coarse-grained brownian dynamics studies. Biophys J. 2008;95:4659–4667. doi: 10.1529/biophysj.108.140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elber R. A milestoning study of the kinetics of an allosteric transition: atomically detailed simulations of deoxy Scapharca hemoglobin. Biophys J. 2007;92:L85–87. doi: 10.1529/biophysj.106.101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sosinsky GE, Deerinck TJ, Greco R, Buitenhuys CH, Bartol TM, Ellisman MH. Development of a model for microphysiological simulations: small nodes of ranvier from peripheral nerves of mice reconstructed by electron tomography. Neuroinformatics. 2005;3:133–162. doi: 10.1385/NI:3:2:133. [DOI] [PubMed] [Google Scholar]
- 29.Hsu ST, Fucini P, Cabrita LD, Launay H, Dobson CM, Christodoulou J. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc Natl Acad Sci U S A. 2007;104:16516–16521. doi: 10.1073/pnas.0704664104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- •• 31.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008 doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors use hydroxyl radical footprinting to inverstigate the ribosome assembly process.
- 32.Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem. 2006;281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 33.Vallurupalli P, Hansen DF, Kay LE. Structures of invisible, excited protein states by relaxation dispersion NMR spectroscopy. Proc Natl Acad Sci U S A. 2008;105:11766–11771. doi: 10.1073/pnas.0804221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugase K, Lansing JC, Dyson HJ, Wright PE. Tailoring relaxation dispersion experiments for fast-associating protein complexes. J Am Chem Soc. 2007;129:13406–13407. doi: 10.1021/ja0762238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 35.Suh JY, Tang C, Clore GM. Role of electrostatic interactions in transient encounter complexes in protein-protein association investigated by paramagnetic relaxation enhancement. J Am Chem Soc. 2007;129:12954–12955. doi: 10.1021/ja0760978. [DOI] [PubMed] [Google Scholar]; In this study, the authors use paramagnetic relaxation enhancement to detect transient macromolecular interactions.
- 36.Chen Y, Muller JD. Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy. Proc Natl Acad Sci U S A. 2007;104:3147–3152. doi: 10.1073/pnas.0606557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sykora J, Kaiser K, Gregor I, Bonigk W, Schmalzing G, Enderlein J. Exploring fluorescence antibunching in solution to determine the stoichiometry of molecular complexes. Anal Chem. 2007;79:4040–4049. doi: 10.1021/ac062024f. [DOI] [PubMed] [Google Scholar]
- 38.Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O'Toole ET, Giddings TH, Jr., Sundin BA, Davis TN. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dange T, Grunwald D, Grunwald A, Peters R, Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443:466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- • 41.Moffitt J, Chemla Y, Smith S, Bustamante C. Recent advances in optical tweezers. Annual Review of Biochemistry. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 42.Moerner WE. New directions in single-molecule imaging and analysis. Proc Natl Acad Sci U S A. 2007;104:12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 44.Scheres SH, Nunez-Ramirez R, Gomez-Llorente Y, San Martin C, Eggermont PP, Carazo JM. Modeling experimental image formation for likelihood-based classification of electron microscopy data. Structure. 2007;15:1167–1177. doi: 10.1016/j.str.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laederach A, Shcherbakova I, Liang MP, Brenowitz M, Altman RB. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J Mol Biol. 2006;358:1179–1190. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 46.Tuma R, Tsuruta H, French KH, Prevelige PE. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J Mol Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; In the paper, time resolved SAXS is used to detect unstable intermediates states during virus capsid formation. By assuming that intermediates are spherically symmetric, the authors are able to determine the structure of the intermediates.
- 47.Chodera JD, Singhal N, Pande VS, Dill KA, Swope WC. Automatic discovery of metastable states for the construction of Markov models of macromolecular conformational dynamics. J Chem Phys. 2007;126:155101. doi: 10.1063/1.2714538. [DOI] [PubMed] [Google Scholar]
- 48.Kasson PM, Pande VS. Control of membrane fusion mechanism by lipid composition: predictions from ensemble molecular dynamics. PLoS Comput Biol. 2007;3:e220. doi: 10.1371/journal.pcbi.0030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 49.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annual Review of Biochemistry. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]; The paper reviews single-molecule fluorescence techniques including FRET and single particle tracking.
