Abstract
This review represents a summary of the development and application of a novel mixed-mode HPLC approach to the separation and analysis of peptides and proteins termed hydrophilic interaction/cation-exchange chromatography (HILIC/CEX). This approach combines the most advantageous aspects of two widely different separation mechanisms, i.e. a separation based on hydrophilicity/hydrophobicity differences between polypeptides overlaid on a separation based on net charge. Applications described include HILIC/CEX separations of cyclic peptides, α-helical peptides, random coil peptides and modified or deletion products of synthetic peptides. In addition, the excellent resolving ability of HILIC/CEX for modified histone proteins is described. This approach is shown to represent an excellent complement to RP chromatography (RPC), as well as being a potent analytical tool in its own right.
Keywords: HILIC/CEX, Mixed-mode, Peptides, Proteins, RPC
1 Introduction
For more than two decades, there has been a tremendous growth in the application of rapid, HPLC methods in peptide and protein chemistry. In its various modes (generally size-exclusion chromatography or SEC; ion-exchange chromatography or IEX; and RP chromatography or RPC), HPLC has clearly demonstrated the ability to achieve high-resolution separations of peptides and proteins within short separation times and with medium to high sample throughputs. This development of HPLC methodology continues to meet a concomitant requirement for rapid and efficient separations of proteins, protein digests and synthetic peptides in such areas as biochemistry, immunology and biotechnology. The complexity of peptide mixtures may vary considerably depending on the source. For instance, peptides obtained from biological tissues are often found in only very small quantities and may require extensive purification, while impurities arising from peptide synthesis are usually closely related to the peptide of interest (deletion, terminated or chemically modified peptides), missing perhaps only one amino acid residue, and may thus be difficult to separate. RPC, a technique that does not have a traditional predecessor, has been the most widely used mode of HPLC for peptide separation [1–9], taking advantage as it does of frequently just subtle variations in hydrophilicity/hydrophobicity between peptides to effect resolution. However, even RPC is not always able to produce complete resolution of peptide mixtures in the course of a single run, with some peptides perhaps showing coelution under all mobile phase conditions employed. With a view to developing a novel separation technique which could complement and/or rival the ubiquitous RP-HPLC mode for peptide separations, this laboratory introduced mixed-mode hydrophilic interaction/cation-exchange chromatography (HILIC/CEX) for such applications over 15 years ago [10]. Thus, HILIC/CEX combines the most advantageous aspects of two widely different separation mechanisms: a separation based on hydrophilicity/hydrophobicity differences between peptides overlaid on a separation based on net charge [6, 10–12]. The present review summarizes work to date on HILIC/CEX of both peptides and proteins.
2 HILIC versus HILIC/CEX
2.1 HILIC
The term hydrophilic interaction chromatography (HILIC) was originally coined by Alpert [13] to describe separations based on solute hydrophilicity, the author noting that increasing the proportion of organic solvent (generally ACN) in the mobile phase during CEX increases the sensitivity of the sorbent to the hydrophilic residues of peptides (low levels of an organic modifier, generally ∼10% ACN, are advantageous for ion-exchange applications to suppress any undesirable hydrophobic interactions between solutes and the stationary phase [14]). A packing that was subsequently introduced for HILIC, termed PolyHydroxyethyl A [13] (Fig. 1, top), was designed as a complement to RPC, whereby the HILIC packing surface was designed to be as hydrophilic as possible in contrast to the hydrophobic stationary phase characteristic of RPC. Thus, separation by HILIC, in a manner similar to normal-phase chromatography (to which it is related), depends on hydrophilic interactions between the solutes and the hydrophilic stationary phase, i.e. assuming the absence of hydrophilic or hydrophobic preferred binding domains (discussed below). Solutes are eluted from a HILIC column in order of increasing overall hydrophilicity (decreasing hydrophobicity), in contrast to RPC where solutes are eluted in order of increasing overall hydrophobicity (decreasing hydrophilicity). Elution is generally effected by a decreasing organic modifier gradient, with the starting eluent containing a high level of ACN (e.g. 70−90%) to induce hydrophilic interactions between the column packing and the solutes. A comparison of RPC and HILIC of a mixture of synthetic peptide standards is shown in Fig. 2 [10]. From Fig. 2A, the five peptides are eluted from an RPC column in order of increasing overall hydrophobicity by a traditional linear AB increasing gradient of ACN in an aqueous mobile phase containing 0.05% TFA. Figure 2B shows the separation of the same peptides on the HILIC column. The peptides were now subjected to a linear AB decreasing gradient of ACN starting from 85% ACN in an aqueous mobile phase containing 0.2% phosphoric acid and were eluted in reverse order to that shown by RPC (Fig. 2A). Note that phosphoric acid instead of TFA was used for HILIC since the negatively charged phosphate ion is hydrophilic (unlike the more hydrophobic trifluoroacetate ion) and, for HILIC, it is desirable to emphasize peptide hydrophilicity as much as possible when positively charged side-chains (Lys and Arg in this case) interact with the negatively charged counterions in the mobile phase. Excellent recent reviews of HILIC applications for a range of solutes and a range of polar stationary phases are represented by refs. [15, 16].
Figure 1.
Top: structure of coating of poly(2-hydroxyethyl aspartamide)-silica (known commercially as PolyHydroxyethyl A HILIC stationary phase). Bottom: structure of coating of poly(2-sulphoethyl aspartamide)-silica (known commercially as PolySulphoethyl A strong-CEX stationary phase).
Figure 2.
Separation of a mixture of synthetic peptide standards by (A) RPC and (B) HILIC. Columns: (A) SynChropak RP-P C18 (250 × 4.6 mm id, 6.5 μm particle size, 300 Å pore size); (B) PolyHydroxyethyl A HILIC column (200 × 4.6 mm id, 5 μm, 300 Å). Conditions: RPC, linear AB (increasing ACN) gradient (1% ACN/min) at a flow-rate of 1 mL/min and a temperature of 26°C, where eluent A is 0.05% aq. TFA and eluent B is 0.05% TFA in ACN; HILIC/CEX, linear AB (decreasing ACN) gradient (1% ACN/min) at a flow-rate of 1 mL/min and a temperature of 26°C, where eluent A is 0.2% H3PO4 in ACN and eluent B is 0.2% aq. H3PO4 (starting conditions of 85% A–15% B). Peptide sequences: Ac-Arg-Gly-X-X-Gly-Leu-Gly-Leu-Gly-Lys-amide, where -X-X- is substituted by -Gly-Gly- (S2), -Ala-Gly- (S3), -Val-Gly- (S4) and -Val-Val- (S5); S1 has the same sequence as S3 except for the presence of a free α-amino group; Ac denotes Nα-acetyl and amide denotes Cα-amide. Reprinted from ref. [10] with permission from Elsevier.
2.2 HILIC/CEX
The HILIC material originally introduced by Alpert [13], i.e. the PolyHydroxyethyl A stationary phase, was designed to be ‘more hydrophilic’ than the strong cation-exchange packing (designated PolySulphoethyl A; Fig. 1, bottom), ‘but lacking its charged character’. However, our laboratory took the view that charged groups on the ion-exchange packing are, of course, also inherently hydrophilic [10] and that hydrophilic interactions superimposed on electrostatic interaction [10, 13] (i.e. a mixed-mode approach whereby the presence of simultaneous mechanisms effects a separation) may have a greater resolving capability than just a single separative mechanism based on only differences in either solute net charge or hydrophilicity/hydrophobicity. In addition, charged solutes will likely be retained strongly enough by an ion-exchange stationary phase to allow an effective HILIC separation of same/similar charged species. It should be noted that, although HILIC can cause solute retention even if the solute and ion-exchange matrix carry the same charge [61], we propose that having opposite charges on the solute and stationary phase leads to a more effective separation by mixed-mode HILIC/CEX. Considering the common occurrence of charged groups in polypeptides (potentially positively charged Lys, Arg and His side-chains as well as a free positively charged α-amino group; potentially negatively charged Glu and Asp side-chain, as well as a free α-carboxyl group) as well as the range of hydrophilicity/hydrophobicity of amino acid side-chains [17], the potential of this mixed-mode approach to polypeptide separations seemed clear. Our studies to date have been carried out on a strong cation-exchange column, hence the term HILIC/CEX (or HILIC/CEC, as it was originally introduced by our laboratory [11] and is denoted as such in early figures from our laboratory included in the present review).
2.2.1 General principles and conditions of HILIC/CEX
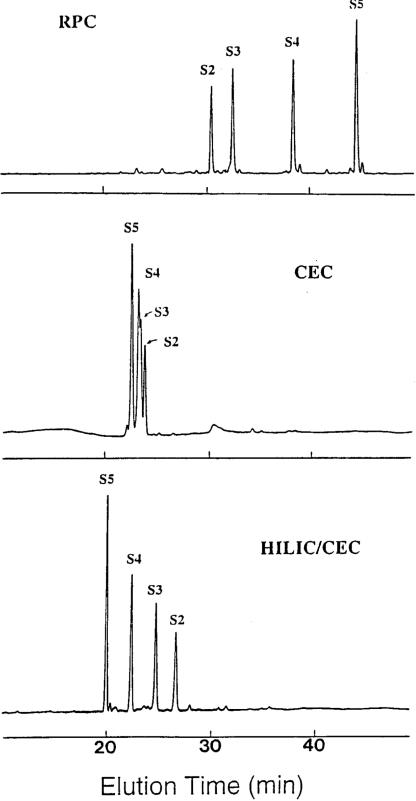
Characteristic of HILIC/CEX separations is the presence of a high organic modifier (generally ACN) concentration to promote hydrophilic interactions between the solute and the hydrophilic/charged cation-exchange stationary phase, with peptides then being eluted from the column with a salt gradient. Peptides are generally eluted in groups in order of increasing net positive charge; within these groups, peptides are eluted in order of increasing hydrophilicity (decreasing hydrophobicity). Indeed, HILIC/CEX is basically CEX in the presence of high concentrations of ACN (60−80%, in general). Figure 3 compares the separation of synthetic peptide RPC standards, S2–S5, by RPC (top), CEX (middle) and HILIC/CEX (bottom) [18]. The lack of secondary structure made these peptides useful to demonstrate the basic principles distinguishing RPC from HILIC/CEX. This four-peptide mixture contains peptides with the same net positive charge (+2) and subtly increasing hydrophobicity (S2 < S3 < S4 < S5). It should be noted that the only difference between the CEX and HILIC/CEX runs was the presence of 10% v/v ACN in the former compared to 80% v/v in the latter. From Fig. 3, peptides were, as expected, eluted from the RPC column (top) in order or increasing hydrophobicity (decreasing hydrophilicity). Under characteristic cation-exchange conditions (middle), the presence of 10% v/v ACN helps to eliminate unwanted hydrophobic interactions between solutes and the column matrix [14]. The four peptides were very poorly resolved, which was expected given the identical net charge on the peptides. Interestingly, the low concentration of ACN (10%) has already induced hydrophilic interactions with the matrix in that the elution order is already opposite to that of RPC, i.e. the most hydrophobic peptide (S5) is eluted first and the most hydrophilic peptide (S2) is eluted last. In contrast, under HILIC/CEX conditions (bottom), the elution order remains the same as the CEX run (middle), but the peptides are now well resolved. Clearly, to effect a separation of these peptides on the cation-exchange column, an elevated concentration (80% v/v) of ACN was required in the mobile phase in order to promote hydrophilic interactions with the column matrix to complement the ionic interactions.
Figure 3.
General principles of RPC versus HILIC/CEX. Columns: RPC, Zorbax SB300-C8 (150 × 4.6 mm id, 5 μm particle size, 300 Å pore size); CEX (denoted CEC in Fig. 3) and HILIC/CEX (denoted HILIC/CEC in Fig. 3), PolySulphoethyl A (200 × 4.6 mm id, 5 μm, 300 Å). Conditions: RPC, linear AB gradient (0.5% ACN/min) at a flow-rate of 1 mL/min, where eluent A is 20 mM aq. triethylammonium phosphate (TEAP), pH 3, and eluent B is eluent A containing 50% v/v ACN, both eluents containing 100 mM NaClO4; CEX, linear AB gradient (5 mM NaClO4/min, following 5-min isocratic elution with eluent A) at a flow-rate of 1 mL/min, where eluent A is 20 mM aq. TEAP, pH 3, containing 10% v/v ACN and eluent B is eluent A containing 400 mM NaClO4; HILIC/CEX, same conditions as for CEX, except for 80% v/v ACN in eluents A and B; all runs carried out at 30°C and peaks detected by absorbance at 210 nm. The sequences of peptides S2–S5 are shown in Fig. 2. Reprinted from ref. 18 with permission from Elsevier.
