Abstract
Despite eradication of smallpox three decades ago, public health concerns remain due to its potential use as a bioterrorist weapon. Smallpox and other orthopoxviruses express virulence factors that inhibit the host’s complement system. In this study, our goals were to characterize the ability of the smallpox inhibitor of complement enzymes, SPICE, to regulate human complement on the cell surface. We demonstrate that SPICE binds to a variety of cell types and that the heparan sulfate and chondroitin sulfate glycosaminoglycans (GAGs) serve as attachment sites. A transmembrane engineered version as well as soluble recombinant SPICE inhibited complement activation at the C3 convertase step with equal or greater efficiency than that of the related host regulators. Moreover, SPICE attached to GAGs was more efficient than transmembrane SPICE. We also demonstrate that this virulence activity of SPICE on cells could be blocked by a mAb to SPICE. These results provide insights related to the complement inhibitory activities of poxviral inhibitors of complement and describe a mAb with therapeutic potential.
Keywords: complement regulation, poxviruses, glycosaminoglycans
Introduction
The employment of smallpox virus as a bioterrorist weapon is a major public health concern. Existence of clandestine stockpiles, lack of immunity by the majority of the population, and complications of the current vaccine have reinvigorated interest in the poxviral field. Additionally, many authorities consider another orthopox virus, monkeypox, to be the most important poxviral infection and an emerging natural threat (1–3). The monkeypox epidemic in the US in 2003 and its potential spread to rodents in the US underpin these concerns.
To establish infection, poxviruses must first subvert the innate immune response of the host. The complement system lies at the interface between innate and adaptive immunity, providing a first line of defense against pathogens. It consists of a family of soluble and cell-surface proteins that recognize pathogen-associated molecular patterns, altered-self ligands, and immune complexes (4, 5). One strategy used by poxviruses to control complement activation is the expression of inhibitors of complement enzymes (PICES) that mimic the host’s complement regulators. PICES from variola, monkeypox, and vaccinia are named SPICE, MOPICE and VCP, respectively (6–8). Structurally, SPICE and VCP consist exclusively of four of the repeating domains called complement control protein (CCP) modules that are the structural mimics (~30% homologous) to human membrane regulators, membrane cofactor protein (MCP; CD46) and decay accelerating factor (DAF; CD55) (9, 10). MOPICE has three CCPs and a small remnant of the fourth (7, 11). Functionally, the PICES also possess the same two functional activities as those of human regulators: Cofactor activity (CA) refers to the limited proteolytic degradation of C3b and C4b that requires a cofactor protein working in concert with the plasma serine protease factor I while Decay-accelerating activity (DAA) refers to the dissociation or decay of the catalytic serine protease domain from complement-activating enzyme complexes or convertases. Utilizing these inhibitory mechanisms, previous studies have established that SPICE inactivates human complement more efficiently (100–1000-fold) than either VCP or MOPICE (6, 7, 12, 13).
Additionally, PICES possess heparin binding sites that are similar to those found in the human plasma complement inhibitors, factor H and C4b-binding protein (7, 14–16). The binding of heparin by factor H enhances cofactor and enzyme dissociating activities (17). Structural investigations suggest that the heparin binding sites may overlap complement inhibitory sites (15). We previously demonstrated that SPICE, MOPICE and VCP bind to heparin with a higher affinity than human factor H (7). Additionally, recombinant VCP can attach to the surface of cells via its interaction with heparan sulfate proteoglycans (16). Binding to heparin and GAGs may be an important functional capability because it provides a mechanism for a secreted protein to anchor to host cells, viruses, or virally-infected cells where it may modulate complement activation (18).
An emerging national priority is development of improved diagnostics and therapeutics to treat smallpox (19, 20). New therapeutic strategies include production of antiviral compounds and therapeutic mAbs that target virulence factors such as the PICES (19–21). Poxviral complement regulators are attractive targets for therapeutic intervention. For example, VCP can inhibit antibody-dependent, complement-enhanced neutralization of vaccinia virus virions (22) and viruses lacking VCP are attenuated (22, 23). These results point to an important role for VCP (and SPICE by inference) in attenuating the host’s complement system and their attractiveness as targets to treat poxviral infections.
Our studies demonstrate that SPICE anchored to cells via a transmembrane domain or through GAGs potently inhibits human complement activation. Further, we identify a mAb that inhibits SPICE function on cells. Thus, these studies establish a mechanism for SPICE attachment to host cells and demonstrate its potent complement inhibitory activity following such binding.
Materials and Methods
Generation of stable lines expressing SPICE-TM
Unless otherwise noted, Chinese hamster ovary cells (CHO) were the CHO-K1 cell line from American Type Culture Collection (Manassas, VA). Generation of the MCP 3–10 CHO cell line was previously described (24). To prepare transmembrane SPICE expressed in CHO, CCPs 1 – 4 were generated by PCR from the previously described SPICE cDNA (7) using the following primers: 5' GCGGATCCGGAATGGGAATGAAGGTGGAGAGCGTG 3' and 5' CCGGAATTCGCGTACACATTTTGGAAGTTC 3'. It was subsequently cloned into the BamH1 and EcoR1 sites of pcDNA3 (Invitrogen). The resulting plasmid was digested with EcoR1 and Not1 and ligated with an MCP-BC1 fragment containing the juxtamembraneous 10 amino acid domain, transmembrane domain and cytoplasmic tail generated from the template MCP-BC1 using the following primers: 5' CCGGAATTCGGATATCCTAAACCTGAGGA 3'and 5'ATAAGAATGCGGCCGCTTAGCATATTCAGCTCCACCATC 3'. Pvu1 linearized DNA was then transfected into CHO cells using FUGENE-6 (Roche), according to the manufacturer’s recommendations. Cells were maintained in Ham’s F12 with 10% heat inactivated FBS. After 48 h, G418 was added at a concentration of 0.5 mg/ml. G418 resistant pools, labeled with a polyclonal Ab that recognizes SPICE (7), were sorted according to expression level. Single cells were deposited onto a 96-well plate using a MoFlo high speed flow cytometer (DAKO Cytomation). A stable line (clone H3) was selected for SPICE surface expression by FACS using a polyclonal Ab and mAb KL5.1 (25). The expression level of this cell line was compared to the MCP clone (3–10) via flow cytometry using MCP polyclonal antibody and mAb TRA-2-10 (24) and analyzed by CellQuest Pro (BD Biosciences).
Cell lines
Cell lines used to assess SPICE binding were obtained from the Washington University Tissue Culture Support Center. The HeLa epithelial cell line was grown in Dulbecco’s Modified Eagles Medium, 2 mM L-glutamine and 10% FBS; HepG2 epithelial cell line was grown in MEM plus Earle’s salts with 2 mM L-glutamine and 10% FBS; the HaCat, keratinocyte line, was grown in Dulbecco’s Modified Eagle’s Medium with 8 mM L-glutamine, 50 mM HEPES and 10% FBS; the HMEC endothelial cell line was grown in MCDB 131 (Invitrogen) supplemented with 10% FBS, 2 mM L-glutamine, 0.3% NaHCO3, 1 µg/ml hydrocortisone (Sigma), 10 ng/ml epidermal growth factor (Sigma); IMR-90, a fibroblast cell line, was grown in Dulbecco’s Modified Eagles Medium, 2 mM L-glutamine and 10% FBS. CHO K1, 745, and M1 cell lines were grown in Ham’s F12 supplemented with 10% heat-inactivated FBS and 2 mM L-glutamine. These cell lines have been previously described (26–31). Media were supplemented with 100 units/ml penicillin G and 100 µg/ml streptomycin sulfate.
Isolation of human peripheral blood subsets
Primary human peripheral blood mononuclear cells (PBMC) were isolated from the buffy coats of healthy volunteers by Ficoll-Hypaque (Pharmacia) density gradient centrifugation per manufacturer’s protocol. Human CD14+ monocytes, CD19+ B cells, CD4+ T and CD8+ T lymphocytes were purified from the PBMC by positive selection using antibody-coated magnetic beads (Miltenyi Biotec) per manufacturer’s direction. Red blood cells were obtained from whole blood from a normal volunteer via protocols (32).
SPICE binding to cells and evaluation by flow cytometry
Cells were obtained by treatment either with 0.05% trypsin/0.53 mM EDTA or 4 mM EDTA (both from Mediatech) and washed with PBS/1% FBS. Typically, 1 × 106 cells were mixed with 20 µg of SPICE in a total volume of 100 µl and incubated at 30°C with gentle shaking in an Eppendorf Thermomixer for 30 min. Following incubation, cells were placed on ice and washed with PBS/1% FBS. SPICE was detected by flow cytometry using a previously prepared rabbit polyclonal Ab against VCP (7) followed by incubation with a FITC-donkey anti- rabbit IgG secondary Ab (Sigma). Cells were fixed in 0.5% paraformaldehyde in PBS.
Sodium chlorate treatment of cultured cells
Sulfation was inhibited by sodium chlorate treatment (33). CHO cells were cultured in sulfate-free Ham’s F12 medium (Washington University Tissue Culture Support Center) supplemented with 10% dialyzed FBS (Fisher Scientific) containing different concentrations (1–25 mM) of sodium chlorate. After overnight culture, cells were processed for SPICE binding as described above.
