Abstract
Previous crystallographic studies of the AppA BLUF domain indicated that Trp104 is capable of undertaking alternate conformations depending on the length of the BLUF domain. A BLUF domain containing a C-terminal deletion (AppA1–126) reveals that Trp104 is partially solvent exposed while a BLUF domain containing a slightly longer carboxyl terminal region (AppA17–133) shows that Trp104 is deeply buried. This observation has led to a model proposing that Trp104 moves from a deeply buried position in the dark state to a solvent-exposed position in the light excited state. In this study we investigated whether there is indeed movement of Trp104 upon light excitation using a combination of NMR and absorption spectroscopy, steady-state fluorescence, and acrylamide quenching of tryptophan fluorescence. Our results indicate that AppA17–133 and AppA1–126 contain Trp104 in distinct alternate conformations in solution and that light absorption by the flavin causes partial movement/uncovering of Trp104. However, we conclude that light exposure does not cause dramatic change of Trp104 from “Trp-in” to “Trp-out” conformations (or vice versa) upon light absorption. These results do not support a model of Trp104 movement as a key output signal.
AppA is a well-characterized member of the “blue-light sensing using FAD” (BLUF)1 class of photoreceptors (1, 2). Details are emerging on the role and mechanism of activity of BLUF proteins in several systems. For example, the BLUF protein AppA regulates photosynthesis in gene expression of the purple bacterium Rhodobacter sphaeroides (1, 2), and the BLUF protein PixD from the cyanobacterium Synechocystis PCC 6803 is involved in phototaxis in response to blue light (3). Structural and mutational studies of these proteins have revealed both similarities and dissimilarities in their photocycles.
Significant progress has been made in determining crystal and solution structures of several BLUF domains (4–9). These studies reveal a conserved βαββαββ fold, as well as several conserved residues that form a flavin-binding pocket. Residues in the flavin-binding pocket form an intricate hydrogen bond network among themselves and to a flavin. Upon light excitation of the flavin, this hydrogen bond network undergoes a poorly defined alteration that leads to a conformational change of the protein. Fast spectroscopic analysis of intermediate steps indicates that fast electron and subsequent proton transfer initially occurs from a conserved Tyr to the light-excited flavin (10–12). Beyond these steps, there are several models to explain a 10 nm spectral shift that subsequently occurs. In one model it is proposed that proton transfer results in release of a hydrogen bond between the oxygen in Tyr to ε2N of a neighboring Gln, allowing the Gln to rotate and form a new hydrogen bond with O4 of the flavin (10, 11). This model is supported by FTIR studies, which indicate that a change occurs in the hydrogen bond state of O4 upon light excitation (14, 15), by theoretical calculations (13), and by an NMR analysis (14). Several alternative models based on theoretical quantum mechanical analysis propose that the conservedGln63 undergoes formation of a light-induced tautomer, including or excludingGln63 rotation (15, 16), leading to a rearrangement of hydrogen bonds between the glutamine tautomer and flavin.
A controversial feature of the AppA BLUF domain is the position of Trp104. In the Anderson et al. (5) BLUF crystal structure, Trp104 is located near the flavin where the ε1N of Trp104 forms a hydrogen bond with ε1O of Gln63. A different AppA BLUF crystal structure solved with a BLUF domain of shorter length has Trp104 swung ~10 Å away from Gln63 to a position where it is partially solvent exposed. In this alternative conformation, a nearby methionine (Met106) occupies a position near that of Gln63 (7). Likewise, a crystal structure of the small BLUF protein PixD from Synechocystis shows that PixD forms a 10-subunit complex in which 9 subunits have the equivalent Trp (Trp91) swung away from the flavin and one subunit where Trp91 is close to the flavin (9). These results have led to a model in which Trp is hydrogen bonded to Gln in the dark state (called the “Trp-in” state) whereas in the light-induced state it is proposed that the Trp is swung away from the flavin (called the “Trp-out” state) (4, 9, 14, 17, 18). An alternative model based on theoretical energetic calculations by Domratcheva et al. (15), Obanayama et al. (19), and Sadeghian et al. (16) suggests that a Trp-out conformation represents the dark state and that light promotes Trp104 to exchange place withMet106 to form the Trp-in state. In these models, Trp104 in the Trp-in state forms transient hydrogen bonds that stabilize Gln63 and a nearby Asn45 residue while also possibly forming a hydrogen bond with 4C=O of the flavin (19). Obanayama et al. (19) suggest that movement of Trp104 from its “out” to “in” conformation is enough to strengthen or weaken H-bonds between flavin and surrounding amino acids to explain the spectroscopic results.