The choice of a relatively low pH (e.g. pH 3 in Fig. 3) is governed by the desire to maximize the basic character of the peptide solutes to enhance ionic interactions with the negatively charged strong-cation-exchange matrix. Thus, at pH 3, any acidic (potentially negatively charged) residue (Asp, Glu) will be protonated, i.e. uncharged. In addition, a full positive charge on the basic residue His (pKa = 6.5) is also assured at low pH). It is also necessary to be cautious with the pH of the mobile phase when considering basic residues such as Lys (pKa ∼10) and Arg (pKa ∼12). Thus, through the use of synthetic peptide models, Sereda et al. [19] demonstrated that the hydrophobic environment characteristic of RPC (hydrophobic packings and organic modifiers) had a profound effect on the pKa values of ionizable groups, these values being decreased to 5.8, 7.4 and 7.3 for His, Lys and Arg, respectively. Although an ion-exchange matrix is clearly far less hydrophobic than that of an RP packing, it was felt that the presence of a high ACN concentration (perhaps as high as 90%) in HILIC/CEX mobile phases necessitates the use of relatively low pH conditions to ensure full protonation (i.e. a full positive charge) of basic side-chains. Also considering the presence of high ACN concentrations, characteristic of HILIC/CEX, NaClO4 is suitable for this mixed-mode approach due to its excellent solubility in aqueous solution even in the presence of high concentrations of organic modifier [11, 20].
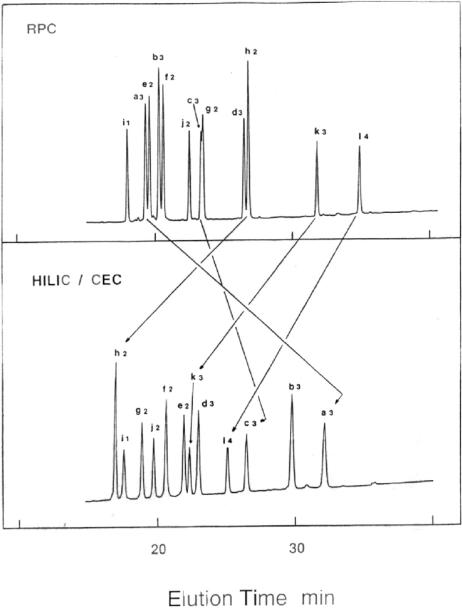
Apart from overall net charge on a specific peptide, the major determinant of how retentive it will be during HILIC/CEX (in the absence of any conformational effects; see Sections 3.1 and 3.2) will depend on the level of ACN in the mobile phase. Thus, Fig. 4 illustrates the effect of ACN concentration on the HILIC/CEX separation of a mixture of 12 peptides of varying net charge and hydrophobicity (note that the numbers in the peptide denotations refers to the number of positive charges the peptides possess; no negative charges are present) [11]. From Fig. 4A, the overall elution order of all 12 peptides was based on increasing net positive charge (albeit with separations within the charge groups based on hydrophilicity/hydrophobicity already apparent), i.e. ionic interactions are dominating the separation process. As the ACN concentration is increased to 50% (Fig. 4B), peptides are still generally eluted in order of increasing net positive charge, except for l4, which was eluted earlier than the lesser charged a3 and b3. The overall hydrophobicity of these three peptides increases (i.e. decreasing hydrophilicity) in the order a3 < b3 << l4, indicating that, at this ACN concentration, more highly charged peptides may be eluted prior to less highly charged peptides if the latter are significantly more hydrophilic than the former. Finally, at a level of 90% ACN (Fig. 4C), peptide i1 (+1 net charge) was eluted after the less hydrophilic h2 (+2); e2 (+2) was eluted after the much less hydrophilic k3 (+3); and, most dramatically, the peptide l4 (+4), was eluted before the more hydrophilic c3, b3 and a3 (all +3), indicating that hydrophilic interactions now dominate the separation process. Figure 4D is an excellent example of how dominant hydrophilic (over ionic) interactions may be maintained while reducing analysis time but retaining good column selectivity. This was achieved by combining the increasing salt gradient with a decreasing ACN gradient. Thus, the lower level of ACN in buffer B (50% as against 90% in buffer A) led to a decrease in peptide retention relative to that obtained by maintaining 90% ACN in both mobile phase buffers (Fig. 4C). The peptide elution order of Fig. 4D was almost identical (except for a reversal of e2 and k3) to that shown in Fig. 4C, but was obtained in about two-third of the time and with sharper peptide peaks.
Figure 4.
Effect of ACN concentration on HILIC/CEX of peptides. Column: same CEX column as Fig. 3. Conditions: panels A–C, linear AB increasing salt gradient (2% B/min, equivalent to 5 mM NaClO4/min, starting with 100% A) at a flow-rate of 1 mL/min, where eluent A is 5 mM aq. TEAP, pH 7, and eluent B is A plus 250 mM NaClO4, both A and B containing 20% (panel A), 50% (panel B) or 90% (panel C) v/v ACN; panel D linear AB gradient (2% B/min, equivalent to a linear increasing salt gradient of 5 mM NaClO4/min and a linear decreasing ACN gradient of 0.8% ACN/min, starting with 100% A) at a flow-rate of 1 mL/min, where eluent A is 5 mM aq. TEAP, pH 7, containing 90% v/v ACN and eluent B is 5 mM aq. TEAP, pH 7, containing 250 mM NaClO4 and 50% v/v ACN; all runs carried out at 30°C. Peptide sequences: peptides e–h are identical to peptides S2–S5, respectively, in Fig. 2 (all +2 net charge); peptides a–d are identical to peptides S2–S5, respectively, in Fig. 2 except they have a free N-terminal α-amino group (all +3 net charge); peptide i, Ac-Gly-Gly-Gly-Leu-Gly-Gly-Ala-Gly-Gly-Leu-Lys-amide (+1); peptide j, same as peptide i except for -Lys-Tyr- at positions 1 and 2 (+2); peptide k, Ac-Gly-Gly-Ala-Leu-Lys-Ala-Leu-Lys-Gly-Leu-Lys-amide (+3); peptide l, same as peptide k except for -Lys-Tyr- at positions 1 and 2 (+4); Ac denotes Nα-acetyl and amide denotes Cα-amide. The numbers in the peptide denotions represent the net positive charge on the peptide. Adapted from ref. [11] with permission from Elsevier.
At this point, it is important to distinguish the difference in using ion-exchange stationary phases in a non-HILIC mode versus a true HILIC mode, such a distinction depending on the level of organic modifier in the run solvents. Thus, in (1), for a non-HILIC separation, the presence of organic modifier may be required simply to eliminate nonspecific hydrophobic interactions with the matrix [14], improve solubility of solutes being separated or perhaps enhance ionic interactions and hydrophilic effects to improve separation of some of the individual components of a solute mixture. Reports of non-HILIC mode separations in the presence of varying levels of organic solvent have been reported for various solutes, including amino acids [21], peptides [22, 23], proteins [24–26] and carbohydrates [27]. However, our definition of (2), a separation carried out on an ion-exchange column operated in HILIC mode is the point at which the minimum organic modifier concentration required to reverse the solute elution order of a particular solute mixture relative to RPC is reached. Below this minimum concentration, hydrophilic effects may be present that affect resolution of just some of the mixture components. However, above this minimum concentration, the resolution of all sample components is affected, with higher concentrations potentially able to improve the separation still further. In fact, at very high concentrations of organic modifier, the HILIC mode is so dominant over the ion-exchange mode that a peptide with a greater net positive charge can be eluted prior to a lesser-charged peptide (Fig. 4) [11, 12].
2.2.2 Selectivity of HILIC/CEX versus RPC
Figure 5 compares the RPC (top) and HILIC/CEX elution profiles of the same mixture of peptides with negligible secondary structure used in Fig. 4. In fact, the HILIC/CEX double gradient system shown in Fig. 4D is here being compared to RPC. From Fig. 5 (top), the RPC elution profile shown represents a relative measure of the hydrophobicity of the 12 peptides, from the least hydrophobic to the most hydrophobic as expressed by their increasing RPC elution times. Under HILIC/CEX conditions (Fig. 5, bottom) instead of a simple reversal of peptide elution, cation-exchange interactions overlaid on interactions involving the hydrophilic/hydrophobic nature of the peptides now lead to useful selectivity differences between the two modes, major examples of which are denoted by arrows in Fig. 5. The resolution of the peptide mixture is satisfactory using RPC or HILIC/CEX, albeit that achieved by HILIC/CEX is superior in this case.
Figure 5.
Selectivity variations of RPC versus HILIC/CEX. Columns: RPC, same as Fig. 3, except for 250 Å pore size; HILIC/CEX (denoted HILIC/CEC in Fig. 5), same as Fig. 3. Conditions: RPC, linear AB gradient (2% B/min, equivalent to 1% ACN/min at a flow-rate of 1 mL/min where eluent A is 10 mM aq. (NH4)2HPO4, pH 7, and eluent B is A containing 50% v/v ACN, both eluents also containing 200 mM NaClO4; HILIC/CEX same as Fig. 4D; runs carried out at 30°C and peaks detected by absorbance at 210 nm. Peptide sequences: same as Fig. 4. The numbers in the peptide denotations represent the net positive charge on the peptides. Arrows denote major selectivity changes between the two HPLC modes.
The key observation from Fig. 5 is that a HILIC/CEX separation is not necessarily a straightforward reversal of RPC elution order, i.e. just another approach which the ubiquitous RPC mode already achieves, but a novel separation approach in its own right which is an excellent complement to RPC. In addition, for some separations, RPC is unable to resolve peptides (and proteins) readily resolved by HILIC/CEX, with examples presented below.
3 Analytical applications of HILIC/CEX for peptide separations
3.1 Cyclic peptides
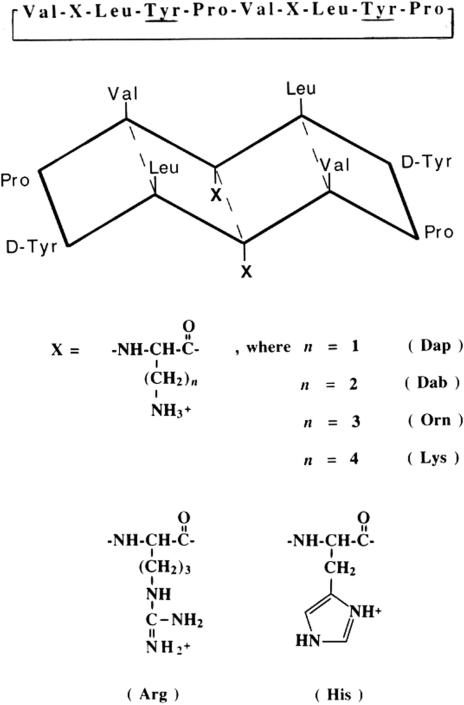
Figure 6 shows the sequence of cyclic 10-residue analogues of the antibiotic peptide gramicidin S (GS) as well as the schematic representation of their amphipathic β-sheet structure: the hydrophobic face of the molecule, made up of Leu and Val residues is constant; only the hydrophilic face made up of two (positively charged) diaminopropionic acid (Dap), diaminobutyric acid (Dab), ornithine (Orn), Lys, His or Arg residues, is varied [28]. Such amphipathic molecules have a preferred face for binding with hydrophobic or hydrophilic/charged stationary phases. Thus, the hydrophobic (nonpolar) face of the cyclic peptides shown in Fig. 6 represents the preferred binding domain for RPC; in contrast, the hydrophilic/charged face of the peptides represents a preferred binding domain for HILIC/CEX.