Production of recombinant SPICE in E. coli
Using SPICE cDNA (7) as a template, the coding sequence of SPICE was generated by PCR using the following oligos: 5'CGCGGATCCATGTGCTGTACTATTCCGTCAC 3' and 5' ATAAGAATGCGGCCGCTTATTTTGGAAGTT 3'. The resulting PCR fragment was ligated into the BamH1 and Not1 sites of pET28a(+)-2, a derivative generated in our laboratory of pET28a(+) (EMD/Novagen). For recombinant protein production a method was developed (modified from (7)). First, 25 ml of an overnight culture of E.coli containing the construct was inoculated into 500 ml of LB containing kanamycin (30 µg/ml) and chloramphenicol (34 µg/ml) and grown to an OD600 of 0.6–0.8 followed by induction with 1 mM IPTG at 37°C for an additional 3–5 hr. Cells were harvested and pellets were frozen at −80°C until needed.
For inclusion body protein purification, pellets were thawed and resuspended in 50 ml of Solution Buffer (50 mM Tris, pH 8.0, 25% sucrose, 1 mM EDTA, 0.01% NaN3, and 10 mM DTT) to which 0.8 ml of freshly prepared 50 mg/ml lysozyme (Sigma), 1250 units of benzonase nuclease (Novagen), and 1 ml of 1M MgCl2 were added. An equal volume of Lysis Buffer (50 mM Tris, pH 8.0, 1% Triton X-100, 0.1 M NaCl, 0.01% NaN3, and 10 mM DTT) was added and the solution was stirred gently at room temperature for one h. After cooling, the suspension was sonicated with three 15s bursts (Fisher Scientific Model 500 Sonic Dismembrator) at 50% amplitude followed by the addition of 5 ml of 0.5 M EDTA. The lysate was then centrifuged at 6000 g for 30 min at 4°C. The resulting inclusion body pellet was washed (50 mM Tris, pH 8.0, 0.5% Triton X-100, 0.1 M NaCl, 1 mM EDTA, 0.01% NaN3, and 1 mM DTT) followed by a second wash with the same buffer, but without Triton X-100.
For solubilization of the inclusion bodies, the pellet was resuspended in 6 M guanidine HCl, 10 mM Tris pH 8.0, and 20 mM β-mercaptoethanol and centrifuged at 14,000 g for 10 min. A second high speed centrifugation at 100,000 g for 30 min at 4°C was performed to remove any insoluble material.
For protein refolding, solubilized inclusion body protein was added dropwise in three injections over 36 h at a final concentration of 10–100 µg/ml in refolding buffer (100 mM Trizma Base, 400 mM L-arginine-HCl (Sigma), 2 mM EDTA, 0.02 M ethanolamine, 0.5 mM oxidized glutathione (Sigma), and 5 mM reduced glutathione (Sigma). The refolding solution was concentrated in a Millipore Stirred Filtration Cell followed by buffer exchange with 20 mM Tris, pH 8.0.
C3b and C4b binding
An ELISA format was used for ligand binding as described (7). Briefly, human C3b or C4b (Complement Technologies) was coated overnight at 4°C on wells at 5 µg/ml in PBS followed by blocking for 1 h at 37°C with 1% BSA, 0.1% Tween 20 in PBS. Proteins were diluted in low salt ELISA buffer (10 mM Tris, pH 7.2, 25 mM sodium chloride, 0.05% Tween 20, 4% BSA, and 0.25% Nonidet P-40) and incubated for 1.5 h at 37°C (7). Rabbit anti-VCP Ab (1/5000) in low salt buffer was then added for 1 h at 37°C. Following washing, a peroxidase-coupled donkey anti-rabbit IgG was added, and OD of the TMB substrate was determined. Binding assays were performed employing serially diluted samples on at least four separate occasions.
Initiation of Complement Activation
The standard procedure for the initiation of the complement pathways has been reported (34). Briefly, CHO cells are grown to ~70% confluency and collected by trypsinization into 1% FCS-PBS. Sensitizing antibody was an IgG prepared from rabbits injected with whole CHO cells (Harlan). The Ab was added to cells and the mixture incubated for 30 min at 4°C. Following two washes with 1% FCS-PBS, 100 µl of C8-deficient (C8d) serum (donated by P. Densen, University of Iowa, Iowa City, IA) in gelatin veronal-buffered saline (GVB++) (Complement Technologies) was added. To block the classical pathway, gelatin veronal-buffered saline (GVB0) (Complement Technologies) was used with added 10 mM EGTA and 7 mM magnesium chloride (Mg2+-EGTA). Cells were harvested at indicated time points and washed twice in PBS containing 1% FCS before C4 and C3 fragment analysis.
FACS analysis of complement fragment deposition
This procedure has been previously described (34). Briefly, following complement deposition and two washes, murine mAbs to the human complement component fragments C3d, C4c, or C4d (Quidel) were added (5 µg/ml). After a 30 min incubation at 4°C, FITC-conjugated goat anti-mouse IgG (Sigma) was added for 30 min at 4°C. Cells were fixed with 0.5% paraformaldehyde and analyzed on a BD Biosciences FACSCalibur system (BD Biosciences).
Soluble GAG Binding Assays
SPICE (10 µg) was preincubated with soluble GAGs (5 µg/ml) in a total volume of 100 µl in PBS at room temperature for 20 min. After preincubation, the mixture was added to 5 × 105 CHO cells (harvested from flasks using 4 mM EDTA) in a total volume of 100 µl and incubated at 30 °C for 30 min with gentle mixing. The cells were washed with PBS. SPICE was detected by flow cytometry using a rabbit polyclonal Ab as described above.
Results
Preparation and isolation of cell lines expressing transmembrane SPICE
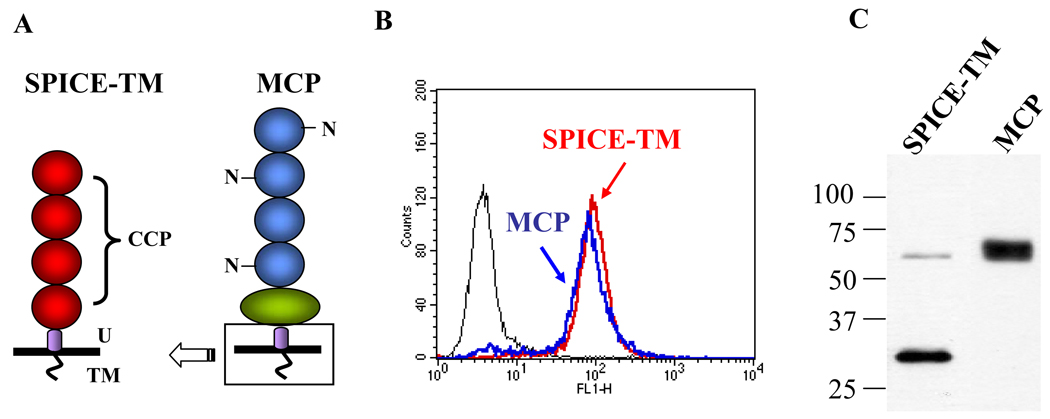
To analyze human complement regulatory activity by membrane-bound SPICE, we fused the sequence for the transmembrane domain of the human complement regulator membrane cofactor protein (MCP) to SPICE to create SPICE-TM (Fig. 1A). Stably transfected Chinese hamster ovary (CHO) clones expressing SPICE-TM were next isolated. Based on flow cytometry, a SPICE clone with equivalent expression (~25,000/cell) to an MCP clone, MCP-3-10 (34), was selected (Fig. 1B and 1C). The Western blot showed the expected Mr (Fig. 1C) for SPICE-TM. Approximately 10% of the protein was expressed as a dimer, as has been previously described (7).
Figure 1.
Characterization of SPICE bearing the transmembrane domain of membrane cofactor protein (MCP; CD46). A, Schematic diagram of SPICE-TM and MCP. Each is composed of four homologous modules called complement control protein repeats (CCPs). The human complement regulator, MCP, has three N-linked sugars (N). The CCP region of MCP is followed by an O-glycosylated domain (green oval), a 12 amino acid segment of unknown significance (U, purple), and a transmembrane domain with cytoplasmic tail (TM). The four CCPs of SPICE were fused to the U-segment, transmembrane domain and cytoplasmic tail of MCP. B, These constructs were expressed in Chinese hamster ovary cells. Clones were selected for similar expression levels via flow cytometry. C, Western blot showing the expected Mr for each protein. Expression level was ~25,000 copies per cell for both SPICE-TM and MCP as determined by flow cytometry and ELISA. Flow cytometry with mAbs to MCP and SPICE and employing the same secondary Ab further established that the expression levels were similar (not shown).
SPICE-TM regulates human complement on CHO cells
We assessed the ability of SPICE-TM to regulate human complement and profiled its activity relative to that of the native regulator MCP. Using the clones described in Fig. 1, we activated the alternative pathway of complement employing a previously described model system (34, 35). In this “complement challenge” design, rabbit anti-CHO Ab is used to sensitize cells. This is followed by exposure of the cells to a source of nonlytic complement (10% C8-deficient serum) diluted in a buffer (Mg++GVB), thereby allowing for only alternative pathway activation. The quantity of C3b deposited was then assessed by FACS with a mAb (anti-C3d) that recognizes cleavage fragments of C3 containing this fragment.