While it is clear from NMR studies that the C-terminal α-helix and a loop containing Trp104 and Met106 are dynamic (4, 20), there is as yet no direct experimental evidence that light excitation actually results in a significant movement of Trp104 from a deeply buried to a solvent-exposed state (or vice versa). Indeed, the NMR study of light versus dark solution BLUF structure by Grinstead et al. (4) shows minimal light-induced chemical shift changes of Trp104. Furthermore, Trp104 is not universally conserved in about 20% of known BLUF domains exhibiting a substitution at this position. Analysis of Trp movement is therefore needed to understand the nature of the changes in hydrogen bonding that occur upon light excitation of the flavin and to test the validity of models invoking movement of Trp104 and Met106. In this study, we utilize a combination of NMR and absorption spectroscopy, steady-state fluorescence, and acrylamide quenching of tryptophan fluorescence as tools to analyze the exposure of tryptophans in AppA. Our results indicate that there is minimal movement of Trp104 in solution, shedding doubt on previous models invoking a role of Trp104 as a key output signal.
METHODS
Mutant Construction and Protein Purification
Clones of different lengths of wild-type AppA (pTY-AppA126, pTY-AppA17– 133, and pTY-AppA) were kindly provided by Dr. Shinji Masuda, Japan. Briefly, the PCR fragments of 1–126 or 17–133 amino acids of AppA were cloned into NdeI and EcoRI sites of pTYB12 vector (New England Biolabs). The PCR fragment of full-length AppA was inserted into the same vector using sites NdeI and NotI. All of these constructs contain a chitin binding domain as an affinity tag, and thus all proteins were purified using chitin beads using the manufacturer’s protocol. The protein was removed from the chitin beads by inducing self-proteolysis by incubation overnight with 50 mM dithiothreitol. Eluted proteins were further purified, and DTT was removed by size-exclusion chromatography on Superose 12 using the AKTA FPLC system in 20 mM Tris-HCl, pH 8.0, and 100 mM NaCl.
The following mutations were constructed in AppA1–126 clone (in pTY-AppA126 plasmid):W64F, W104A, M106A, and a double mutant W104M-M106W. In full-length AppA, we constructed W104A, M106A, and W302F mutants. W64F mutant was also constructed in AppA17–133. The mutations were introduced using a QuikChange kit (Stratagene) with appropriate primers and their complements. Mutations were confirmed by sequencing.
Flavin Composition of BLUF Domains and FAD Reconstitution in Full-Length AppA
It has been shown previously that BLUF domains expressed in Escherichia coli have a heterogeneous flavin composition (21). In our expression system, the BLUF domains are composed of mostly FAD and FMN, with only ~5% content of riboflavin in fresh samples (5), and are flavin saturated with protein/flavin absorption ratios of 4.2–4.7. FMN replacing FAD in the BLUF domains has been shown to possess basically the same spectral characteristics, including the photocycle rates and fluorescence properties (21). Therefore, we used freshly isolated BLUF domain samples for fluorescence and quenching experiments. To ensure full 100%saturation of FAD in full-length AppA, we incubated cell extracts containing full-length AppA with 1 mM FAD for 30 min on ice prior to chitin bead purification.
Spectral Analysis
Absorption spectra were recorded on a Beckman DU-640 spectrophotometer using quartz cuvettes with 1 cm path. Fluorescence emission spectra between 310 and 570 nm were recorded on a PerkinElmer LS50B spectrofluorometer using diluted samples (absorbance <1 at 280 nm), with excitation wavelength at 295 nm.