Figure 6.
Sequences and structure of synthetic amphipathic 10-residue GS analogues. At top is the linear sequence of the cyclic peptides (underlined residues represent d-amino acids); below is a schematic representation of the β-sheet structure of the peptides; X denotes the substitution site on the hydrophilic face of these amphipathic β-sheet analogues. The peptides are denoted by the three-letter abbreviations of the side-chains at the substitution site: Dap refers to the peptide analogue with diaminopropionic acid substituted at both sites; Dab, diaminobutyric acid; Orn, ornithine; Lys, lysine; Arg, arginine; His, histidine. Reprinted from ref. [28] with permission from Elsevier.
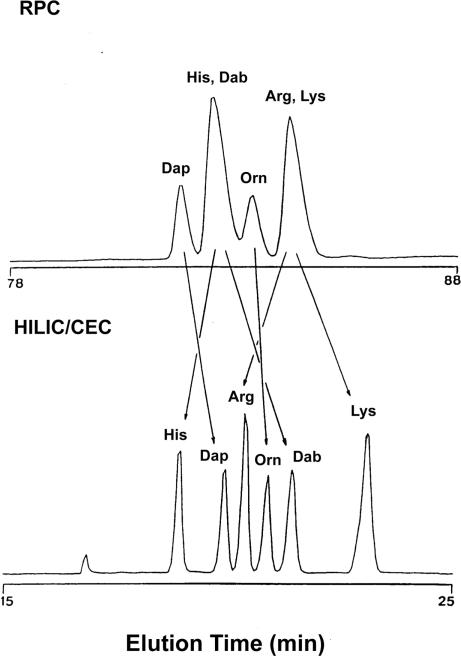
Figure 7 shows the effect of mobile phase ACN concentration on the HILIC/CEX elution profiles of the six peptide analogues at pH 3, where all six different basic side-chains should be fully protonated, i.e. positively charged [28]. From Fig. 7, it can be seen that the ACN concentration in the mobile phase has a profound effect on peptide elution behaviour, ranging from generally poorly resolved, broad peaks with relatively long retention times at 10% ACN (likely due to hydrophobic interactions between the column matrix and the highly nonpolar face of these amphipathic, cyclic peptides) to baseline resolution of all six peptides at 90% ACN (just 80% ACN was used in eluent B, due to NaClO4 solubility concerns at 90% ACN), concomitant with well-defined peaks and shorter retention times. It is interesting to note the reduction in retention time of the peptides in the 80% ACN run compared to the 40% ACN run, which is unlikely to be due to the suppression of any further hydrophobic interactions at these high ACN concentrations. Rather, they highlight the often complex nature of chromatography where not only interactions between the solute and the column matrix must be considered, but also interactions between the solute and the mobile phase must be taken into account. This is particularly true of highly amphipathic peptide solutes such as the analogues shown in Fig. 6 and employed in Fig. 7, where the interaction of the preferred hydrophilic and hydrophobic binding domains with not only the stationary phase but also the mobile phase may vary depending on the concentration of ACN in the mobile phase and give rise to the profiles illustrated in Fig. 7.
Figure 7.
Effect of ACN concentration on HILIC/CEX elution profiles of cyclic, amphipathic β-sheet peptides. Column: same as Fig. 3. Conditions: linear AB gradient (2.5 mM NaClO4/min, following 5-min isocratic elution with 100% eluent A) at a flow-rate of 1 mL/min and a temperature of 30°C, where eluent A is 20 mM aq. TEAP, pH 3, and eluent B is buffer A containing 400 mM NaClO4; ACN is present in both eluents at concentrations v/v of 10, 40, or 80% (denoted as such on the elution profiles), or present at a concentration of 90% v/v in eluent A and 80% v/v in eluent B (denoted 90/80%). Peptides were detected by their absorbance at 210 nm. Peptide sequences are shown in Fig. 6. Reprinted from ref. [28] with permission from Elsevier.
Figure 8, which compares RPC and HILIC/CEX of the six cyclic peptides, shows useful selectivity differences between RPC (top) and HILIC/CEX (bottom) for peptide mixtures, thus highlighting again the complementarity of the two approaches [28]. In addition, the improved separation of the analogues by HILIC/CEX compared to RPC is quite clear, due to side-chain substitutions being made in the hydrophilic face of the peptides, i.e. the preferred binding domain for HILIC/CEX.
Figure 8.
RPC versus HILIC/CEX of cyclic, amphipathic β-sheet peptides. Columns: RPC and HILIC/CEX (denoted HILIC/CEC in Fig. 8), same as Fig. 3. Conditions: RPC, linear AB gradient (1% B/min, equivalent to 0.5% ACN/min) at a flow-rate of 1 mL/min, where eluent A is 0.05% aq. TFA and eluent B is 0.05% TFA in ACN; HILIC/CEX, linear AB gradient (2.5 mM NaClO4/min) at a flow-rate of 1 mL/min, where eluent A is 20 mM aq. TEAP, pH 3, and eluent B is eluent A containing 400 mM NaClO4, eluents A and B also containing 90% v/v and 80% v/v ACN, respectively. Runs were carried out at 30°C and peaks detected by absorbance at 210 nm. Peptide sequences are shown in Fig. 6. Reprinted from ref. [28] with permission from Elsevier.
3.2 α-Helical peptides
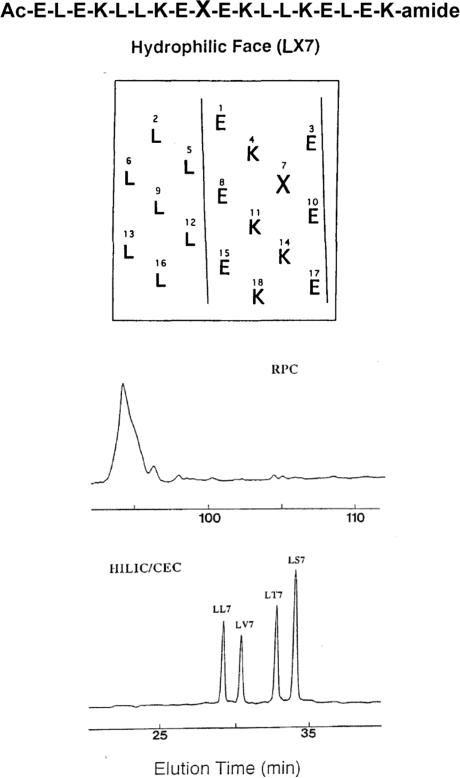
Figure 9 (top) shows the sequence of a series of model amphipathic α-helical peptides, denoted LX7, where position X (position 7 of the sequence) is substituted by polar residues Ser or Thr (LS7 and LT7, respectively) or nonpolar residues Val or Leu (LV7 and LL7, respectively) [18]. Figure 9 (top) also shows a helical net representation of the peptide sequence. The area between the solid lines represents the hydrophilic face (made up of Lys and Glu residues) of the peptides; the remaining residues (all Leu) comprise the hydrophobic face. The substitution position, X, on the hydrophilic face is surrounded by polar Lys and Glu residues. The amino acid sequence shown in Fig. 9 (top) is known to have a high potential to form α-helices [29, 30]; in addition, the considerable amphipathic character of such peptides has also been reported previously [29]. It has also been shown that high concentrations of organic modifiers such as ACN can induce helix formation in a potentially helical peptide [31, 32]. Thus, under characteristic conditions of HILIC/CEX (high concentrations of ACN in the mobile phase), the peptides would be expected to be α-helical, allowing interaction of the charged, hydrophilic face with the cation-exchange matrix; indeed, the presence of high concentrations of ACN (80% in Fig. 9, bottom) serve to enhance such interactions.
Figure 9.
RPC versus HILIC/CEX for separation of amphipathic α-helical peptides with amino acid side-chains substituted in the hydrophilic face of the α-helix. At top is the linear sequence of the α-helical peptides, together with a helical net representation of the peptide structure; X denotes the substitution site on the hydrophilic face of these amphipathic α-helical analogues. At bottom are the RPC and HILIC/CEX (denoted HILIC/CEC in Fig. 9) elution profiles of a mixture of four peptide analogues, substituted with Leu (LL7), Val (LV7), Thr (LT7) or Ser (LS7) at position X. Columns: RPC and HILIC/CEX, same as Fig. 3. Conditions: RPC, same as Fig. 8; HILIC/CEX, same as Fig. 3. Adapted from ref. [18] with permission from Elsevier.
Figure 9 (bottom) compares RPC and HILIC/CEX separations of the four α-helical peptides [18]. As would be expected, the identical (all Leu residues) hydrophobic preferred binding domains of the peptides bind to the hydrophobic matrix in RPC, resulting in coelution of all four peptides, i.e. substitutions in the hydrophilic face had little effect on the RPC retention behaviour of the peptides. In contrast, all four peptides were well resolved by HILIC/CEX at pH 3, even though all peptides have the same net positive charge (+5 from 5 Lys; Glu residues are protonated, i.e. neutral under the conditions employed), with the substitution sites in the hydrophilic faces of the peptides able to interact intimately with the cation-exchange matrix and, hence, influence the retention behaviour of the four analogues. The HILIC/CEX elution order is in order of decreasing hydrophobicity (increasing hydrophilicity) of the substituted side-chain (Ser < Thr < Val < Leu; [17]), with the Leu analogue being eluted first followed by the Val, Thr and Ser analogues.
Figure 10 compares RPC and HILIC/CEX separations of amphipathic α-helical peptides with amino acid substitutions made in the centre of the hydrophobic face or hydrophilic face of the helix: Ac-KWKSFLKTFK-X1-A-X2-KTVLHTALKAI-SS-amide, where position X1 (in the centre of the hydrophilic face; X1 = Ser) and position X2 (in the centre of the hydrophobic face; X2 = Val) are substituted by various l-amino acids [33]. The native peptide sequence, with Ser and Val at positions X1 and X2, respectively, is a biologically active amphipathic α-helix (denoted V681) with potent antimicrobial and hemolytic properties [34, 35]. As shown in Fig. 10, RPC (left) and HILIC/CEX (right) are best suited for resolving amphipathic peptides where substitutions are made in the nonpolar and polar faces, respectively. The ability to monitor the hydrophilicity/hydrophobicity effects of amino acid substitutions in both the nonpolar and polar faces of potentially useful antimicrobial amphipathic α-helical (and, indeed, amphipathic cyclic) peptides is critical in the design process of such molecules. Thus, in addition to demonstrating yet again the complementarity of the RPC and HILIC/CEX modes, Fig. 10 concomitantly illustrates that RPC and HILIC/CEX are best suited as monitors of hydrophilicity/hydrophobicity variations where amino acid substitutions are made in the nonpolar and polar faces, respectively, of amphipathic peptides.
Figure 10.
RPC versus HILIC/CEX for separation of amphipathic α-helical peptides with amino acid side-chains substituted in the hydrophilic or hydrophobic face of the α-helix. Columns: RPC (denoted RP-HPLC in Fig. 10) and HILIC/CEX, same as Fig. 3 except both columns have an id of 2.1 mm. Conditions: RPC, linear AB gradient (1% B/min, equivalent to 1% ACN/min) at a flow-rate of 0.3 mL/min, where eluent A is 0.05% aq. TFA and eluent B is 0.05% TFA in ACN; HILIC/CEX, linear AB gradient (5 mM NaClO4 to 250 mM NaClO4 in 60 min) at a flow-rate of 0.3 mL/min, where eluent A is 5 mM aq. TEAP, pH 4.5, and eluent B is 5 mM aq. TEAP containing 250 mM NaClO4, pH 4.5, both eluents also containing 70% v/v ACN; runs carried out at 25°C (RPC) or 65°C (HILIC/CEX). Peptide sequences: Ac-Lys-Trp-Lys-Ser-Phe-Leu-Lys-Thr-Phe-Lys-X-Ala-Y-Lys-Thr-Val-Leu-His-Thr-Ala-Leu-Lys-Ala-Ile-Ser-Ser-amide, where position X (Y = Val) is substituted by Gly, Lys, Ser, Ala, Val or Leu for hydrophilic face substitutions or position Y (X = Ser) is substituted by the same amino acids for hydrophobic face substitutions. The peptides are denoted by the substituted amino acid (all l-isomers plus Gly). Adapted from ref. [33] with permission from Elsevier.