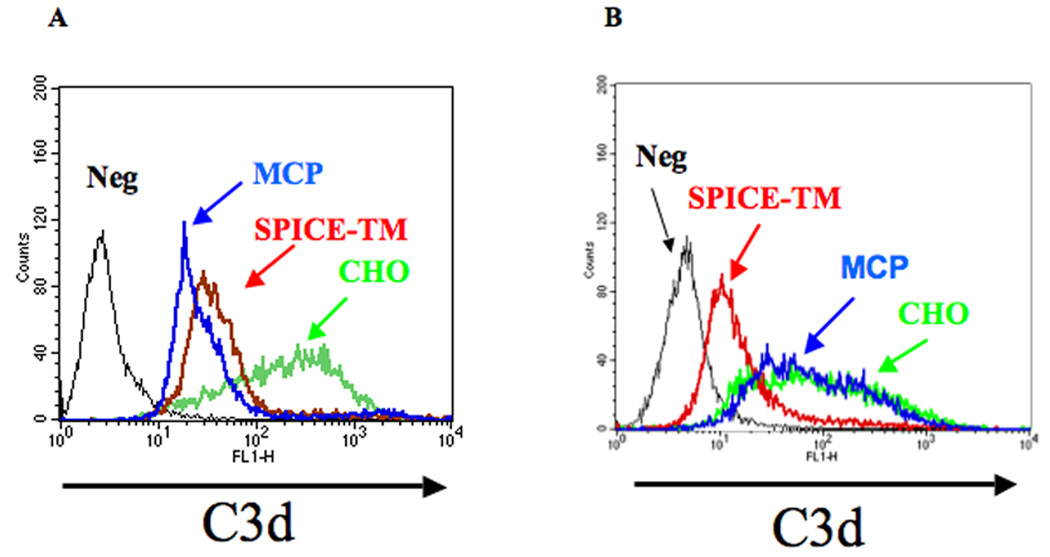
Fig. 2A demonstrates that SPICE-TM inhibits C3 fragment deposition similarly to on MCP-expressing clone (shown in Fig. 1). Following alternative pathway (AP) activation, large quantities of C3 fragments deposit on CHO cells that do not express a regulatory protein for human complement. In contrast, both SPICE-TM and MCP clones, carrying approximately equal copy number of each inhibitor, decrease C3 deposition by ≥ 90% [MFIs of 22 (MCP), 31 (SPICE-TM) vs 307 (CHO)]. Thus, transmembrane SPICE regulates the alternative pathway with similar efficiency to that of the native regulator MCP. Also, the regulation is primarily mediated by cofactor activity since MCP has no (10) and SPICE barely detectable (7) decay accelerating activity (DAA) for the alternative pathway.
Figure 2.
Alternative pathway complement challenge of CHO cells expressing SPICE or MCP. A, Transmembrane SPICE reduces C3 deposition similarly to MCP after alternative pathway challenge. CHO control, transmembrane SPICE and transmembrane MCP expressing cells were sensitized with 0.5 mg/ml of anti-CHO Ab followed by 10% C8-deficient serum for 45 min at 37°C in GVB-MgEGTA. Deposition of C3 fragments was measured by FACS using a mAb to C3d and a FITC rabbit anti-mouse IgG for detection. Negative (Neg) control used an isotypic mAb. B, Complement inhibitory activity of SPICE-TM is blocked by mAb KL5.1. CHO cells, with or without transmembrane SPICE, were sensitized with 0.5 mg/ml anti-CHO Ab followed by incubation with 6.5 µg/ml of mAb KL5.1. Cells were subsequently complement “challenged” as described above. Negative (Neg) described above. Results shown are a representative experiment of three or four.
mAb KL5.1 blocks SPICE complement regulatory activity
In a report to be published elsewhere, we describe a mAb that binds soluble SPICE and blocks its C3b and C4b binding and cofactor activity (25). To assess its ability to inhibit the function of cell-bound SPICE, we incubated this mAb with the SPICE-TM clone and then challenged the cells as described above. SPICE-TM complement regulatory activity (Fig. 2B) was abrogated as the profile relative to C3 fragment deposition was similar to CHO cells lacking a regulator. An isogenic IgG control did not reduce SPICE’s inhibitory activity (‘Neg’ profile, Fig. 2B).
Expression and characterization of SPICE in E. coli
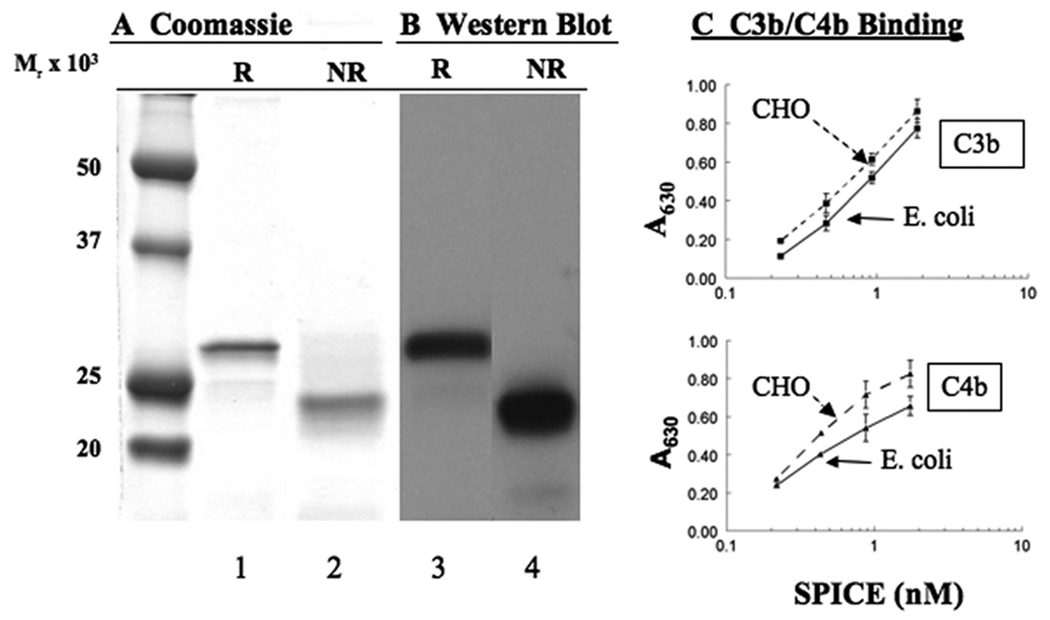
We next asked if soluble SPICE could attach to cells and regulate complement activation. Previous studies established that SPICE binds heparin, providing a possible mechanism for its attachment to membranes and extracellular matrices (7). To assess whether SPICE can bind to cells, we prepared the soluble, recombinant protein in an E. coli expression system. SPICE migrated predominantly as a single band on reducing (R) and nonreducing (NR) gels (SDS-PAGE), as assessed by Coomassie blue staining (Fig. 3A) and Western blotting (Fig. 3B). The slower Mr on reducing gels is characteristic of CCP containing proteins as each module contains two disulfide bridges (36). Because this protein was produced in E. coli and requires refolding from inclusion bodies, as part of its characterization we performed functional analyses. SPICE produced by E. coli bound human C3b/C4b analogous to what we observed with mammalian expression systems (Fig. 3C) (7) and had comparable cofactor activity for C3b (not shown). Also, the absence of disulfide-dependent dimer formation (compare to Fig. 1C) is consistent with expression in the E. coli system (7, 12).
Figure 3.
Characterization of SPICE produced recombinantly in an E. coli expression system. Electrophoresis on a 12% SDS-PAG (A, B). Reducing (lanes 1 and 3) and non-reducing (lanes 2 and 4) samples were analyzed by either (A) Coomassie blue staining or (B) by Western blotting with a polyclonal Ab. C, To assess activity, SPICE binding to C3b or C4b was characterized in ELISA. Mean ± SEM for three experiments.