- •• 50.Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2008 doi: 10.1038/nature07600. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study a mimic of the central channel of the nuclear pore complex in a membrane was designed. This artificial system reproduces key features of trafficking through the NPC, including transport-factor-mediated cargo import.
- 51.Nishizaka T, Oiwa K, Noji H, Kimura S, Muneyuki E, Yoshida M, Kinosita K. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nature Structural and Molecular Biology. 2004;11:142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- • 52.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 54.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proc Natl Acad Sci U S A. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Three-color alternating-laser excitation of single molecules: monitoring multiple interactions and distances. Biophys J. 2007;92:303–312. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokokawa M, Wada C, Ando T, Sakai N, Yagi A, Yoshimura SH, Takeyasu K. Fast-scanning atomic force microscopy reveals the ATP/ADP-dependent conformational changes of GroEL. EMBO J. 2006;25:4567–4576. doi: 10.1038/sj.emboj.7601326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr., Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 59.Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors used optical tweezers to monitor translation in the ribosome. The resolution of the experiment was sufficient to observe three substeps in each translocation and conclude that there are two rate-limiting steps in the process.
- 60.Kern D, Eisenmesser EZ, Wolf-Watz M. Enzyme dynamics during catalysis measured by NMR spectroscopy. Methods Enzymol. 2005;394:507–524. doi: 10.1016/S0076-6879(05)94021-4. [DOI] [PubMed] [Google Scholar]
- • 61.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 62.Vestergaard B, Groenning M, Roessle M, Kastrup JS, van de Weert M, Flink JM, Frokjaer S, Gajhede M, Svergun DI. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 64.Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wuthrich K. Direct NMR observation of a substrate protein bound to the chaperonin GroEL. Proc Natl Acad Sci U S A. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 65.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci U S A. 2005;102:16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study TROSY NMR experiments were used to describe the interactions of inhibitors with the 670 kDa 20S proteasome.
- •• 66.Sanbonmatsu KY, Tung CS. High performance computing in biology: multimillion atom simulations of nanoscale systems. J Struct Biol. 2007;157:470–480. doi: 10.1016/j.jsb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; A review of the current capabilities of molecular dynamics and the challenges in the field.
- 67.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 68.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 69.Ayton GS, Voth GA. Multiscale simulation of transmembrane proteins. J Struct Biol. 2007;157:570–578. doi: 10.1016/j.jsb.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 70.Nakamura T, Sasa S-i. Systematic derivation of coarse-grained fluctuating hydrodynamic equations for many Brownian particles under nonequilibrium conditions. Physical Review E (Statistical, Nonlinear, and Soft Matter Physics) 2006;74:031105–031114. doi: 10.1103/PhysRevE.74.031105. [DOI] [PubMed] [Google Scholar]
- 71.Tozzini V, Trylska J, Chang CE, McCammon JA. Flap opening dynamics in HIV-1 protease explored with a coarse-grained model. J Struct Biol. 2007;157:606–615. doi: 10.1016/j.jsb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 72.Arkhipov A, Yin Y, Schulten K. Four-scale description of membrane sculpting by BAR domains. Biophys J. 2008;95:2806–2821. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 73.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 74.West AM, Elber R, Shalloway D. Extending molecular dynamics time scales with milestoning: example of complex kinetics in a solvated peptide. J Chem Phys. 2007;126:145104. doi: 10.1063/1.2716389. [DOI] [PubMed] [Google Scholar]
- • 75.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci U S A. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang YM, Rader AJ, Bahar I, Jernigan RL. Global ribosome motions revealed with elastic network model. J. Struct. Biol. 2004;147:302. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 77.Tama F, Valle M, Frank J, Brooks CL. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:9319. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rader AJ, Vlad DH, Bahar I. Maturation dynamics of bacteriophage HK97 capsid. Structure. 2005;13:413. doi: 10.1016/j.str.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 79.Tama F, Brooks CL., III Diversity and identity of mechanical properties of icosahedral viral capsids studied with elastic network normal mode analysis. J. Mol. Biol. 2005;345:299. doi: 10.1016/j.jmb.2004.10.054. [DOI] [PubMed] [Google Scholar]