In their extensive review of HILIC, Hemström and Irgum [15] noted that the retention properties of large peptides in both HILIC and RPC can be understood by the contact region concept of Regnier and coworkers [36], specifically noting that the work of our laboratory in comparing the separation of amphipathic peptides by RPC and HILIC/CEX investigated the contact region concept. In the case of the results shown in Figs. 7–10, we are, of course, defining these (major) contact regions as preferred binding domains of amphipathic cyclic (Figs. 7 and 8) and α-helical (Figs. 9 and 10) peptides.
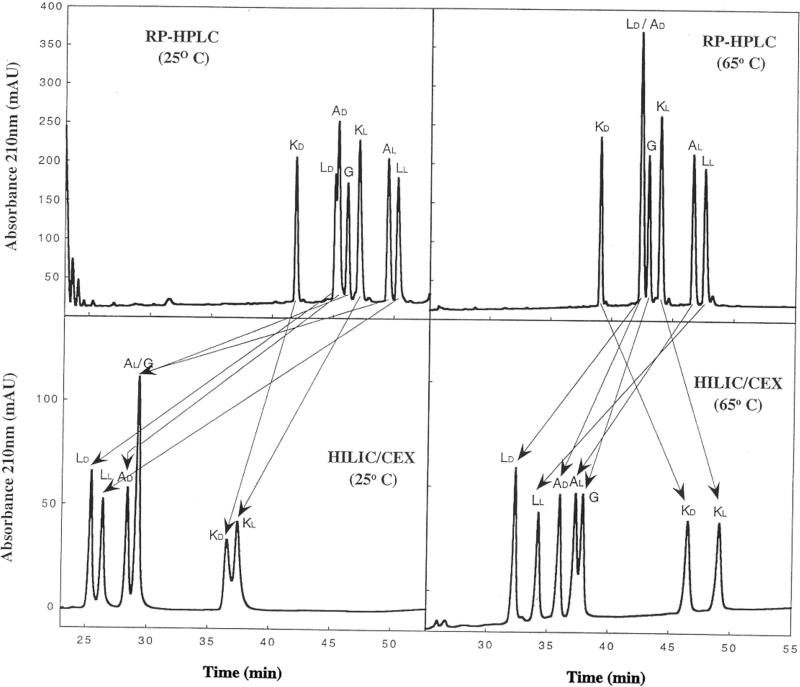
Figure 11 now compares the effect of temperature on RPC and HILIC/CEX of amphipathic α-helical peptides of the same sequence as for Fig. 10 but where l- or d-amino acid substitutions of Lys, Ala, Leu (+Gly) are made in the hydrophilic face only [37]. From Fig. 11 (left), RPC and HILIC/CEX again show useful selectivity differences between the two modes at 25°C. The d-amino acid substituted peptides are always eluted prior to their l-counterparts for both separation modes. This retention time decrease of the d-analogues compared to the l-analogues can be rationalized as being due to disruption of the amphipathic α-helix due to the introduction of the d-amino acid [38–42], this disruption affecting both the hydrophobic face of the helix as well as the hydrophilic face where the substitution has been made. The overall effect on the nonpolar face would be a decrease in the apparent hydrophobicity or hydrophilicity of the nonpolar or polar faces, respectively, when the helix is substituted with a d-amino acid relative to its l-diastereomer and, hence, a decrease in retention time of the former compared to the latter.
Figure 11.
Effect of temperature on RPC versus HILIC/CEX of amphipathic α-helical peptides. Columns: RPC (denoted RP-HPLC in Fig. 11) and HILIC/CEX, same as Fig. 10. Conditions: RPC and HILIC/CEX, same as Fig. 10 with runs carried out at 25°C (left panels) and 65°C (right panels). Peptide sequences: same as Fig. 10 for hydrophilic face (position X) substitutions. The peptides are denoted by the substituted amino acid (either the l- or d-isomer plus Gly). Reprinted from ref. [37] with permission from Elsevier.
From Fig. 11 (right), the effect of temperature on the HILIC/CEX elution behaviour of the peptides is quite distinct from that of its effect during RPC, with the HILIC/CEX retention times of all peptides increasing (as opposed to the decrease seen for RPC) on raising the temperature from 25°C (Fig. 11, lower left) to 65°C (Fig. 11, lower right). In addition, resolution of the peptides is greatly improved at the higher temperature of HILIC/CEX. Thus, AL and G are coeluted at 25°C (Fig. 11, lower left) but mainly resolved at 65°C (Fig. 11, lower right); the LL/LD, AL/AD and KL/KD peptide pairs are also better separated at 65°C (Fig. 11, lower right) compared to 25°C (Fig. 11, lower left). In addition, the effect of the rise in temperature from 25 to 65°C during HILIC/CEX had a greater effect on the Lys-substituted analogues KL and KD than the other peptide analogues, suggesting that the presence of an extra positive charge (which is concomitantly a significant increase in hydrophilicity of the polar face of the helix) enhances the effect of temperature during HILIC/CEX. The complementarity of the two HPLC modes is still apparent at this higher temperature (as denoted by the arrows), albeit the superior resolution of the peptide mixture by HILIC/CEX (all seven peptide peaks visible, with five resolved to baseline plus a major doublet) compared to RPC (six peaks visible with just five resolved and two coeluted) is also clear.
3.3 Purification and analysis of solid-phase synthesis products
3.3.1 Serine acetylation
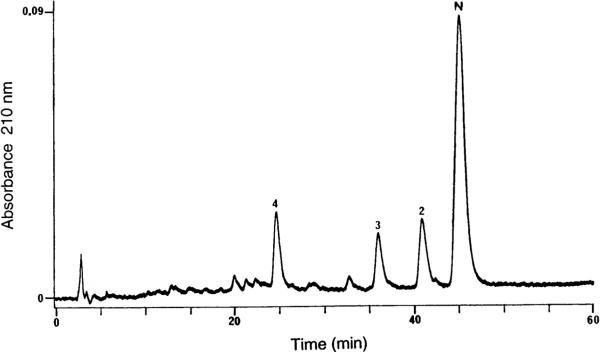
Figure 12 shows the HILIC/CEX analysis of a synthetic 21-residue amphipathic α-helical peptide, denoted N, which had already been through a single RPC purification step to produce a semipurified peptide product with closely-related impurities [43]. Subsequent analysis of the four peaks (N, 2−4) confirmed a lysine-deletion peptide (peak 4) as well as two acetylated impurities (peaks 2 and 3). With the exception of the lysine-deletion peptide, these impurities were only poorly resolved from the main peptide peak, N, by RPC and proved difficult to remove by preparative RPC while maintaining satisfactory yield of the desired product. A similar result was seen with CEX, with a satisfactory separation of the lysine-deletion peptide but complete coelution of acetylated impurities 2 and 3 (from Fig. 12) with peptide N. Figure 13 (top) represents the peptide as an α-helical net, with the wide hydrophilic face bordered by the two solid lines and the narrow hydrophobic face made up of Ile and Leu residues. The three potential acetylation sites (Ser residues) are boxed. Their presence in the hydrophilic face explains why the acetylated impurities are only poorly resolved by RPC since they are not in the preferred (hydrophobic) binding domain of the peptide for this HPLC mode. The nonresolution of these impurities from peptide N by CEX is due to the fact that the serine-acetylated peptides had the same overall net charge as peptide N, unlike the lysine-deletion peptide which contained one lesser positive charge than the desired peptide product. A subsequent purification protocol carried out an initial HILIC/CEX step of the crude peptide to remove major peptide contaminants, followed by a RPC step of the main peak obtained from the HILIC/CEX step as a final purification step with the concomitant use of a volatile mobile phase prior to lyophilization. Thus, this was an excellent example of the complementarity of HILIC/CEX and RPC for the design of a two-step purification protocol.
Figure 12.
HILIC/CEX of a semipurified synthetic amphipathic α-helical peptide (N). Column: same as Fig. 3. Conditions: linear AB gradient (2.5 mM NaClO4/min, following 10-min isocratic elution with 10% eluent B) at a flow-rate of 1 mL/min, where eluent A is 10 mM aq. TEAP, pH 6.5, and eluent B is 10 mM aq. TEAP containing 350 mM NaClO4, pH 6.5, both eluents also containing 65% v/v ACN; temperature 25°C. Peaks 2 and 3 represent acetylated peptide impurities; peak 4 represents a Lys-deletion peptide. The sequence of peptide N is shown in Fig. 13. Reprinted from ref. [43] with permission from Munksgaard.
Figure 13.
Top: Sequence of peptide N presented as an α-helical net; the area between the solid lines represent the more hydrophilic face of the peptide (Leu and Ile residues represent the narrow hydrophobic face) with the boxed Ser residues representing potential sites of side-chain acetylation; the peptide is also Nα-acetylated and contains a Cα-amide to avoid any peptide orientation issues or any effects of charged peptide termini. Bottom: effect of ACN on HILIC/CEX separation of acetylated peptides; column same as Fig. 12; conditions, same as Fig. 12 except the pH was changed to pH 3.0, with just 40% v/v ACN in eluents A and B. Ac3, Ac10 and Ac17 denote peptide N-acetylated at position 3, 10 or 17 (top). Adapted from ref. [43] with permission from Munksgaard.
It should be noted that, although there are three potential acetylation sites on the hydrophilic face of the α-helical peptide (Fig. 13, top), only two peaks containing acetylated impurities were observed (Fig. 12). Thus, either only two of the serine side-chains were acetylated or, where three acetylated derivatives were formed, two were coeluted by HILIC/CEX. Although this had no bearing on the subsequent two-step purification protocol, it was intriguing that HILIC/CEX was able to separate acetylated peptides of the same charge and extremely similar, if not identical, hydrophilicities and it was decided to explore further the sensitivity of this HPLC mode for such subtle differences between peptides. Thus, a series of peptide N analogues was made with acetylated serines introduced at one of the three serine positions: positions 3, 10 and 17 for peptide analogues denoted Ac3, Ac10 and Ac17, respectively. From Fig. 13 (top), all three serines are in the middle of the hydrophilic face of the amphipathic helix. Ser 10 and Ser 17 are surrounded by four potentially charged residues (two Lys and two Glu); Ser 3, in contrast, is near the N-terminus of the peptide and has only two potentially charged residues (one Lys and one Glu) as neighbours. It was felt that it was possible that this difference in environment may cause subtle differences in the relative hydrophilicities of acetylated Ser residues at position 3 compared to positions 10 and 17 which could be exploited by HILIC/CEX. Figure 13 (bottom) shows the effect of ACN concentration in the mobile phase on the HILIC/CEX separation of the three acetylated peptide analogues. It should be noted that the run was carried out at pH 3.0 to ensure protonation of the Glu residues, i.e. only positively charged (Lys) residues are present on the hydrophilic face (Fig. 13, top). Over an ACN range of 25−65%, optimum resolution of the peptides was obtained at 40% ACN (Fig. 13, bottom), with four peaks apparent. Peak N is the original desired peptide product (Figs. 12 and 13, top). From Fig. 13 (bottom), Ac3 is eluted first and corresponds to peak 3 in Fig. 12, supporting the aforementioned view that an acetylated serine at position 3 experiences a different environment and so has a different relative hydrophilicity compared to the same residues at position 10 or 17. Remarkably, it was possible to show a separation between Ac10 and Ac17, which were resolved halfway to baseline under these conditions, despite the essentially identical environment surrounding the acetylated serine residues, i.e. two Glu and two Lys residues. It should be noted that these two peptides were partially resolved at a level of 65% ACN in the mobile phase (not shown), i.e. under the same ACN (and gradient, 2.5 mM NaClO4/min) conditions as resulted in the HILIC/CEX separation shown in Fig. 12. Thus, peak 2 in Fig. 12 represents only one acetylated product (Ac10 or Ac17) and not coelution of two peptides.