Recombinant SPICE binds to CHO cells
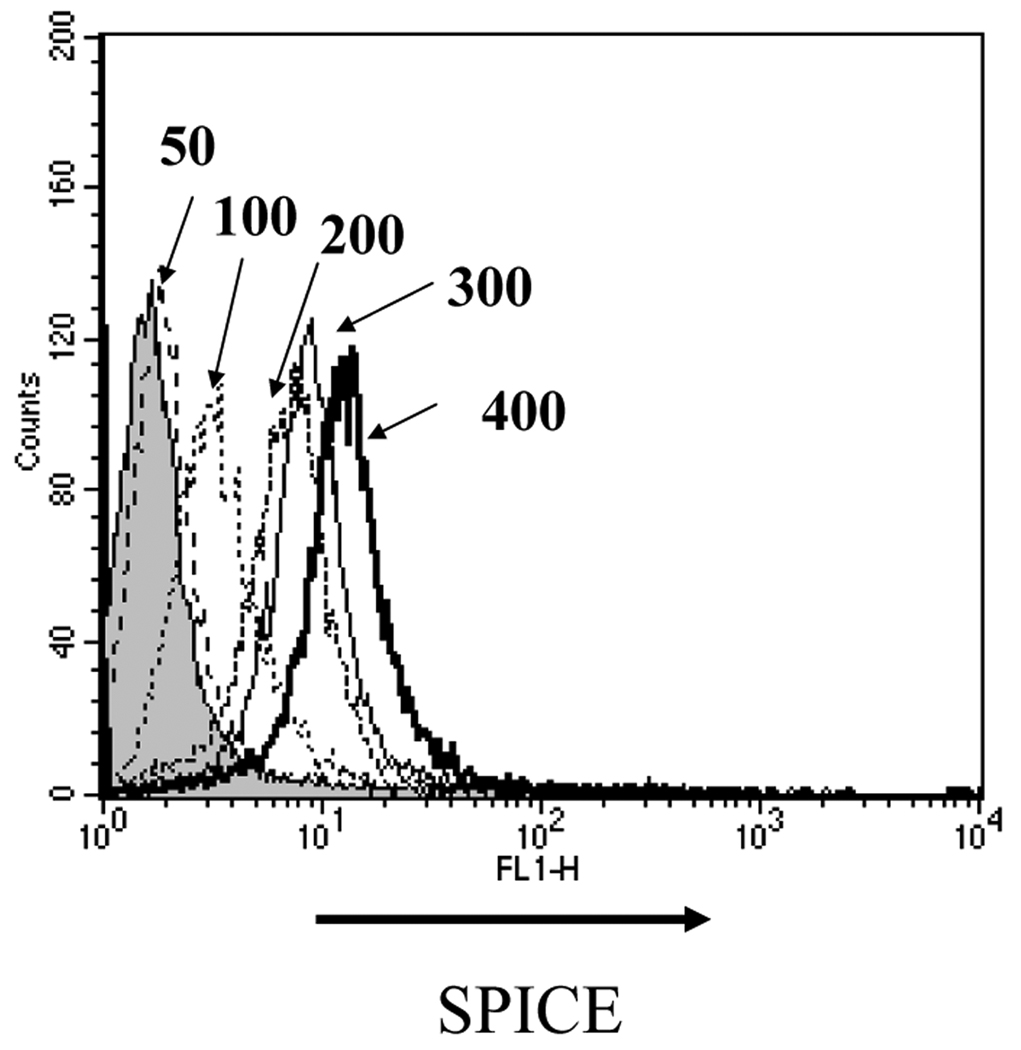
To assess binding of SPICE to CHO cells, we incubated SPICE (50–400 µg/ml) with 5 × 105 cells and monitored attachment with a cross-reacting rabbit polyclonal anti-VCP Ab (Fig. 4). Increasing the SPICE concentration led to greater quantities being deposited on the surface. In multiple such experiments, saturation was not routinely achieved, as the increase in binding between 200 and 400 µg/ml was not linear and in some experiments approached saturability. This binding behavior of SPICE suggests that SPICE oligomerizes on the cell membrane for which we have obtained preliminary evidence (LZ, MKL and JPA, unpublished data).
Figure 4.
Recombinant SPICE produced in an E. coli expression system binds to CHO cells. To 5 × 105 cells, purified SPICE was incubated at 50, 100, 200, 300 and 400 µg/ml in 50 µL at 37°C for 30 min. SPICE binding was detected by FACS using a polyclonal Ab and a FITC-labeled secondary Ab. An isogenic IgG control is indicated by the shaded area. A representative experiment of three is shown.
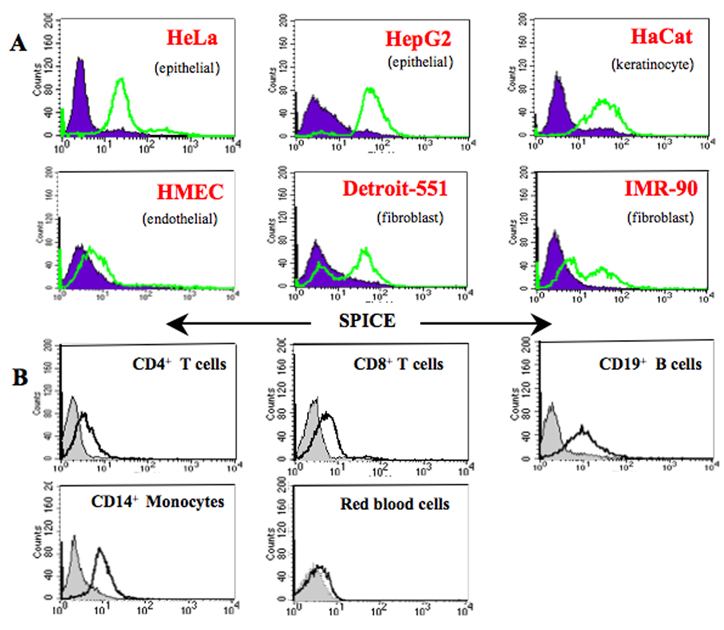
Recombinant SPICE binds to human cells
We next characterized the ability of SPICE to bind of human cells and cell lines (Fig. 5). SPICE attached to all six human cell lines shown (Fig. 5A), although there was minimal binding to HMEC. On human peripheral blood cells, SPICE bound to a similar extent to B lymphocytes and monocytes, to a lesser degree to CD4+ and CD8+ T lymphocytes and minimally to red blood cells (Fig. 5B). These data suggest that different types of heparin or heparin-like constituents on a given cell membrane influence the ability of SPICE to attach to cells.
Figure 5.
SPICE attaches to multiple human cell types. A, Cells were incubated with recombinant SPICE (200 µg/ml) for 1 h, harvested from flasks by treatment with EDTA (for adherent cells), washed, and assessed for SPICE binding using a rabbit polyclonal Ab and a FITC secondary Ab (light line). An IgG isotype control is shown (shaded). B, CD4+ T cells, CD8+ T cells, CD19+ B cells, CD14+ monocytes, and red blood cells were purified from human peripheral blood and incubated with SPICE as in A. B cells and monocytes were pre-incubated with human IgG to block Fc receptors and subsequently analyzed with a polyclonal Ab to SPICE. Detection was with a FITC-conjugated F(ab′)2 secondary Ab for FACS analysis. SPICE bound to T cells was detected similarly, without preincubation with human IgG. Representative experiments of three or four are shown.
SPICE binding to cells is GAG dependent
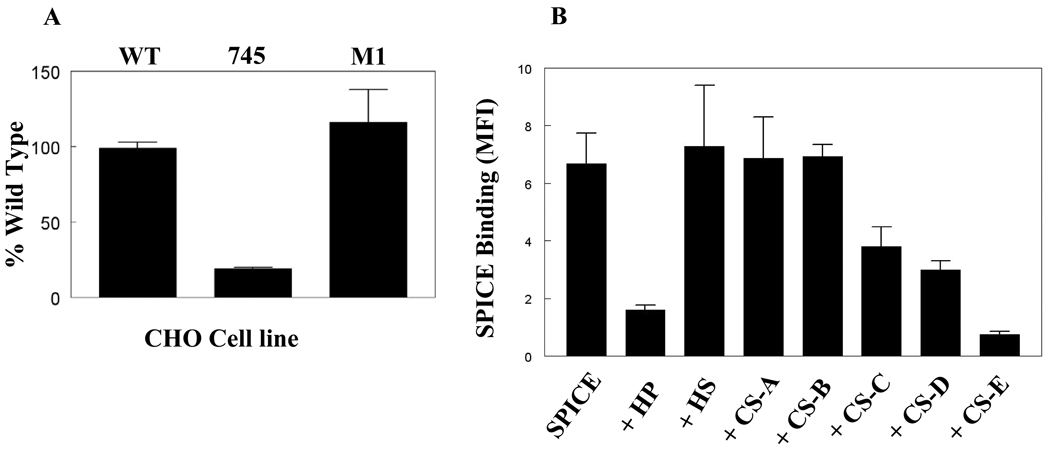
To determine if the binding of SPICE is dependent on glycosaminoglycans (GAGs), we compared the binding of SPICE on wild-type CHO cells to that of two GAG-deficient CHO cell lines (Fig. 6A). The cell line 745-CHO lacks xylosyltransferase, the enzyme required for biosynthesis of both heparan sulfate (HS) and chondroitin sulfate (CS) (26). Cell line M1-CHO lacks surface heparan sulfate (30). SPICE bound to wild-type CHO and M1-CHO but had reduced (~90%) binding activity to 745-CHO (Fig. 6A). These results suggest that the mechanism for SPICE binding is likely to be GAG-dependent.
Figure 6.
Binding of SPICE to cells is GAG-dependent. A, Soluble SPICE was incubated with wild-type (WT-CHO) and mutant CHO cell lines defective for specific enzymes in GAG synthesis (745-CHO or M1-CHO, see text). Cells were detached with EDTA. Binding was detected by FACS analysis with a polyclonal Ab followed by incubation with a FITC-conjugated anti-rabbit IgG. B, Heparin (HP) and chondroitin sulfate-E (CS-E) block binding of SPICE to CHO cells. Soluble SPICE (200 µg/ml) was preincubated with soluble GAGs (5 µg/ml) followed by binding to CHO cells. The ability of soluble GAGs to block SPICE binding was assessed by FACS with a polyclonal Ab followed by staining with a FITC-anti rabbit IgG. HP, heparin; HS, heparan sulfate, CS-A through CS-E are types of chondroitin sulfates. Data from A and B represent the mean ± SEM for 3 to 5 experiments.