Fluorescence Quenching
Fluorescence quenching experiments were performed according to the method described by Eftink et al. (22) using acrylamide as a quencher. This quencher can decrease fluorescence emission from Trp by collisional (or dynamic) quenching which involves diffusional collision of acrylamide with light-excited Trp (Trp*) that effectively quenches Trp* (23). A second quenching process is static quenching in which a “static” Trp*–acrylamide complex is present. Trp* quenching then occurs nearly instantaneously (23).
Quenching experiments were performed on a PerkinElmer LS50B spectrofluorometer with excitation at 295 nm and single point emission recorded at 360 nm, with temperature controlled at 15 °C. A sample volume of 2 mL was stirred at all times, and the additions of 8 M acrylamide were made in regular intervals up to 250 μL (0.88 M acrylamide). An inner-filter effect caused by acrylamide absorption at 295 nm and the dilution factor caused by adding acrylamide solution were corrected according to the method described by Coutinho et al. (24). Both flavin and tryptophan fluorescence spectra were recorded before and after the quenching experiment to monitor possible spectral change caused by acrylamide. For generating light-excited protein, a strong white light was applied to the sample for ~30 s, and the fluorescence signal was recorded immediately following irradiation.
For proteins with a homogeneous fluorophore, the fluorescence quenching by acrylamide can be described by the Stern–Volmer relationship (25):
| (1) |
in which F0 is the fluorescence intensity in the absence of quencher and F is the fluorescence intensity in the presence of various concentrations of quencher [Q]. Ksv and V are Stern–Volmer quenching constants used to describe dynamic and static quenching, respectively.
For protein that contains heterogeneous fluorophores (different fluorophores or the same fluorophore with different exposing conformations), the Stern–Volmer relationship can also be written as
| (2) |
where fi is the maximum fractional fluorescence for fluorophore i and Ksvi and Vi are corresponding collisional or static quenching constants (26). If only collisional quenching is considered, eq 3 can be written as
| (3) |
or
| (4) |
where KQeff=ΣKsvi is taken as the “effective” quenching constant and faeff = ΣfiKsvi/ΣKsvi is taken as “effective” fractional maximum accessible fluorescence of which 100% indicates a homogeneous fluorophore conformation (27).
NMR Chemical Shift Perturbation