3.3.2 Cysteine deletion
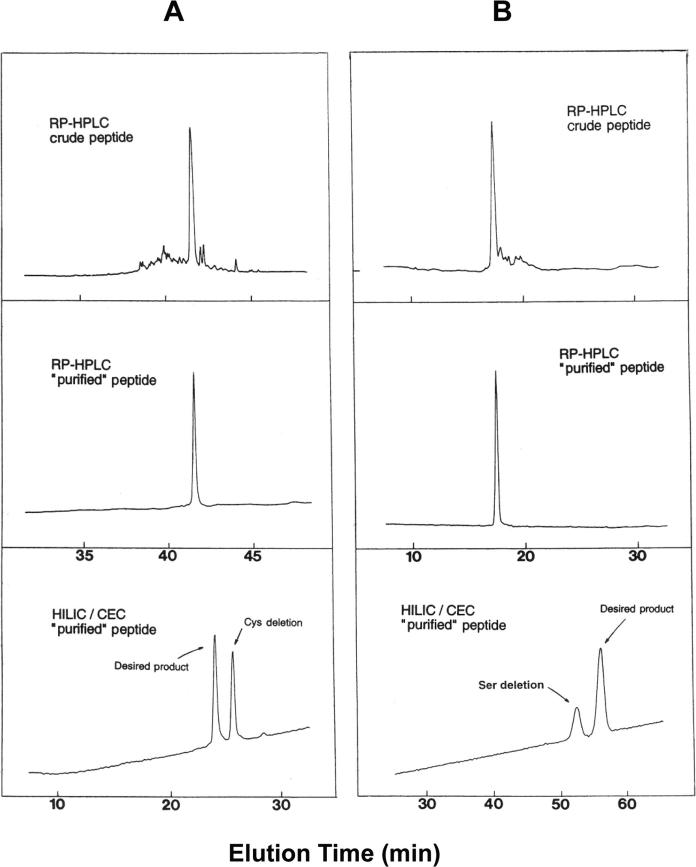
Figure 14A shows the purification and analysis of a 35-residue amphipathic α-helical synthetic peptide with a Cys residue at position 2 [6]. At a first glance, the crude peptide elution profile (Fig. 14A, top) indicated a successful synthesis with relatively few impurities, enabling an easy purification of the reduced peptide (to prevent Cys oxidation) by RPC. The resulting analytical RPC run of the purified peptide (Fig. 14A, middle) showed a single symmetrical peak and the lack of any shoulder on the peak, obtained on a very efficient RPC column, suggested excellent peptide purity. However, MS of this peptide showed not only the expected product mass, but a second strong signal exhibiting a peptide mass 103 U less than the expected product, indicating deletion of the Cys residue at position 2 of the peptide. The Cys side-chain is relatively hydrophobic [17] and it may have been expected that RPC would have easily detected such a deletion product. However, the contribution which a residue makes to the overall hydrophilicity/hydrophobicity of a peptide diminishes with increasing peptide length [44]. In addition, even though Cys is relatively hydrophobic compared to a smaller nonpolar side-chain such as Ala (RPC-derived values of 7.6 and 2.8, respectively [17]), its overall contribution to the nonpolar face of the α-helix, the remainder of which is made up of five Leu residues (a hydrophobicity value of 23.3) and three Val residues (13.4) (for a total hydrophobicity value of the Leu and Val residues of 156.7) is very small. Also, it has been shown that the contribution of a hydrophobe at the end of an amphipathic α-helix is much less than its contribution in the centre of the hydrophobic surface to interaction of this nonpolar surface with another hydrophobic surface [62]. Taken together, these factors have served to mask the loss of a Cys residue when applying RPC for purification and as a purity check. However, Fig. 14A (bottom) illustrates the excellent separation of the two peptides when HILIC/CEX was applied to the RPC ‘purified’ peptide (Fig. 14A, middle). The loss of the Cys residue would make the peptide more hydrophilic (less hydrophobic), hence the observation that the Cys-deletion impurity (which represents a very considerable 47% of the RPC purified peptide peak) is eluted later than the desired peptide during HILIC/CEX. It should be noted that CEX alone failed to separate this impurity due to the identical net charge on this peptide and the desired peptide product.
Figure 14.
Analysis and purification of synthetic peptides by RPC and HILIC/CEX. (A) 35-residue Cys-containing amphipathic α-helical peptide; (B) 17-residue intrachain disulphide-bridged peptide. Columns: RPC and HILIC/CEX (denoted HILIC/CEC in Fig. 14), same as Fig. 3. Conditions: RPC, linear AB gradient (1% B/min, equivalent to 1% ACN/min) at a flow-rate of 1 mL/min, where eluent A is 0.05% aq. TFA and eluent B is 0.05% TFA in ACN; HILIC/CEX, linear AB gradient (2.5 mM NaClO4/min from 30 mM NaClO4, following 10-min isocratic elution with 30 mM NaClO4) at a flow-rate of 1 mL/min, where eluent A is 5 mM aq. TEAP, pH 7, and eluent B is 5 mM aq. TEAP containing 400 mM NaClO4, pH 7, both eluents also containing 65% v/v ACN; runs carried out at 26°C and peaks detected by absorbance at 210 nm. Peptide sequences: (A) Ac-Gln-Cys-Gly-Ala-Leu-Gln-Lys-Gln-Val-Gly-Ala-Leu-Glu-Lys-Glu-Glu-Gly-Ala-Leu-Glu-Lys-Gln-Val-Gly-Ala-Leu-Gln-Lys-Gln-Val-Gly-Ala-Leu-Gln-Lys-amide; (B) Ac-Lys-Cys-Lys-Ser-Thr-Gln-Asp-Glu-Gln-Phe-Ile-Pro-Lys-Gly-Cys-Ser-Lys (intrachain disulphide bridge between two Cys residues; Ac denotes Nα-acetyl and amide denotes Cα-amide). Reprinted from ref. [6] with permission of Academic Press.
3.3.3 Serine deletion
Figure 14B outlines the purification and analysis of a 17-residue synthetic peptide containing an intrachain disulphide bridge [6]. Following purification of the crude oxidized peptide mixture Fig. 14B (top) by RPC, analysis of the purified product showed a single, symmetrical peak Fig. 14B (middle), indicating, as above (Fig. 14A), excellent peptide purity. However, MS of this single peak again showed, in addition to the expected mass, a second signal; in this case, this second signal exhibited a peptide mass 87 U less than the desired product, indicating deletion of one of the Ser residues. This deletion was almost certainly at position 4 of the sequence, because the Ser residue at position 16 was part of a core peptide common to other peptide analogues and this observed Ser deletion was only detected when a second Ser was added during elongation of this core peptide. On the basis of side-chain hydrophobicity coefficients [17], the polar Ser side-chain contributes little to the retention behaviour of a peptide during RPC and is classed, in RPC terms, as only a slightly hydrophilic group. Thus, it is not surprising that the deletion product would be difficult to detect, let alone resolve by RPC, particularly considering the length of the peptide (see above comments concerning peptide chain length effects) and the position of the residue within the intrachain disulphide bridge. Due to the neutrality of the Ser side-chain, CEX is again not suitable for purification of the desired peptide product. However, Fig. 14B (bottom) again illustrates the efficacy of the HILIC/CEX approach to resolving two peptides inseparable by RPC Fig. 14B (middle). This mixed-mode approach appeared to enhance the hydrophilic contribution of a Ser residue, as the Ser-deletion impurity is now baseline-resolved from the desired product. The Ser-deletion peptide is eluted prior to the desired product because loss of this residue has made the peptide less hydrophilic (i.e. more hydrophobic) than the native peptide.
3.4 Random coil peptides with the same composition but different sequences (SCDS)
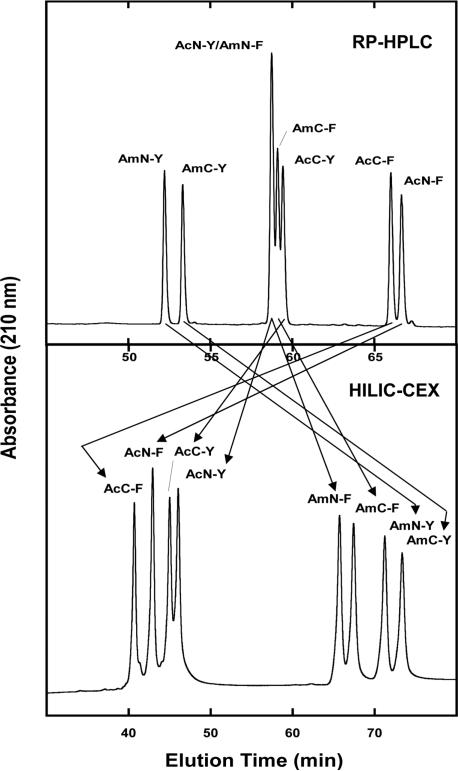
Figure 15 compares RPC and HILIC/CEX for the separation of four pairs of peptides with the SCDS (Table 1): peptide pairs denoted AmN-Y/AmC-Y, AmN-F/AmC-F, AcN-Y/AcC-Y, AcN-F/AcC-F; note that peptides containing the ‘Am’ (for ‘amino’) denotation exhibit a net charge of +3, whilst those with the ‘Ac’ (for ‘acetylated’) denotation exhibit a net charge of +2 [45]. These peptides differ only in the substitution position of a single amino acid (Phe or Tyr), either towards the N-terminal end of the 12-residue sequence (position 2) or towards the C-terminal end of the sequence (position 9). The adjacent residues to these substitution positions are always the same, i.e. Gly-X-Leu, where X is the position of substitution, assuring that the environment around the substituted region does not change. The presence of Gly residues throughout the sequences ensures the peptides possess no secondary structure during chromatography [46, 47]. Such peptide pairs frequently offer a challenge to the resolution ability of RPC, particularly in the case of peptides with negligible secondary structures. From Fig. 15, the two elution profiles are markedly different, reflecting profound differences in the selectivity (as highlighted by the arrows) of the two HPLC modes. It should be noted that RPC (Fig. 15, top), like HILIC/CEX (Fig. 15, bottom), is able to separate all four SCDS peptide pairs. However, coelution of various Phe and Tyr analogues in this particular peptide mixture makes RPC (Fig. 15, top), unlike HILIC/CEX (Fig. 15, bottom), unsuitable for resolution of all eight peptides of this particular mixture. Overall, however, Fig. 15 again demonstrates the excellent complementary nature of the two HPLC modes.
Figure 15.
RPC versus HILIC/CEX separation of peptides with the SCDS. Columns: RPC (denoted RP-HPLC in Fig. 15), Luna C8 (250 × 3 mm id, 5 μm particle size, 100 Å pore size); HILIC/CEX, same as Fig. 10. Conditions: RPC, linear AB gradient (0.5% ACN/min) at a flow-rate of 0.5 mL/min, where eluent A is 0.2% aq. TFA and eluent B is 0.18% TFA in ACN; HILIC/CEX, linear AB gradient (1 mM NaClO4/min), at a flow-rate of 0.3 mL/min, where eluent A is 5 mM aq. TEAP, pH 4.5, and eluent B is eluent A plus 250 mM NaClO4, both eluents also containing 90% v/v and 80% v/v ACN, respectively; runs were carried out at 25°C. Peptide sequences: shown in Table 1. Arrows denote selectivity variations between the two separation modes. Reprinted from ref. [45].
Table 1.