SPICE binding is inhibited by heparin and chondroitin sulfate-E
To define the nature of the GAGs responsible for SPICE binding, we performed competitive inhibition binding assays with soluble GAGs (Fig. 6B). There was significant inhibition by heparin (HP) (75 %) and chondroitin sulfate-E (CS-E) (90%) and a lesser degree of inhibition by CS-D (54%) and CS-C (44%). CS-A and CS-B did not modulate SPICE binding. Taken together, these results suggest that SPICE interacts strongly with heparin and CS-E but less avidly with CS-C and -D. Interestingly, CS-E is enriched in disulfated disaccharides and is unique compared to other CS in that two sulfates are present in the same GalNAc residue. Therefore, clustered sulfates rather than net negative charge on the disaccharide backbone enhance SPICE binding.
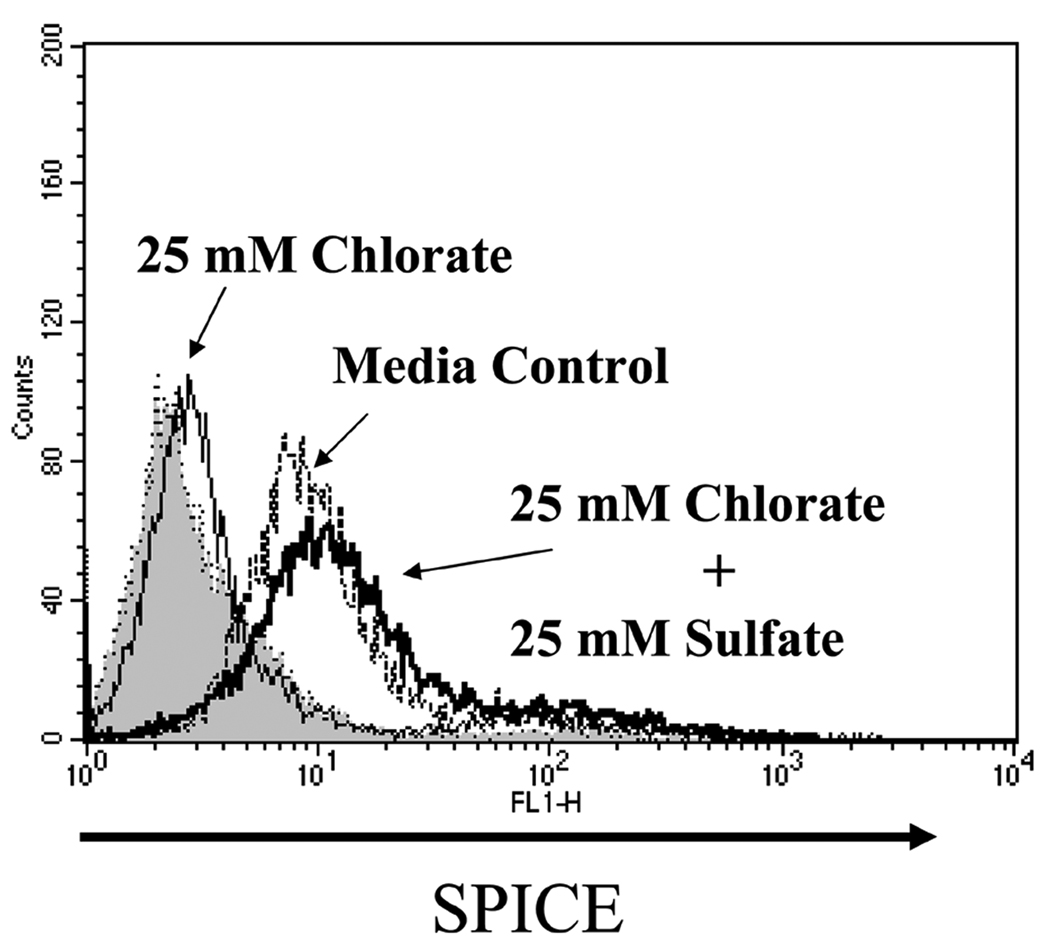
GAG sulfation is critical for SPICE binding to cells
To further establish the necessity of clustered sulfates, we treated CHO cells with the reversible sulfation inhibitor, sodium chlorate (33) (Fig. 7). Binding of SPICE was decreased by ~90% following the addition of 1– 25 mM sodium chlorate. This inhibition was completely reversed by the exogenous addition of sodium sulfate and sodium chlorate (25 mM each). Thus these studies establish that sulfated GAGs are ligands involved in binding of SPICE to cells.
Figure 7.
GAG sulfation is critical for SPICE binding to CHO cells. CHO cells were incubated in sulfate-free Ham’s F12 containing dialyzed FBS with 25 mM chlorate, 25 mM chlorate plus 25 mM sulfate, or normal Ham’s F12 (media control). After harvesting cells by EDTA treatment, they were incubated with SPICE (as described in Fig. 5). SPICE binding was detected by FACS analysis using a polyclonal Ab and a FITC-labeled secondary Ab. Representative experiment of three is shown.
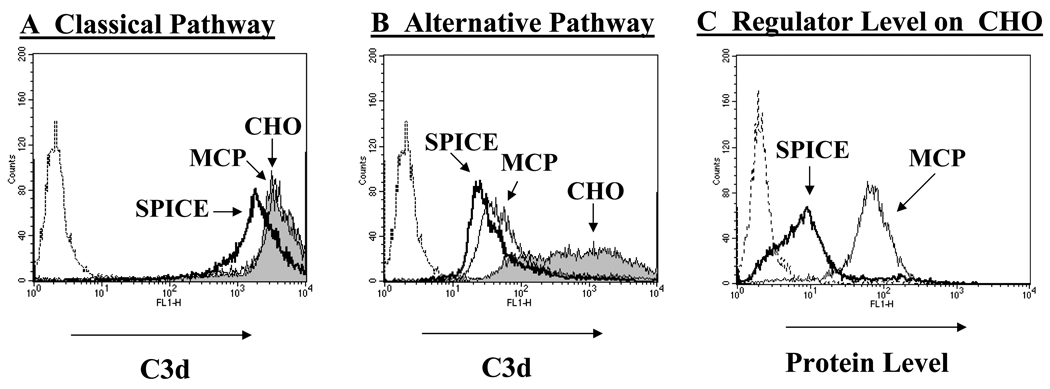
rSPICE inhibits C3 deposition on CHO cells
To determine if SPICE bound to cells via GAGs inhibits complement activation, we sensitized CHO cells with Abs to activate the classical or alternative pathway (34, 37). Relative to classical pathway activation, SPICE decreased C3b deposition by 50% [Fig. 8A, compare dark line of SPICE (MFI 2,106) to the shaded area of CHO (MFI 4,350)]. This result is consistent with SPICE possessing DAA for the classical pathway convertase (7) and comparable to decay accelerating factor’s (DAF) activity in this same experimental system (24). As expected, MCP did not reduce C3b deposition (MFI 4290), which is consistent with previous findings that MCP controls the alternative but not classical pathway on the cell surface (34). Relative to alternative pathway activation (Fig. 8B), both regulators decreased C3b deposition to a similar extent (96% for SPICE, 94% for MCP; MFIs for CHO, SPICE, and MCP were 630, 28 and 39, respectively). Of note, there was a similar degree of inhibition of SPICE and MCP, yet SPICE was present at a 5–10 fold less copy number/cell than MCP (Fig. 8C). Table 1 compares the percent inhibition of C3b deposition on CHO cells expressing SPICE, SPICE-TM or MCP. All three decrease deposition similarly. From these data, we conclude that SPICE is an efficient inhibitor of the alternative pathway, especially if attached to cells via GAGs. Additionally, since SPICE lacks DAA for the alternative pathway (7), these results are consistent with SPICE attached via GAGs being more efficient in mediating cofactor activity.
Figure 8.
SPICE reduces complement deposition following activation of the classical and alternative pathways. A, C3 fragment deposition by classical pathway activation. CHO cells, preincubated with SPICE (dark line) or without (shaded) and an MCP-expressing stable line (light line, see Fig. 1), were sensitized with 1 mg/ml anti-CHO Ab and subsequently challenged with 10% C8-deficient serum for 45 min at 37°C in GVB++ (method per Fig. 5). C3 fragment deposition was measured with a mAb to C3d. The dotted line to the left is a control in which there was no serum added. MFI for C3b deposition for CHO control, SPICE, and MCP was 4351, 2106, and 4290, respectively. Shown is a representative experiment of three. B, C3 fragment deposition by alternative pathway activation. Same designations as in (A). Cells were sensitized with 0.5 mg/ml anti-CHO Ab followed by incubation with 10% C8d serum for 45 min at 37°C in GVB-MgEGTA buffer. MFI are: CHO, 630; SPICE, 28; MCP, 39).C, The quantity of SPICE and MCP present on CHO cells was monitored using a rabbit polyclonal Ab and mAbs (not shown) and each demonstrated ~ 10 fold more MCP than SPICE-GAG. Shown for A, B, and C are representative experiments of three or four conducted.