- 80.van Vlijmen HWT, Karplus M. Normal mode calculations of icosahedral viruses with full dihedral flexibility by use of molecular symmetry. J. Mol. Biol. 2005;350:528. doi: 10.1016/j.jmb.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 81.Li GH, Cui Q. Analysis of functional motions in Brownian molecular machines with an efficient block normal mode approach: myosin-II and Ca2+-ATPase. Biophys. J. 2004;86:743. doi: 10.1016/S0006-3495(04)74152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Navizet I, Lavery R, Jernigan RL. Myosin flexibility: structural domains and collective vibrations. Proteins. 2004;54:384. doi: 10.1002/prot.10476. [DOI] [PubMed] [Google Scholar]
- 83.Keskin O, Bahar I, Flatow D, Covell DG, Jernigan RL. Molecular mechanisms of chaperonin GroEL-GroES function. Biochemistry. 2002;41:491. doi: 10.1021/bi011393x. [DOI] [PubMed] [Google Scholar]
- 84.Tama F, Brooks CL. SYMMETRY, FORM, AND SHAPE: Guiding Principles for Robustness in Macromolecular Machines. Annual Review of Biophysics and Biomolecular Structure. 2006;35:115–133. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- 85.Enosh A, Raveh B, Furman-Schueler O, Halperin D, Ben-Tal N. Generation, comparison and merging of pathways between protein conformations: Gating in K-channels. Biophys J. 2008 doi: 10.1529/biophysj.108.135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomas S, Song G, Amato NM. Protein folding by motion planning. Phys Biol. 2005;2:S148–155. doi: 10.1088/1478-3975/2/4/S09. [DOI] [PubMed] [Google Scholar]
- 87.Tang X, Kirkpatrick B, Thomas S, Song G, Amato NM. Using motion planning to study RNA folding kinetics. J Comput Biol. 2005;12:862–881. doi: 10.1089/cmb.2005.12.862. [DOI] [PubMed] [Google Scholar]
- 88.Cortes J, Simeon T, Ruiz de Angulo V, Guieysse D, Remaud-Simeon M, Tran V. A path planning approach for computing large-amplitude motions of flexible molecules. Bioinformatics. 2005;21(Suppl 1):i116–125. doi: 10.1093/bioinformatics/bti1017. [DOI] [PubMed] [Google Scholar]
- 89.Hershko D, Bornstein G, Ben-Izhak O, Carrano A, Pagano M, Krausz MM, Hershko A. Inverse relation between levels of p27(Kip1) and of its ubiquitin ligase subunit Skp2 in colorectal carcinomas. Cancer. 2001;91:1745–1751. doi: 10.1002/1097-0142(20010501)91:9<1745::aid-cncr1193>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 90.Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, Erdjument-Bromage H, Tempst P, Buranda T, Sklar LA, Baumler J, et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 91.Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 92.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 93.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 95.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 97.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walz J, Erdmann A, Kania M, Typke D, Koster AJ, Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 99.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrinhomology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 102.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 103.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 104.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 105.Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 106.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 108.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 109.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]; In this study, a special isotope labeling scheme is used in combination with methyl-TROSY NMR spectroscopy to reveal functionally important motions of the 670 kDa 20S proteasome. These results demonstrate that NMR spectroscopy can provide detailed insight into assemblies over an order of magnitude larger than those routinely studied using methodology that is generally applicable.
- 110.Baldwin RL. Protein folding. Matching speed and stability. Nature. 1994;369:183–184. doi: 10.1038/369183a0. [DOI] [PubMed] [Google Scholar]
- 111.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 112.Ohki S, Dohi K, Tamai A, Takeuchi M, Mori M. Stable-isotope labeling using an inducible viral infection system in suspension-cultured plant cells. J Biomol NMR. 2008;42:271–277. doi: 10.1007/s10858-008-9283-x. [DOI] [PubMed] [Google Scholar]
- 113.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 114.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci U S A. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Wynsberghe AW, Cui Q. Interpreting correlated motions using normal mode analysis. Structure. 2006;14:1647–1653. doi: 10.1016/j.str.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 117.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc. Natl. Acad. Sci. USA. 2003;100:12570. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Latombe JC. Probabilistic Roadmaps: A Motion Planning Approach Based on Active Learning. Cognitive Informatics, 2006. ICCI 2006. 5th IEEE International Conference.2006. pp. 1–2. [Google Scholar]
- 119.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- • 120.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 121.Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]