Synthetic peptides with the SCDS
| Peptide | Peptide sequence | Net charge |
|---|---|---|
| AcN-F | Ac-Gly-Phe-Leu-Gly-Leu-Ala-Leu-Gly-Gly-Leu-Lys-Lys-amide | +2 |
| AcC-F | Ac-Gly-Gly-Leu-Gly-Leu-Ala-Leu-Gly-Phe-Leu-Lys-Lys-amide | +2 |
| AmN-F | Gly-Phe-Leu-Gly-Leu-Ala-Leu-Gly-Gly-Leu-Lys-Lys-amide | +3 |
| AmC-F | Gly-Gly-Leu-Gly-Leu-Ala-Leu-Gly-Phe-Leu-Lys-Lys-amide | +3 |
| AcN-Y | Ac-Gly-Tyr-Leu-Gly-Leu-Ala-Leu-Gly-Gly-Leu-Lys-Lys-amide | +2 |
| AcC-Y | Ac-Gly-Gly-Leu-Gly-Leu-Ala-Leu-Gly-Tyr-Leu-Lys-Lys-amide | +2 |
| AmN-Y | Gly-Tyr-Leu-Gly-Leu-Ala-Leu-Gly-Gly-Leu-Lys-Lys-amide | +3 |
| AmC-Y | Gly-Gly-Leu-Gly-Leu-Ala-Leu-Gly-Tyr-Leu-Lys-Lys-amide | +3 |
Ac denotes Nα-acetyl; amide denotes Cα-amide. Peptides with +3 net charge have a free α-amino group. Positions of substituted amino acid (Phe and Tyr) are in bold.
4 HILIC/CEX peptide standards
Table 2 shows the sequences of synthetic HILIC/CEX peptide standards, together with properties of charge and carbon atom content [45]. From Table 2, the 27 decapeptides making up the peptide mixture possess only two distinct properties with which they may be resolved, i.e. charge and hydrophobicity. The 10-residue length of the peptides was designed to reflect the average size of cleavage fragments from proteolytic digests of proteins. The presence of only basic amino acid residues (Lys), i.e.no acidic, potentially negatively charged residues, and the blocked C-terminus for all peptides, and the use of a HILIC/CEX mobile phase at a pH value (pH 4.5) far below the pKa values of the N-terminal amino group and the Lys side-chain amino group, ensures a constant net positive charge on the peptides. Finally, in a similar manner to that noted above for the SCDS peptides, the random coil nature of the peptides is ensured by the inclusion of helix-disrupting glycine residues throughout the peptide sequences, thus ruling out separations based on conformational differences between peptides [46, 47]. From Table 2, the peptides are divided into three groups of nine peptides with nominal charges of +1 (peptides 1a–1i), +2 (peptides 2a–2i) and +3 (peptides 3a–3i). Within each peptide group, there is only a subtle hydrophobicity variation between adjacent peptides – generally just a single CH2 group change from peptide to peptide – the only exceptions being the 1e/1f, 2e/2f and 3e/3f adjacent peptide pairs which have the same number of carbon atoms and the same mass (the Val-Ala sequence is changed to an Ile-Gly sequence) as well as the same net positive charge. The peptides shown in Table 2 are listed in order of increasing hydrophobicity within each peptide group as expressed by their RPC retention behaviour at pH 2. These standards were designed to serve a double purpose, both as proteomic peptide standards to help develop multidimensional HPLC protocols (CEX and RPC) and as a potent test mixture for the mixed-mode HILIC/CEX approach. Thus, such a test mixture can be employed for evaluating CEX, stationary phases and optimization of HILIC/CEX mobile phases for peptide applications.
Table 2.
Sequences of HILIC/CEX synthetic peptide standards
| Peptide standard | Peptide sequence | Net charge | Change in carbon atom content |
|---|---|---|---|
| 1a | Ac-Gly-Gly-Gly-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 0 |
| 1b | Ac-Gly-Gly-Ala-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 1 |
| 1c | Ac-Gly-Gly-Ala-Ala-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 2 |
| 1d | Ac-Gly-Gly-Val-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 3 |
| 1e | Ac-Gly-Gly-Val-Ala-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 4 |
| 1f | Ac-Gly-Gly-Ile-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 4 |
| 1g | Ac-Gly-Gly-Ile-Ala-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 5 |
| 1h | Ac-Gly-Gly-Ile-Val-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 7 |
| 1i | Ac-Gly-Gly-Ile-Ile-Gly-Leu-Gly-Leu-Gly-Lys-amide | +1 | 8 |
| 2a | Gly-Gly-Gly-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 0 |
| 2b | Gly-Gly-Ala-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 1 |
| 2c | Gly-Gly-Ala-Ala-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 2 |
| 2d | Gly-Gly-Val-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 3 |
| 2e | Gly-Gly-Val-Ala-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 4 |
| 2f | Gly-Gly-Ile-Gly-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 4 |
| 2g | Gly-Gly-Ile-Ala-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 5 |
| 2h | Gly-Gly-Ile-Val-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 7 |
| 2i | Gly-Gly-Ile-Ile-Gly-Leu-Gly-Leu-Gly-Lys-amide | +2 | 8 |
| 3a | Gly-Gly-Gly-Gly-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 0 |
| 3b | Gly-Gly-Ala-Gly-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 1 |
| 3c | Gly-Gly-Ala-Ala-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 2 |
| 3d | Gly-Gly-Val-Gly-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 3 |
| 3e | Gly-Gly-Val-Ala-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 4 |
| 3f | Gly-Gly-Ile-Gly-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 4 |
| 3g | Gly-Gly-Ile-Ala-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 5 |
| 3h | Gly-Gly-Ile-Val-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 7 |
| 3i | Gly-Gly-Ile-Ile-Lys-Leu-Gly-Leu-Gly-Lys-amide | +3 | 8 |
Ac denotes Nα-acetyl; amide denotes Cα-amide. Standards in the +2 and +3 groups have a free α-amino group. Variations in composition of the peptide analogues are indicated in bold where the amino acid residues differ from 1a, 2a or 3a. The change in carbon atom content in the +1, +2 and +3 groups are compared to peptides 1a, 2a and 3a, respectively.
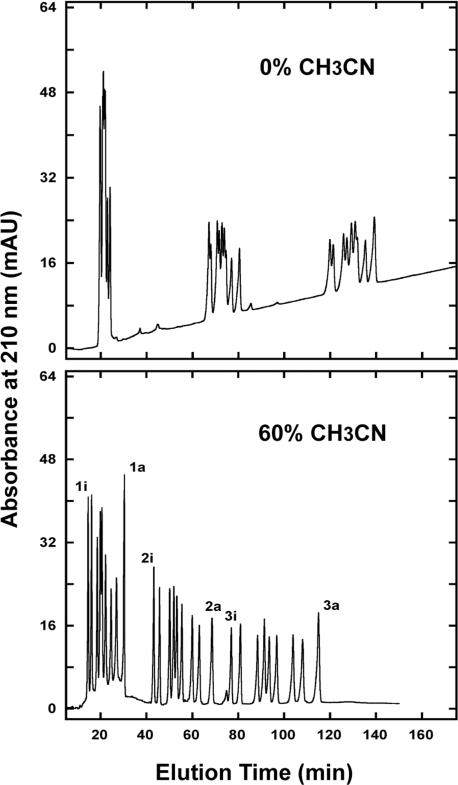
Figure 16 shows the separation of the 27-peptide standards mixture in the presence of 0% ACN (top; CEX separation only) and 60% ACN (bottom; HILIC/CEX separation). At a level of 0% ACN (Fig. 16, top), the three groups of peptides are widely separated by charge (CEX) separation, but are only poorly resolved within the three charge groups. However, the presence of 60% ACN in the mobile phase (HILIC/CEX separation; Fig. 16, bottom) dramatically improves resolution with all 27 peptides essentially separated to baseline, including the 2e/2f and 3e/3f peptide pairs which contain the same number of carbon atoms (Table 2). It should be noted that while the three peptide groups are eluted in order of +1 < +2 < +3 via a cation-exchange mechanism, within these three groups the peptides are eluted in order of increasing hydrophilicity (decreasing hydrophobicity) i.e. via a HILIC mechanism.
Figure 16.
HILIC/CEX of 27 peptide standards. Column: same as Fig. 10. Conditions: linear AB gradient (1 mM NaClO4/min) at a flow-rate of 0.3 mL/min, where eluent A is 5 mM aq. TEAP, pH 4.5, and eluent B is eluent A plus 500 mM NaClO4 (0% ACN run) or 250 mM NaClO4 (60% ACN run), both eluents also containing the corresponding 0% v/v (top) or 60% v/v (bottom) ACN; runs were carried out at 25°C. Peptide sequences: shown in Table 2. Adapted from ref. [45].
5 HILIC/CEX of proteins
Histones are the most extensively studied group of basic nuclear proteins and are of great importance with regard to the organization of chromatin structure and gene activity. Histones can be resolved into five main classes (H1, H2A, H2B, H3 and H4) and, in mammals, each class of these nuclear proteins, with the exception of H4, is represented by primary sequence variants or subtypes which differ only slightly in molecular size and primary structure. In addition, all histone proteins are known to be post-translationally modified, e.g. acetylation, phosphorylation, methylation, etc. Thus, efficient resolution of histone variants (traditionally by gel electrophoresis, conventional CEX and RPC) has represented and remains a significant challenge. Fortunately, as described below, HILIC/CEX has provided a novel and important alternative to traditional separation techniques for such proteins, as described by excellent work by Lindner and coworkers [48–56] as well as other researchers [57–60].
5.1 Separation of acetylated core histones
One of the major types of post-translational modifications of histones is reversible acetylation of lysine residues within the N-terminal domain of the core histones (H2A, H2B, H3, H4). Figure 17A shows the multistep gradient RPC resolution of whole histones isolated from erythroleukaemic cells [48]. From Fig. 17A, 11 histone fractions were obtained: linker histone subtypes H1°, H1a, H1b, H1C, H1d + H1e and the core histones H2B, H2A.2, H4, H2A.1, H3.2 + H3.3 and H3.1. Subsequent application of H4 to a HILIC/CEX separation (Fig. 17B) produced three peaks which were coeluted during RPC: ac0, ac1 and ac2, representing nonoacetylated H4, monoacetylated H4 and diacetylated H4, respectively. The higher the degree of acetylation, the lower the net positive charge on the histone and, of course the lower the overall hydrophilicity of the histone; hence, the order of elution is ac2 < ac1 < ac0. In addition, histones display multidomain structures, where an extended hydrophobic domain is flanked by one or two hydrophilic regions. In an analogous manner to amphipathic peptides described above, these hydrophilic regions should bind preferentially to the charged/hydrophilic CEX stationary phase. Since acetylation occurs exclusively in the hydrophilic N-terminal region of histones, thereby changing the hydrophilicity of their contact regions (or preferred binding domains), mixed-mode HILIC/CEX is able to discriminate between individual modified proteins. Note that a cation-exchange separation alone could not be achieved due to irreversible absorption of the histone via, assumedly, strong hydrophobic interactions which are subsequently overcome by the high levels of ACN (70% in the present example) characteristic of HILIC/CEX. It should also be noted that modification of the hydrophilic domain while the hydrophobic domain is unchanged would also account for the inability of RPC to resolve these modified histones. Overall, the results in Fig. 17 show the excellent complementarity of RPC and HILIC/CEX for complete resolution of this histone mixture.
Figure 17.
HILIC/CEX separation of acetylated core histones. Columns (A) RPC, Nucleosil 300−5 C4 (125 × 8 mm id, 5 μm particle size, 300 Å pore size); (B) HILIC/CEX (denoted HILIC in Fig. 17), SynChropak CM300 weak cation-exchange column (250 × 4.6 mm id, 6.5 μm, 300 Å). Conditions: (A) RPC, multistep gradient, at a flow-rate of 1 mL/min, starting at 40% B and increasing linearly in the order of 40% B to 61% B in 45 min, to 64% B in 35 min, to 74% B in 2 min (maintained at 74% B for 9 min) and, finally, to 100% B in 10 min, where eluent A is 0.1% aq. TFA and eluent B is 0.1% TFA in 70% aq. ACN, both eluents also containing 10% ethylene glycol monomethyl ether (EGME); (B) HILIC/CEX, two-step gradient, at a flow-rate of 1 mL/min, starting at 0% B and increasing linearly to 10% in 2 min and to 40% in 30 min, where eluent A is 15 mM aq. TEAP, pH 3, and eluent B is eluent A containing 680 mM NaClO4, both eluents also containing 70% v/v ACN; the gradients for both the RPC and HILIC/CEX runs are denoted by the dotted lines on the elution profiles. The histone H4 fraction isolated by RPC (A) was subsequently applied to HILIC/CEX (B); ac0, ac1 and ac2 denote nonacetylated, monoacetylated and diacetylated histone H4, respectively. Adapted from ref. [48] with permission from Elsevier.