Table I.
Heparin binding and complement regulatory profile of SPICE HS mutantsa
| Protein | Peak Elutionb (mM NaCl) |
C3b Bindingc (% Wild type) |
C4b Bindingc (% Wild type) |
C3b Cofactord (% Wild type) |
C4b Cofactord (% Wild type) |
|---|---|---|---|---|---|
| SPICE | 447 ± 5 | 100 ± 5 | 100 ± 4 | 100 ± 7 | 100 ± 8 |
| HS1 | 384 ± 3 | 80 ± 2 | 46 ± 2 | 42 ± 4 | 48 ± 3 |
| HS2 | 328 ± 3 | 116 ± 3 | 87 ± 4 | 54 ± 5 | 88 ± 6 |
| HS3 | 388 ± 4 | 59 ± 4 | 95 ± 6 | 54 ± 2 | 76 ± 8 |
| HS1-2-3 | 239 ± 6 | 11 ± 1 | 4 ± 1 | 8 ± 5 | None detected |
SPICE with mutated heparin binding sites (see Fig. 1 and Fig. 2A) were expressed in 293T cells and the supernatants were evaluated.
SPICE supernatants were chromatographed over a heparin column, eluted with increasing salt concentrations and fractions monitored via Western blot with a cross-reacting polyclonal Ab to VCP (see Fig. 3A). Peak elution of monomer represents the mean ± SEM of three or four experiments.
C3b and C4b binding were performed in an ELISA format in which human C3b or C4b were adsorbed to microtiter plates. SPICE and mutant supernatants (10 ng/ml) were applied and then detected with a polyclonal antibody. SPICE binding was set at 100%. Data are % of wild-type SPICE and represent mean ± SEM for four experiments.
For the C3b and C4b cofactor assays, biotinylated C3b or C4b was incubated with human factor I and transfectant supernatants (1.7 ng/ml of SPICE or mutant for C3b assays and 6.7 ng/ml for C4b assays) and evaluated on Western blots using HRP-Extravidin. For C3b cofactor activity, comparisons were made based upon the loss of the α′ chain and development of the α1 cleavage fragment. For C4b cofactor activity, comparisons were made based on density of the α′ band relative to the β chain. Data are % of wild-type SPICE and represent mean ± SEM for four experiments.
SPICE cleaves C4b on cells more efficiently than MCP
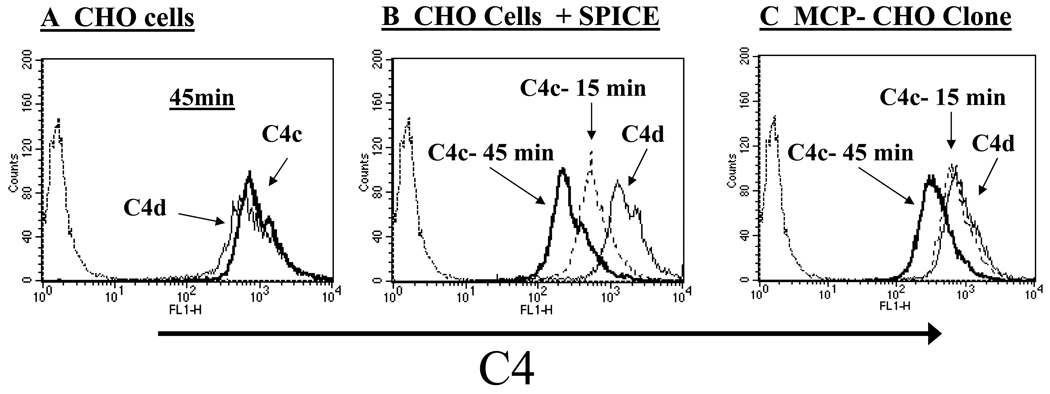
SPICE, like MCP, promotes cleavage of C4b to C4c and C4d by serving as a cofactor protein (6, 7, 12). Since soluble SPICE and MCP have similar C4b cofactor activity (6, 7), we compared their ability to cleave C4b deposited on cells following complement activation (Fig. 9). C4b degradation by SPICE and MCP in conjunction with factor I was monitored utilizing mAbs to C4d. This fragment remains covalently bound to cells following cleavage of C4b and release of the C4c fragment. As anticipated, C4d and C4c levels are similar on control CHO cells, since these cells lack an endogenous cofactor protein for human C4b degradation (Fig. 9A). In the presence though of SPICE, C4c levels are progressively reduced over a 45 min period (Fig. 9B). MCP also showed increased C4b cleavage consistent with previous findings (38). However, via its cofactor activity, SPICE cleaves C4b more quickly and to a greater degree (despite being present at a 5 to 10-fold reduced copy number per cell than MCP, see Fig. 8C). The cleavage of C4b at 15 and 45 min is 57% and 82% for SPICE and 13% and 58% for MCP (see MFIs in Fig. 8 legend). From these data, we conclude that SPICE is an efficient cofactor for the factor I mediated degradation of human C4b deposited on cells. Further, SPICE is more efficient in this regard than MCP, possibly secondary to its enhanced membrane mobility.
Figure 9.
SPICE cleaves C4b deposited on complement challenged CHO cells. Comparison of C4b cleavage by SPICE (conditions per Fig. 5) versus an MCP-expressing CHO clone (described in Fig. 1). C4b cleavage was analyzed via FACS using a mAb to C4c (heavy and dashed lines) and C4d (light line). To activate complement, cells were sensitized with 1 mg/ml of anti-CHO Ab followed by incubation with 10% C8-deficient serum for 15 or 45 min in GVB++ buffer. The dotted line on the left in the histograms indicates a condition without serum exposure. A, CHO cells lacking an inhibitor show no cleavage of C4b after 45 min. B, CHO cells with deposited SPICE demonstrate dose-dependent loss of the C4c fragment. C, MCP-expressing CHO clone also shows dose-dependent loss of C4c fragment, but to a lesser extent than SPICE. Representative experiment of three is shown.
Discussion
In this report we have characterized the inhibitory profile of a smallpox virulence protein that regulates the human complement system. The goal was to assess the ability of SPICE to regulate complement on the cell surface. To do this we prepared both a transmembrane engineered version as well as soluble recombinant SPICE. The latter studies focused on the mechanism of SPICE binding to uninfected cells and subsequent ability in situ to inhibit activation on cells undergoing a complement attack. The principle findings were that SPICE attached to cells in a GAG-dependent manner and then became an exceptionally potent down-modulator of the alternative complement pathway. Specifically, SPICE attached via GAGs was a more efficient inhibitor than transmembrane SPICE, being at least equivalent and probably many fold more potent than its human counterparts. Below, we will further address SPICE’s mechanism of cellular attachment and highlight its efficiency as a complement inhibitor.
SPICE interacts with GAGs on human cells
In preparation for these cell-binding experiments, recombinant SPICE was produced in an E. coli expression system. The material was >90% pure by Coomassie blue staining and contained no detectable fragments by Western blotting. It had equivalent C3b and C4b binding and cofactor activity to that produced by mammalian cells. This functional profile coupled with the expected interactions with monoclonal and polyclonal Abs and mobility on reducing vs non-reducing gels indicated that the recombinant material was properly folded. Also, there were no detectable dimers, in contrast to SPICE synthesized by CHO cells, which is ~10% dimers (7).
Recombinant SPICE bound to multiple human cell lines including those of epithelial, endothelial and fibroblastic lineage. Binding though to human microvascular endothelial cells was minimal. A similar copy number attached to human peripheral blood B lymphocytes and monocytes but there was less binding to T lymphocytes and none to erythrocytes.
An unexpected feature of SPICE’s attachment to CHO cells was an inability to demonstrate typical saturation binding kinetics. While there was the anticipated linear increase in binding from 50 to ~200 µg/ml of added protein, there was a variable and non-linear increase in binding at higher concentrations. These results may be explained by oligomerization of SPICE on the membrane upon its interaction with GAGs. In preliminary studies at the higher inputs of SPICE, we identified multimers by gel shift analysis (LZ, MKL and JPA, unpublished data). We are further analyzing the behavior of the SPICE interaction with cells with alterations in surface charge characteristics (see below).
The binding to many different types of cells, the unusual saturation kinetics and prior reports (14–16) pointed to an interaction with GAGs. To examine this, GAG deficient CHO cell lines, competitive inhibition with purified GAG species and biosynthetic inhibitors of GAG synthesis were employed (28, 32, 33). Thus, binding of SPICE was reduced ~80% in a CHO cell line lacking GAGs, blocked >90% by chlorate (an inhibitor of GAG synthesis) and reduced by >70% in competition experiments with HP or CS-E. There was also ~50% inhibition of binding by CS-C and CS-D but no effect of HS, CS-A, or CS-B. Taken together, these results establish an interaction between SPICE and membrane GAGs. Further, the GAG-SPICE binding profile suggests that precisely clustered sulfates on a disaccharides backbone enhance SPICE binding (32).