All NMR spectra were recorded on a Varian Inova 500 MHz spectrometer equipped with an inverse probe at 35 °C. 1H–15N HSQC spectra were recorded for AppA variants with a 160 × 512 t1 and t2 data points, respectively. The backbone assignments for AppA1–126 were obtained from previously published spectra of AppA5–125 (BMRP accession number 692 (4)).
RESULTS
Structural Differences in AppA-BLUF Crystal and NMR Structures
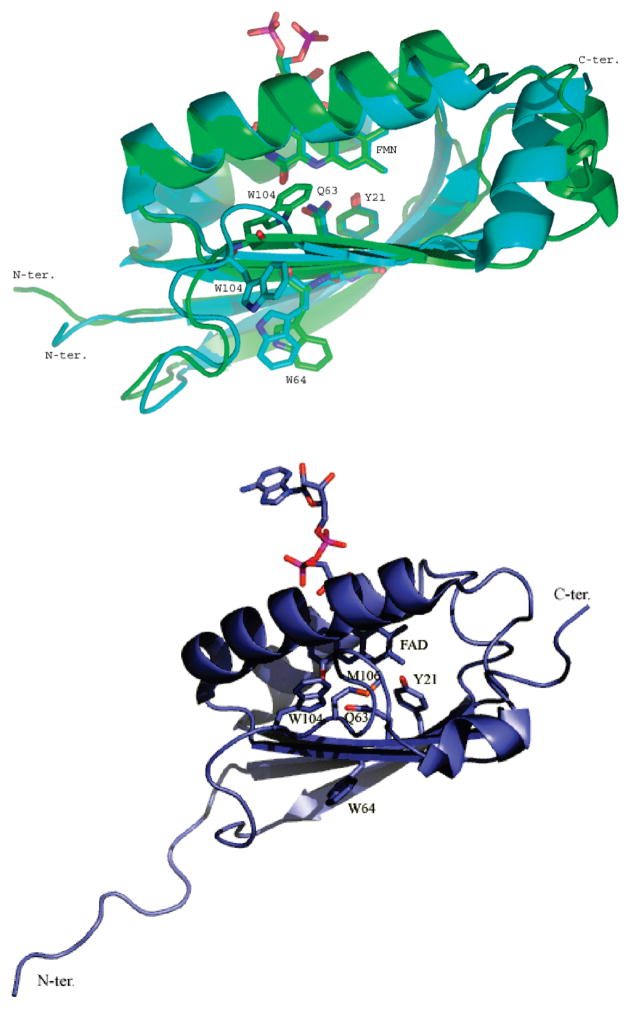
There are two published crystal structures of AppA BLUF domains that have slightly different N- and C-termini. The AppA BLUF structure (1YRX) by Anderson et al. (5) was obtained from an AppA BLUF domain comprised of amino acids 17 through 133. In this structure, Trp104 occupies a deeply buried position close to the flavin ring within hydrogen-bonding distance to Gln63 (Figure 1 top, green structure). The second AppA BLUF structure by Jung et al. (7) (2IYG) was obtained from an AppA C20S mutant domain that extended from amino acids 1 through 124. This second structure is quite similar to the Anderson et al. structure (Figure 1 top, blue structure) with the exception that Trp104 is located away from the flavin and solvent exposed while Met106 occupies a space nearGln63. The Jung et al. (7) structure also contains a molecule of DTT attached to C19. An NMR structure of AppA5–125 by Grinstead et al. (4) was modeled with Trp104 inside the flavin-binding pocket (Figure 1 bottom), which differs from the crystallographic structure of the AppA1–124 C20S mutant. Interestingly, the indole proton of Trp104 has weak NMR intensity, likely due to amide exchange broadening even in the dark state, indicating that this residue occupies various conformations in solution.
Figure 1.
Comparing the three structures of the AppA BLUF domain. Top:Crystallographic structures ofAppA17–133 (green) 1YRX(5) and AppA124 C20S (blue) 2IYG (7). Bottom: NMR structure of AppA5–125 2BUN (4). Side chains important for the photocycle and discussed in this work are labeled.
NMR Spectral Perturbations in BLUF Domains of Different Lengths
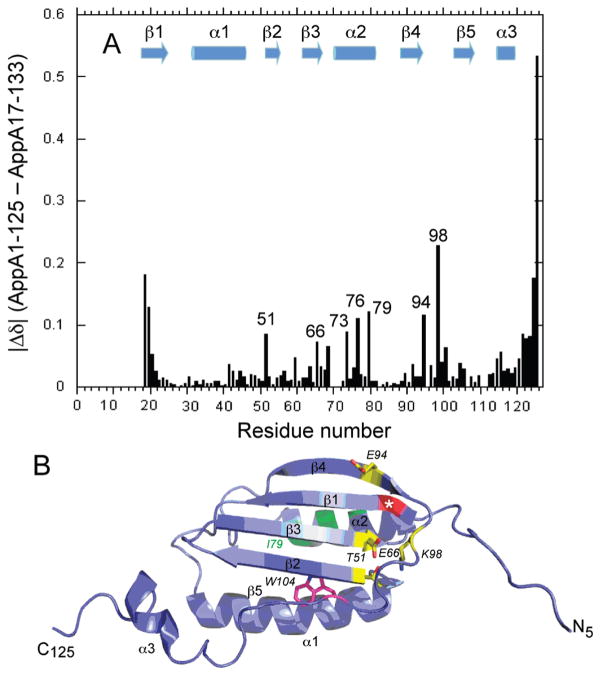
Two-dimensional 1H–15N HSQC spectra of 15N-labeled AppA1–126 and AppA17–133 were acquired in the dark in an effort to assess the degree to which these AppA constructs differed structurally from one another in solution. 1H–15N backbone cross-peak assignments were taken from previously published spectra obtained for AppA5–125 (4) acquired under similar solution conditions since these two 1H–15N HSQC spectra are virtually identical (spectra not shown). A direct comparison of 1H–15N HSQC spectra acquired for AppA1–126 and AppA17–133 readily revealed a number of differences in cross-peaks corresponding to residues common to the two constructs, i.e., residues 17–126, and outside of the extreme N- and C-terminal edges of this range (Figure 2A).