A separate study by the same group [52] also demonstrated how avian histone H5, which was eluted as a single peak in RPC was resolved into two fractions by HILIC/CEX: unblocked H5 versus N-acetylated H5 histone.
5.2 Separation of methylated forms of histone H4
Lindner's group also succeeded in an interesting separation whereby acetylated (at Lys 16) and nonacetylated versions of variously methylated forms of the 102-residue rat kidney histone H4 were separated into distinct fractions by HILIC/CEX from a single eluted peak during RPC [53]. Methylation is a post-translational modification of histone proteins at Lys and/or Arg residues with a site selectivity for Lys methylation at specific positions in the N-termini of histones H3 and H4. Thus, HILIC/CEX of the single H4 peak from RPC produced two major groups of eluted solutes during HILIC/CEX, the two groups representing acetylated (eluted first) and nonacetylated (eluted last) groups of methylated (at Lys 20) forms of H4. The two groups contained, in order of elution, trimethylated < dimethylated < monomethylated forms of the protein, with the dimethylated forms being the most abundant. Note that this elution order within the acetylated/nonacetylated groups would be expected since progressive addition of the nonpolar methyl group would make the protein (or specifically, the hydrophilic preferred binding N-terminal domain where Lys 20 is situated) more hydrophobic (less hydrophilic) with each methyl group addition.
5.3 Separation of phosphorylated H1 histones
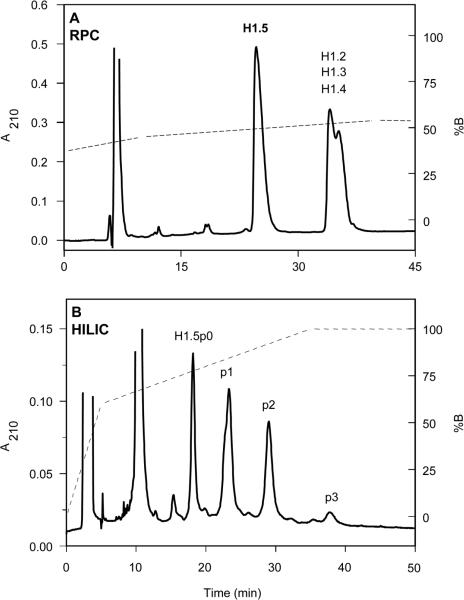
In addition to the heterogeneity of their primary sequences, H1 histones are also post-translationally modified, being phosphorylated at Ser and Thr residues located in the N- and C-terminal domains of the protein. Figure 18A shows the RPC separation of whole linker histones from cultures of Raji cells, originally derived from patients with Burkitt's lymphoma [49]. From Fig. 18A, just two subfractions were obtained: histone H1.5 and a second fraction consisting of coeluted H1 subtypes. Application of H1.5 from the RPC run (Fig. 18A) to a HILIC/CEX separation (Fig. 18B) resolved this peak into four subfractions: nonphosphorylated H1.5 (designated H1.5p0), monophosphorylated (p1), diphosphorylated (p2) and triphosphorylated (p3) forms of H1.5. The shoulder seen on p1 is likely due to phosphorylation at two different locations. Again, CEX alone was not able to produce this separation. In a similar manner to the acetylation of histones described above (Fig. 17), phosphorylation occurs almost exclusively in the hydrophilic N- and C-domains (i.e. the preferred hydrophilic binding domains or contact regions) of the histones; hence, the inability of RPC (Fig. 18A) to resolve these modified histones in contrast to HILIC/CEX (Fig. 18B). Similar phosphorylated histone separations were reported in a later study by the same researchers [56].
Figure 18.
HILIC/CEX separation of phosphorylated H1 histones. Columns: (A) RPC, same as Fig. 17; (B) HILIC/CEX (denoted HILIC in Fig. 18), PolyCAT A weak cation-exchange column (200 × 4.6 mm id, 5 μm particle size, 300 Å pore size). Conditions: (A) RPC, linear AB gradient at a flow-rate of 1.5 mL/min, starting at 47% B and increasing to 55% B in 35 min, where eluent A is 0.1% aq. TFA and eluent B is 0.1% TFA in 70% v/v ACN; (B) HILIC/CEX, two-step gradient, at a flow-rate of 1 L/min, starting from 0% B and increasing linearly to 60% B in 5 min and to 100% B in 30 min, where eluent A is 10 mM TEA-methanephosphonic acid (MPA), pH 3, and eluent B is eluent A containing 1000 mM NaClO4, both eluents also containing 70% v/v ACN; the gradients for both the RPC and HILIC/CEX runs are denoted by the dotted lines on the elution profiles; the histone H1.5 fraction isolated by RPC (A) was subsequently applied to HILIC/CEX (B); histone peaks H1.5p0, p1, p2 and p3 denote nonphosphorylated, monophosphorylated, diphosphorylated and triphosphorylated histone H1.5, respectively. Adapted from ref. [49] with permission from Elsevier.
A very interesting observation from Fig. 18B is the relative elution order of H1.5 and its phosphorylated counterparts, i.e. H1.5p0 < p1 < p2 < p3. Thus, the proteins are being eluted in order of decreasing net positive charge: phosphate groups are negatively charged and, hence, the more extensive the phosphorylation of the protein, the lower its overall net positive charge. In fact, despite poor resolution, these peaks were eluted from the CEX column, when used in CEX mode only, in reverse order to that of HILIC/CEX, i.e. in order of increasing net positive charge, as would be expected. The explanation for the separation seen in Fig. 18B lies in the increasing overall hydrophilicity of the proteins under HILIC/CEX conditions, i.e. high concentrations (70% in this case) of ACN in the mobile phase. As a charged group, the phosphate group is inherently very hydrophilic and, as the ACN concentration is raised, hydrophilic interactions dominate over electrostatic interactions. Thus, the more extensive the phosphorylation of a protein, the greater its overall hydrophilicity and, hence, the elution order under HILIC/CEX conditions is in the expected order of increasing overall protein hydrophilicity or, perhaps more specifically, increasing overall hydrophilicity of the preferred hydrophilic binding domains of the histones. A similar effect was seen in Fig. 4 where, at high ACN concentrations, peptides of a certain net positive charge were eluted prior to peptides of lower net positive charge due to hydrophilic interactions dominating over those of electrostatic interactions with the CEX stationary phase. Alpert [61] noted that such observations reflect the fact that hydrophilic interactions are independent of electrostatic effects.
5.4 Characterization of sequence variations in histones by HILIC/CEX
Protein deamidation is a well-documented nonenzymatic process and includes age-dependent deamidation of histones, including mammalian H1° (Asn → Asp conversion at position 3 of this 193-residue protein). Figure 19A shows the RPC separation of perchloric-acid-extracted linker histones from human placenta [50]. The H1° histone fraction, eluted as a single peak in RPC but known to consist of two subfractions (H1°a and H1°b), was now applied to HILIC/CEX (Fig. 19B). From Fig. 19B, four H1° subcomponents were resolved, denoted peaks 1−4. Subsequent characterization revealed that peaks 1 and 2 correspond to aforementioned subfraction H1°a and peaks 3 and 4 to that of H1°b. Eventual analysis of peaks 1−4 confirmed that peaks 1 and 3 contained Asn at position 3 of the sequence while peaks 2 and 4 contained Asp (i.e. deamidated Asn) at position 3; further, peaks 1 and 2 were blocked (acetylated) at their N-termini, while the later eluted peaks 3 and 4 had free α-amino groups (i.e. an extra positively charged group). This separation of the four subfractions of H1° by HILIC/CEX, with modifications occurring in the hydrophilic N-terminal domain of the protein, again underlines the excellent resolving capability of this mixed-mode approach both in its own right and as a complement to RPC.
Figure 19.
HILIC/CEX separation of amidated and deamidated forms of histone. Columns: (A) RPC, same as Fig. 17; (B) HILIC/CEX (denoted HILIC in Fig. 19), PolyCAT A weak cation-exchange column (250 × 4.6 mm id, 5 μm particle size, 1000 Å pore size). Conditions: (A) RPC, two-step gradient, at a flow-rate of 1.5 mL/min, starting at 37% B and increasing linearly to 45% B in 10 min and to 54% B in 30 min, where eluent A is 0.1% aq. TFA and eluent B is 0.1% TFA in 70% aq. ACN; (B) HILIC/CEX, two-step gradient, at a flow-rate of 1 mL/min, starting at 0% B and increasing linearly to 65% B in 5 min, to 100% B in 45 min and then maintained at 100% B for 30 min, where eluent A is 15 mM TEAP, pH 3, and eluent B is eluent A containing 680 mM NaClO4, both eluents also containing 70% v/v ACN; the gradients for both the RPC and HILIC/CEX runs are denoted by the dotted lines on the elution profiles. The histone H1° fraction isolated by RPC (A) was subsequently applied to HILIC/CEX (B); histone peak pairs 1/2 and 3/4 represent histone fractions with blocked N-termini and unblocked (free α-amino group) N-termini, respectively; histone peak pairs 1/3 and 2/4 represent amidated (Asn at position 3 of the histone sequence) and deamidated (Asp at position 3) histone fractions, respectively. Reprinted from ref. [50] with permission from ASBMB.
Another study by Lindner's group utilized HILIC/CEX to detect the occurrence of microsequence variations of H1 subtypes isolated from various human tumour cell lines [55]. Thus, these researchers were able to identify a Lys → Arg replacement at position 173 of histone H1.4 from Raji cells by HILIC/CEX. This result was particularly impressive considering that these two subfractions were not separated by RPC, high-performance CE (HPCE) or various gel-electrophoretic methods.
5.5 Separation of allelic and nonallelic variants of histone H1
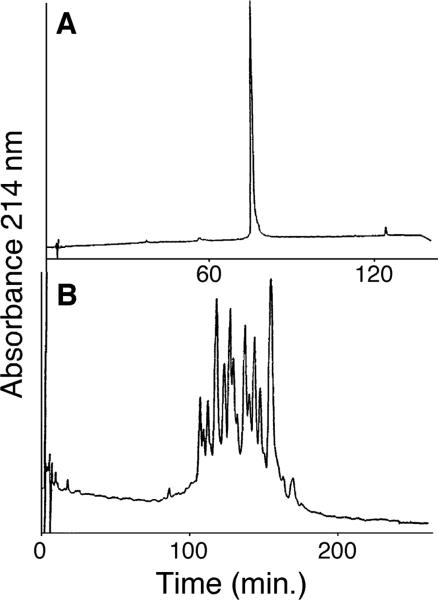
The H1 group is the most heterogeneous of all the histones and the limited resolution of H1 variants (all >200 residues) by traditional separation approaches had hindered comprehensive structural and functional analyses of these proteins. Thus, H1 from pooled chicken blood is eluted as a single peak in conventional RPC (Fig. 20A) [52, 53]. However, a total of 21 peaks representing all 6 nonallelic variants and allelic forms of 5 of these variants are resolved from the same sample by HILIC/CEX (Fig. 20B), although not shown here, CEX alone (0% ACN) or even CEX in the presence of 40% v/v ACN showed only poor resolution of the proteins (only 3−4 peaks observed), i.e. significant hydrophilic interactions in the presence of 70% v/v ACN were required to produce the HILIC/CEX elution profile shown in Fig. 20B.
Figure 20.
Resolution of pooled chicken erythrocyte H1 variants by HILIC/CEX. Columns: (A) RPC, Chromegabond MC-18 (250 × 4.6 mm id, 5 μm particle size, 300 Å pore size); (B) HILIC/CEX, same as Fig. 19. Conditions: (A) RPC, linear AB gradient of 5−60% v/v ACN in 0.1% aq. TFA over 120 min at a flow-rate of 0.8 mL/min; (B) HILIC/CEX, linear AB gradient of 380−590 mM NaClO4 in 10 mM aq. propionic acid, pH 6.5, containing 70% v/v ACN over 240 min at a flow-rate of 0.8 mL/min. Reprinted from ref. [58] with permission of Academic Press.