Complement Inhibitory Profile of SPICE
SPICE has substantially (10- to 1000-fold depending upon the particular regulatory activity being assessed) greater inhibitory capability for human complement than its >90% identical homologs, the vaccinia complement control protein (VCP) and MOPICE (from the Central African/Congo strains of monkeypox) (6, 7, 13). Human membrane regulator MCP (CD46) is ~35% homologous to SPICE, also possesses four complement control repeating modules, binds C3b and C4b and possesses cofactor activity (6, 7, 12, 39). SPICE and MCP have nearly identical binding characteristics for C3b or C4b in ELISA as well as in fluid phase cofactor assays (6, 7, 12, 40). Also, MCP has minimal DAA for C3 convertases while SPICE has DAA primarily for the classical pathway C3 convertase. A question that arose in these comparative analyses was the efficiency of the regulatory activity in situ of SPICE vs that of MCP.
To address this issue, CHO cell lines were isolated expressing SPICE carrying a transmembrane domain and cytoplasmic tail of MCP. These SPICE-TM cells were compared to MCP-expressing CHO cell lines in a complement challenge assay model system in which the alternative pathway is activated by anti-CHO Ab and human serum. In this system, the level of Ab sensitization, the complement source and the buffer system can be varied. At comparable expression levels, the two proteins bearing the same carboxyl-terminus displayed a similar ability to control the alternative pathway.
Having established that SPICE binds to cells, we next asked if SPICE, bound to membrane GAGs such as would occur in a variola infection, could inhibit complement activation. Interestingly, SPICE attached via GAGs was exceptionally effective at blocking the alternative pathway of complement activation. CHO cells with ~2,500 copies of SPICE/cell were as effective as 25,000 copies of MCP/cell. Thus, despite equivalent fluid-phase activities and similar in situ inhibitory profiles, SPICE bound via GAGs was a more efficient inhibitor than MCP. GAGs are expressed on most cell types and provide a relatively simple and energy efficient mechanism of host cell attachment (as opposed to attachment via GPI or transmembrane domain) to mediate complement inhibition and thereby enhance viral virulence.
Two other results from these challenge experiments in which the goal was to mimic an in vivo infection are worth emphasizing. One is that, upon classical complement pathway activation by Abs, complement inhibitors are not particularly effective (34). Thus, in a challenge model, DAF reduced C3b deposition by ~50% while MCP had no blocking activity (24). SPICE possesses classical pathway DAA for C3 convertase and like DAF reduced C3b deposition. However, in view of the large quantity of C3b deposited on these cells (see Fig. 8A), this reduction is unlikely to prevent opsonic or lytic consequences. An interpretation of these and similar experiments is that classical pathway activation triggered by Ab is so efficient that inhibitors are relatively ineffective in blocking complement activation, especially in comparison to their effects on the slower acting alternative pathway. This strategy makes sense as the alternative pathway engages a feedback loop that requires strict regulation to prevent excessive complement activation. In contrast, antibody selects the target in classical pathway activation and this reaction should be allowed to go unimpeded till the target is adequately opsonized or lysed.
Second, we investigated a putative mechanism to account for why SPICE-GAG might be more efficient at complement inhibition. We have previously shown that C4b deposited by classical pathway activation undergoes cleavage mediated by MCP and factor I (34, 37). This limited proteolytic reaction, known as cofactor activity, converts C4b to C4c (liberated) and C4d (remains covalently bound to target). The reaction is linear over a 30 to 45 min period and 70 to 90% of the deposited C4b is cleaved (34, 37). As hypothesized, SPICE-GAG cleaved the deposited C4b more efficiently than endogenous MCP. One interpretation of these data is that SPICE-GAG’s membrane mobility is such that it can more quickly locate the deposited C4b to perform cofactor activity than does a transmembrane protein. Other interpretations include a higher affinity for C4b cross-linking of MCP by the deposited C4b and signaling upon MCP activation by its ligand (41). We plan to dissect this issue further by comparing DAF, MCP and SPICE attached by GPI, transmembrane or GAGs in complement challenge assays.
Comparison of Poxviral Inhibitors of Complement
Our data have relevance to understanding complement regulation by other poxviral inhibitors of complement enzymes (PICES) such as from monkeypox (MOPICE) and vaccinia (VCP, the vaccine strain). While PICES are highly homologous, they differ in key amino acids that are likely to confer host-specificity and result in functional differences for interacting with the human complement system (6, 7, 9, 12, 13). Relative to binding C3b, SPICE was the most potent, MOPICE intermediate and VCP least (7). This translates into SPICE being ~100-fold more efficient than either MOPICE or VCP in cleaving human C3b (7). Relative to C4b, SPICE was 16-fold better than MOPICE and 4-fold more efficient than VCP (7). These data are consistent with variola being a human-specific pathogen. Additionally, SPICE and VCP, but not MOPICE, possess DAA for the C3 and C5 convertases of the classical pathway. An unexpected finding was that all three of these inhibitors have undetectable or very weak DAA for the AP C3 convertases (7). These results suggest that it is the inhibition of the CP via cofactor activity that is particularly targeted by poxviral complement inhibitors.
The PICES mimic host regulators structurally and functionally (7, 9). They are 30–40% homologous, consist of four CCP modules (analogous to MCP and DAF) and, as summarized above, possess C3b/C4b binding, cofactor activity and DAA capabilities. PICES are secreted proteins that bind to human cells and tissue matrices via heparin binding sites (7, 11, 16) as do the plasma proteins factor H and C4b binding protein (42, 43). On the other hand, MCP is a transmembrane protein and DAF is anchored via a glycosylphosphatidyl inositol (GPI) linkage (10). The inhibitory potential of SPICE, therefore, appears to be an amalgamation of the four human inhibitors that control complement activation at the level of the convertase. Thus, the poxviruses evolved a single protein that encompasses features of all four of the closely-related host proteins in order to block complement activation during an infection.
Therapeutic Implications of PICES
Functional differences among poxviral inhibitors may translate into the clinic since PICES are virulence factors. For example, two genetically distinct viral isolates of monkeypox virus have been identified (44–46). A more virulent strain from the Central African Republic of Congo (formerly Zaire) expresses MOPICE while a less virulent strain from Western Africa does not (47). The monkeypox epidemic that occurred in the United States in the summer of 2003 fortunately had no fatalities, likely in part because the infecting strain was a less virulent one lacking MOPICE (1, 47). However, since it was outside of Africa, the epidemic raised concerns that the more virulent monkeypox strain could be used in a bioterrorist attack or pose a threat as a naturally emerging poxviral infection (1, 2).
Thus, the ability of smallpox and monkeypox to downregulate the human complement system via PICES makes these virulence factors attractive targets for therapeutic intervention by mAbs that block their function (25). For example, viruses lacking VCP are attenuated (22, 23). Our studies herein describe the ability of a mAb to SPICE to inhibit its activity in situ and suggest its suitability to decrease activity of a poxviral virulence factor. One could envision a cocktail of such mAbs in which each inhibits a virulence factor that subverts players in the innate or adaptive immune response.
Acknowledgments
We thank Elizabeth Moulton for helpful discussions and Madonna Bogacki and Lorraine Schwartz for excellent secretarial and administrative assistance. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Mo., for the use of the High Speed Cell Sorter Core. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant #P30 CA91842.
Abbreviations
- SPICE
smallpox inhibitor of complement enzymes
- rSPICE
recombinant SPICE
- MOPICE
monkeypox inhibitor of complement enzymes
- VCP
vaccinia complement control protein
- PICES
poxviral inhibitor of complement enzymes
- GAG
glycosaminoglycans
- MCP
membrane cofactor protein (CD46)
- DAF
decay-accelerating factor (CD55)
- CA
cofactor activity
- DAA
decay accelerating activity
- CHO
Chinese hamster ovary cells
Footnotes
Grant Support
This work was supported by National Institutes of Health grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE).
Disclosures
The authors have no financial conflict of interest.
References
- 1.Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, Wegner MV, Kazmierczak JJ, Stratman EJ, Li Y, Fairley JA, Swain GR, Olson VA, Sargent EK, Kehl SC, Frace MA, Kline R, Foldy SL, Davis JP, Damon IK. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [see comment] [DOI] [PubMed] [Google Scholar]
- 2.Di Giulio DB, Eckburg PB. Human monkeypox: an emerging zoonosis. Lancet Infect Dis. 2004;4:15–25. doi: 10.1016/S1473-3099(03)00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jezek Z, Marennikova SS, Mutumbo M, Nakano JH, Paluku KM, Szczeniowski M. Human monkeypox: a study of 2,510 contacts of 214 patients. J. Infect. Dis. 1986;154:551–555. doi: 10.1093/infdis/154.4.551. [DOI] [PubMed] [Google Scholar]
- 4.Walport MJ. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 5.Walport MJ. Complement. Second of two parts. N. Engl. J. Med. 2001;344:1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 6.Rosengard AM, Liu Y, Nie Z, Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8808–8813. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liszewski MK, Leung MK, Hauhart R, Buller RM, Bertram P, Wang X, Rosengard AM, Kotwal GJ, Atkinson JP. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 2006;176:3725–3734. doi: 10.4049/jimmunol.176.6.3725. [DOI] [PubMed] [Google Scholar]
- 8.Kotwal GJ, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 9.Kotwal GJ. Poxviral mimicry of complement and chemokine system components: what's the end game? Immunol. Today. 2000;21:242–248. doi: 10.1016/s0167-5699(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 10.Liszewski MK, Farries TC, Lublin DM, Rooney IA, Atkinson JP. Control of the complement system. Adv Immunol. 1996;61:201–283. doi: 10.1016/s0065-2776(08)60868-8. [DOI] [PubMed] [Google Scholar]
- 11.Uvarova EA, Shchelkunov SN. Species-specific differences in the structure of orthopoxvirus complement-binding protein. Virus Res. 2001;81:39–45. doi: 10.1016/S0168-1702(01)00332-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sfyroera G, Katragadda M, Morikis D, Isaacs SN, Lambris JD. Electrostatic modeling predicts the activities of orthopoxvirus complement control proteins. J. Immunol. 2005;174:2143–2151. doi: 10.4049/jimmunol.174.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav VN, Pyaram K, Mullick J, Sahu A. Identification of hot spots in the variola virus complement inhibitor (SPICE) for human complement regulation. J. Virol. 2008 doi: 10.1128/JVI.01935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganesh VK, Smith SA, Kotwal GJ, Murthy KHM. Structure of vaccinia complement protein in complex with heparin and potential implications for complement regulation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8924–8929. doi: 10.1073/pnas.0400744101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Murthy KH, Smith SA, Ganesh VK, Judge KW, Mullin N, Barlow PN, Ogata CM, Kotwal GJ. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell. 2001;104:301–311. doi: 10.1016/s0092-8674(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 16.Smith SA, Mullin NP, Parkinson J, Shchelkunov SN, Totmenin AV, Loparev VN, Srisatjaluk R, Reynolds DN, Keeling KL, Justus DE, Barlow PN, Kotwal GJ. Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J. Virol. 2000;74:5659–5666. doi: 10.1128/jvi.74.12.5659-5666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meri S, Pangburn MK. Regulation of alternative pathway complement activation by glycosaminoglycans: specificity of the polyanion binding site on factor H. Biochem. Biophys. Res. Commun. 1994;198:52–59. doi: 10.1006/bbrc.1994.1008. [DOI] [PubMed] [Google Scholar]
- 18.Blom AM, Mark L, Spiller OB. Viral heparin-binding complement inhibitors--a recurring theme. Adv Exp Med Biol. 2007;598:105–125. doi: 10.1007/978-0-387-71767-8_9. [DOI] [PubMed] [Google Scholar]
- 19.Fauci AS, Challberg MD. Host-based antipoxvirus therapeutic strategies: turning the tables. J. Clin. Invest. 2005;115:231–233. doi: 10.1172/JCI24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison SC, Alberts B, Ehrenfeld E, Enquist L, Fineberg H, McKnight SL, Moss B, O'Donnell M, Ploegh H, Schmid SL, Walter KP, Theriot J. Discovery of antivirals against smallpox. Proc. Natl. Acad. Sci. U. S. A. (Early Edition) 2004:1–15. doi: 10.1073/pnas.0403600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacs SN, Argyropoulos E, Sfyroera G, Mohammad S, Lambris JD. Restoration of complement-enhanced neutralization of vaccinia virus virions by novel monoclonal antibodies raised against the vaccinia virus complement control protein. J. Virol. 2003;77:8256–8262. doi: 10.1128/JVI.77.15.8256-8262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaacs SN, Kotwal GJ, Moss B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. U. S. A. 1992;89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotwal GJ, Isaacs SN, McKenzie R, Frank MM, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 24.Liszewski MK, Leung MK, Schraml B, Goodship TH, Atkinson JP. Modeling how CD46 deficiency predisposes to atypical hemolytic uremic syndrome. Mol. Immunol. 2007;44:1559–1568. doi: 10.1016/j.molimm.2006.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liszewski MK, Leung MK, Bertram P, Atkinson JP. Dissecting complement regulatory sites of the smallpox inhibitor of complement and abrogating function with a monoclonal antibody. J. Immunol. 2007;178:44.37. [Google Scholar]
- 26.Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Beeler DL, Lawrence R, Lech M, Liu J, Davis JC, Shriver Z, Sasisekharan R, Rosenberg RD. 6-O-sulfotransferase-1 represents a critical enzyme in the anticoagulant heparan sulfate biosynthetic pathway. J. Biol. Chem. 2001;276:42311–42321. doi: 10.1074/jbc.M101441200. [DOI] [PubMed] [Google Scholar]
- 28.Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmstrom A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J. Biol. Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 29.Duncan MB, Chen J, Krise JP, Liu J. The biosynthesis of anticoagulant heparan sulfate by the heparan sulfate 3-O-sulfotransferase isoform 5. Biochim. Biophys. Acta. 2004;1671:34–43. doi: 10.1016/j.bbagen.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Broekelmann TJ, Kozel BA, Ishibashi H, Werneck CC, Keeley FW, Zhang L, Mecham RP. Tropoelastin interacts with cell-surface glycosaminoglycans via its COOH-terminal domain. J. Biol. Chem. 2005;280:40939–40947. doi: 10.1074/jbc.M507309200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Lawrence R, Frazier BA, Esko JD. CHO glycosylation mutants: proteoglycans. Methods Enzymol. 2006;416:205–221. doi: 10.1016/S0076-6879(06)16013-9. [DOI] [PubMed] [Google Scholar]
- 32.Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, Malasit P, Atkinson JP, Diamond MS. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLOS Pathogens. 2007;3:1798–1812. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baeuerle PA, Huttner WB. Chlorate--a potent inhibitor of protein sulfation in intact cells. Biochem. Biophys. Res. Commun. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- 34.Barilla-LaBarca ML, Liszewski MK, Lambris JD, Hourcade D, Atkinson JP. Role of membrane cofactor protein (CD46) in regulation of C4b and C3b deposited on cells. J. Immunol. 2002;168:6298–6304. doi: 10.4049/jimmunol.168.12.6298. [DOI] [PubMed] [Google Scholar]
- 35.Fang CJ, Fremeaux-Bacchi V, Liszewski MK, Pianetti G, Noris M, Goodship THJ, Atkinson JP. Membrane cofactor protein mutations in atypical hemolytic uremic syndrome (aHUS), fatal Stx-HUS, C3 glomerulonephritis and the HEELP syndrome. Blood. 2008;111:624–632. doi: 10.1182/blood-2007-04-084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu. Rev. Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- 37.Liszewski MK, Atkinson JP. Membrane cofactor protein (MCP; CD46). Isoforms differ in protection against the classical pathway of complement. J. Immunol. 1996;156:4415–4421. [PubMed] [Google Scholar]
- 38.Liszewski MK, Leung MK, Atkinson JP. Membrane cofactor protein (CD46): importance of N- and O-glycosylation for complement regulatory function. J. Immunol. 1998;161:3711–3718. [PubMed] [Google Scholar]
- 39.Liszewski MK, Kemper C, Price JD, Atkinson JP. Emerging roles and new functions of CD46. Springer Semin. Immunopathol. 2005;27:345–358. doi: 10.1007/s00281-005-0002-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Morikis D. Immunophysical properties and prediction of activities for vaccinia virus complement control protein and smallpox inhibitor of complement enzymes using molecular dynamics and electrostatics. Biophys. J. 2006;90:3106–3119. doi: 10.1529/biophysj.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemper C, Atkinson JP. T-cell regulation: with complements from innate immunity. Nat Rev Immunol. 2007;7:9–18. doi: 10.1038/nri1994. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt CQ, Herbert AP, Hocking HG, Uhrin D, Barlow PN. Translational mini-review series on complement factor H: structural and functional correlations for factor H. Clin. Exp. Immunol. 2008;151:14–24. doi: 10.1111/j.1365-2249.2007.03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blom AM, Villoutreix BO, Dahlback B. Complement inhibitor C4b-binding protein-friend or foe in the innate immune system? Mol. Immunol. 2004;40:1333–1346. doi: 10.1016/j.molimm.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Mukinda VB, Mwema G, Kilundu M, Heymann DL, Khan AS, Esposito JJ Monkeypox Epidemiologic Working Group. Re-emergence of human monkeypox in Zaire in 1996. Lancet. 1997;349:1449–1450. doi: 10.1016/S0140-6736(05)63725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito JJ, Knight JC. Orthopoxvirus DNA: a comparison of restriction profiles and maps. Virology. 1985;143:230–251. doi: 10.1016/0042-6822(85)90111-4. [DOI] [PubMed] [Google Scholar]
- 46.Esposito JJ, Fenner F. Poxviruses. In: Knipe DM, Howley PM, editors. Virology. New York: Lippincott, Williams & Wilkins; 2001. pp. 2885–2921. [Google Scholar]
- 47.Chen N, Li G, Liszewski MK, Atkinson JP, Jahrling PB, Feng Z, Schriewer J, Buck C, Wang C, Lefkowitz EJ, Esposito JJ, Harms T, Damon IK, Roper RL, Upton C, Buller RML. Virulence differences between monkeypox virus isolates from West Africa and the Congo Basin. J. Virol. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]