Acknowledgments
This work was supported by an NIH grant (RO1 GM61855) to R. S. H. and the John Stewart Chair in Peptide Chemistry to R. S. H. The content is solely the responsibility of the authors and does not necessarily represent the offical view of NIH.
Abbreviations
- CEX
cation-exchange chromatography
- HILIC
hydrophilic interaction chromatography
- RPC
RP chromatography
- SCDS
same composition but different sequence
Footnotes
The authors declared no conflict of interest.
6 References
- 1.Mant CT, Hodges RS, editors. HPLC of Peptides and Proteins: Separation, Analysis and Conformation. CRC; Boca Raton, FL, USA: 1991. [Google Scholar]
- 2.Hearn MTW, editor. HPLC of Proteins, Peptides and Polynucleotides: Contemporary Topics and Applications. VCH; New York, NY, USA: 1991. [Google Scholar]
- 3.Mant CT, Zhou NE, Hodges RS. In: Chromatography. 5th Edn. Heftmann E, editor. Elsevier; Amsterdam, The Netherlands: 1992. pp. B75–B150. [Google Scholar]
- 4.Dorsey JG, Foley JP, Cooper WT, Barford RA, Barth HG. Anal. Chem. 1992;64:353–389. doi: 10.1021/ac00036a020. [DOI] [PubMed] [Google Scholar]
- 5.Mant CT, Hodges RS. Methods Enzymol. 1996;271:3–50. doi: 10.1016/s0076-6879(96)71003-0. [DOI] [PubMed] [Google Scholar]
- 6.Mant CT, Kondejewski LH, Cachia PJ, Monera OD, Hodges RS. Methods Enzymol. 1997;289:426–469. doi: 10.1016/s0076-6879(97)89058-1. [DOI] [PubMed] [Google Scholar]
- 7.Cunico RL, Gooding KM, Wehr T, editors. Basic HPLC and CE of Biomolecules. Bay Bioanalytical Laboratory; Richmond, CA, USA: 1998. [Google Scholar]
- 8.Gooding KM, Regnier FE, editors. HPLC of Biological Macromolecules. 2nd Edn. Marcel Dekker; New York, NY, USA: 2002. [Google Scholar]
- 9.Mant CT, Chen Y, Yan Z, Popa TV, Kovacs JM, Mills JB, Tripet BP, Hodges RS. In: Methods in Molecular Biology. Fields G, editor. Humana Press; Totowa, NJ, USA: 2007. pp. 3–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu B-Y, Mant CT, Hodges RS. J. Chromatogr. 1991;548:13–24. doi: 10.1016/s0021-9673(01)88590-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhu B-Y, Mant CT, Hodges RS. J. Chromatogr. 1992;594:75–86. [Google Scholar]
- 12.Mant CT, Hodges RS. In: Encyclopedia of Separation Science. Wilson ID, Adland TR, Poole CF, Cook M, editors. Academic Press; London: 2000. pp. 3615–3626. [Google Scholar]
- 13.Alpert AJ. J. Chromatogr. 1990;499:177–196. doi: 10.1016/s0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- 14.Burke TWL, Mant CT, Black JA, Hodges RS. J. Chromatogr. 1989;476:377–389. doi: 10.1016/s0021-9673(01)93883-x. [DOI] [PubMed] [Google Scholar]
- 15.Hemström P, Irgum K. J. Sep. Sci. 2006;29:1784–1821. doi: 10.1002/jssc.200600199. [DOI] [PubMed] [Google Scholar]
- 16.Ikegami T, Tomomatsu K, Takubo H, Horie K, Tanaka N. J. Chromatogr. A. 2008;1184:474–503. doi: 10.1016/j.chroma.2008.01.075. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs JM, Mant CT, Hodges RS. Biopolymers (Pept. Sci.) 2006;84:298–309. doi: 10.1002/bip.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mant CT, Litowski JL, Hodges RS. J. Chromatogr. A. 1998;816:65–78. doi: 10.1016/s0021-9673(98)00507-x. [DOI] [PubMed] [Google Scholar]
- 19.Sereda TJ, Mant CT, Quinn AM, Hodges RS. J. Chromatogr. 1993;646:17–30. doi: 10.1016/s0021-9673(99)87003-4. [DOI] [PubMed] [Google Scholar]
- 20.Sereda TJ, Mant CT, Hodges RS. J. Chromatogr. 1997;776:153–165. doi: 10.1016/s0021-9673(97)00150-7. [DOI] [PubMed] [Google Scholar]
- 21.Hodges RS, Merrifield RB. J. Biol. Chem. 1975;250:1231–1241. [PubMed] [Google Scholar]
- 22.Doris PA. J. Chromatogr. 1984;336:392–396. doi: 10.1016/s0378-4347(00)85166-x. [DOI] [PubMed] [Google Scholar]
- 23.Crimmins DL, Thoma RS, McCourt DW, Schwartz BD. Anal. Biochem. 1989;176:255–260. doi: 10.1016/0003-2697(89)90305-9. [DOI] [PubMed] [Google Scholar]
- 24.McGregor JL, Clezardin P, Manach M, Gronlund S, Dechavanne M. J. Chromatogr. 1985;326:179–190. doi: 10.1016/s0021-9673(01)87444-6. [DOI] [PubMed] [Google Scholar]
- 25.Tandy NE, Dilley RA, Regnier FE. J. Chromatogr. 1983;266:599–607. [Google Scholar]
- 26.Bietz JA. In: HPLC of Biological Macromolecules. Gooding KM, Regnier FE, editors. Marcel Dekker; New York, NY, USA: 2002. pp. 547–587. [Google Scholar]
- 27.Tiihonen J, Sainio T, Kärki A, Paatero E. J. Chromatogr. A. 2002;982:69–84. doi: 10.1016/s0021-9673(02)01522-4. [DOI] [PubMed] [Google Scholar]
- 28.Mant CT, Kondejewski LH, Hodges RS. J. Chromatogr. A. 1998;816:79–88. doi: 10.1016/s0021-9673(98)00507-x. [DOI] [PubMed] [Google Scholar]
- 29.Sereda TJ, Mant CT, Sönnichsen FD, Hodges RS. J. Chromatogr. A. 1994;676:139–153. doi: 10.1016/0021-9673(94)00371-8. [DOI] [PubMed] [Google Scholar]
- 30.Mant CT, Zhou NE, Hodges RS. In: The Amphipathic Helix. Epand RE, editor. CRC Press; Boca Raton, FL, USA: 1993. pp. 39–64. [Google Scholar]
- 31.Zhou NE, Mant CT, Hodges RS. Pept. Res. 1990;3:8–20. [PubMed] [Google Scholar]
- 32.Lau SYM, Taneja AK, Hodges RS. J. Chromatogr. 1984;317:129–140. [Google Scholar]
- 33.Hodges RS, Chen Y, Kopecky E, Mant CT. J. Chromatogr. A. 2004;1053:161–172. [PubMed] [Google Scholar]
- 34.Zhang L, Benz R, Hancock REW. Biochemistry. 1998;38:8102–8111. doi: 10.1021/bi9904104. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Falla T, Wu M. Biochem. Biophys. Res. Commun. 1999;247:674–680. doi: 10.1006/bbrc.1998.8848. [DOI] [PubMed] [Google Scholar]
- 36.Fausnaugh-Pollitt J, Thevenon G, Janis L, Regnier FE. J. Chromatogr. 1988;443:221–228. doi: 10.1016/s0021-9673(00)94795-2. [DOI] [PubMed] [Google Scholar]
- 37.Hartmann E, Mant CT, Jungbauer A, Hodges RS. J. Chromatogr. A. 2003;1009:61–71. doi: 10.1016/s0021-9673(03)00620-4. [DOI] [PubMed] [Google Scholar]
- 38.Aguilar MI, Mougos S, Boublik J, Rivier J, Hearn MTW. J. Chromatogr. 1993;646:53–65. doi: 10.1016/s0021-9673(99)87007-1. [DOI] [PubMed] [Google Scholar]
- 39.Rothemund S, Beyermann M, Krause E, Krause G, Bienert M, Hodges RS, Sykes BD, Sönnichsen FD. Biochemistry. 1995;34:12954–12962. doi: 10.1021/bi00040a005. [DOI] [PubMed] [Google Scholar]
- 40.Rothemund S, Krause E, Beyermann M, Dathe M, Bienert M, Hodges RS, Sykes BD, Sönnichsen FD. Pept. Res. 1996;9:79–87. [PubMed] [Google Scholar]
- 41.Krause E, Bienert M, Schmieder P, Wenschuh H. J. Am. Chem. Soc. 2000;122:4865–4870. [Google Scholar]
- 42.Chen Y, Mant CT, Hodges RS. J. Pept. Res. 2002;59:18–33. doi: 10.1046/j.1397-002x.2001.10994.x. [DOI] [PubMed] [Google Scholar]
- 43.Litowski JR, Semchuk PD, Mant CT, Hodges RS. J. Pept. Res. 1999;54:1–11. doi: 10.1034/j.1399-3011.1999.00066.x. [DOI] [PubMed] [Google Scholar]
- 44.Mant CT, Burke TWL, Black JA, Hodges RS. J. Chromatogr. 1988;458:193–205. doi: 10.1016/s0021-9673(00)90564-8. [DOI] [PubMed] [Google Scholar]
- 45.Mant CT, Hodges RS. J. Sep. Sci. 2008;31:1573–1584. doi: 10.1002/jssc.200700619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou NE, Monera OD, Kay CM, Hodges RS. Protein Pept. Lett. 1994;1:114–119. [Google Scholar]
- 47.Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. J. Pep. Sci. 1995;1:319–329. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 48.Lindner H, Sarg B, Meraner C, Helliger W. J. Chromatogr. A. 1996;743:137–144. doi: 10.1016/0021-9673(96)00131-8. [DOI] [PubMed] [Google Scholar]
- 49.Lindner H, Sarg B, Helliger W. J. Chromatogr. A. 1997;782:55–62. doi: 10.1016/s0021-9673(97)00468-8. [DOI] [PubMed] [Google Scholar]
- 50.Lindner H, Sarg B, Hoertnagl B, Helliger W. J. Biol. Chem. 1998;273:13324–13330. doi: 10.1074/jbc.273.21.13324. [DOI] [PubMed] [Google Scholar]
- 51.Lindner H, Sarg B, Grunicke H, Heliger W. J. Cancer Clin. Oncol. 1999;125:182–186. doi: 10.1007/s004320050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarg B, Helliger W, Hoertnagl B, Puschendorf B, Lindner H. Arch. Biochem. Biophys. 1999;372:333–339. doi: 10.1006/abbi.1999.1503. [DOI] [PubMed] [Google Scholar]
- 53.Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. J. Biol. Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 54.Sarg B, Helliger W, Talasz H, Koutzamani E, Lindner HH. J. Biol. Chem. 2004;279:53458–53464. doi: 10.1074/jbc.M409099200. [DOI] [PubMed] [Google Scholar]
- 55.Sarg B, Gráen A, Söderkvist P, Helliger W, Rundquist I, Lindner HH. FEBS J. 2005;272:3673–3683. doi: 10.1111/j.1742-4658.2005.04793.x. [DOI] [PubMed] [Google Scholar]
- 56.Sarg B, Helliger W, Talasz H, Förg B, Lindner HH. J. Biol. Chem. 2006;281:6573–6580. doi: 10.1074/jbc.M508957200. [DOI] [PubMed] [Google Scholar]
- 57.Mizzen CA, Alpert AJ, Lévesque L, Kruck TPA, McLachlan DR. J. Chromatogr. B. 2000;744:33–46. doi: 10.1016/s0378-4347(00)00210-3. [DOI] [PubMed] [Google Scholar]
- 58.Mizzen CA. Methods Enzymol. 2004;375:278–297. doi: 10.1016/s0076-6879(03)75019-8. [DOI] [PubMed] [Google Scholar]
- 59.Pesavento JJ, Mizzen CA, Kelleher NL. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 60.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Nat. Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 61.Alpert AJ. Anal. Chem. 2008;80:62–76. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
- 62.Zhou NE, Kay CM, Hodges RS. J. Biol. Chem. 1992;267:2664–2670. [PubMed] [Google Scholar]