Figure 2.
(A) Chemical shift perturbation map of AppA1–126 vs AppA17–133 represents the combined 1H and 15N chemical shift, where Δδ (ppm)=(Δδ2H + (ΔδN/7)2)1/2 (31). (B) Representation of the NMR structure of AppA5–125 (4) with larger chemical shift differences between AppA1–126 andAppA17–133 highlighted. The asterisk indicates the position of V17 that would define the N-terminus inAppA17–133. Yellow highlights local perturbations that occur in the immediate vicinity of V17. In green are residues that exhibit chemical shift differences in the α2 helix.
The largest chemical shift differences involve residues E94 and K98 (Figure 2A) which define the C-terminus of the β4 strand and the immediately adjacent loop region N-terminal to Trp104 in what is considered a structurally dynamic β5 strand (residues 103–108) (Figure 2B) (4). This β5 region contains Trp104 and is the same region that shows differences in the two published AppA BLUF crystal structures. In addition, several single residues near this loop and K98, e.g., T51 and E66 at the N-terminus and C-terminus of the β2 and β3 strands, respectively, also exhibit detectable differences in the two spectra. All of these residues are in close proximity to V17 (the N-terminus of the AppA17–133, white asterisk, and first residue in the β1 strand) (Figure 2B) and thus can be readily explained as a local perturbation resulting from the N-terminal truncation in AppA17–133. Residues A73, M76, and the highly conserved I79 in the α2-helix inAppA1–126 vsAppA17–133 domains also reside in distinct environments, a finding not anticipated on the basis of the two crystallographic structures but potentially also explicable on the basis of the N-terminal truncation. Finally, although the short C-terminal α3 helix (residues 114–119) is found in distinct structural environments in the two crystal structures, the chemical shift differences observed in the two AppA constructs studied here are rather small (Figure 2A) and may be influenced by the distinct C-termini in the two molecules. It is interesting to note that this region (residues 109–125) is not as well ordered in the NMR solution structure of AppA5–125 and is not well packed against the core domain of AppA (4). Taken collectively, these spectra are consistent with the same overall fold in the core domains of AppA1–126 and AppA17–133 but reveal small differences in the local microenvironment of residues close to the site of N-terminal truncation in AppA17–133. We have not attempted to make assignments for side chain protons of Trp104 or Met106 side chains in the NMR spectra of either BLUF domain and therefore cannot make conclusions about their relative positions in solution from NMR.
Changes in Length of the AppA BLUF Domain Affect Spectral Characteristics
Table 1 shows the absorption maxima and photocycle lifetimes of several wild-type and mutant proteins that were constructed and isolated in this study. AppA1–126 and AppA17–133 have similar absorption maxima at 446 nm in the dark and a similar ~12 nm spectral shift to 458 nm when excited with blue light. Both of these values are slightly red shifted in comparison to full-length wild-type AppA that has a dark maximum at 443 nm and a light-excited maximum at 457 nm. Changes of ~20–30% are observed in the dark recovery rate for different proteins with τ1=690 s for AppA17–133, τ1= 948 s for AppA1–126, and τ1 = 795 s for full-length AppA.
Table 1.
Absorption Maxima and Photocycle Lifetimes of AppA and Its Various Clones and Mutants
| mutant | flavin absorbance max dark (light) ± 0.5 nm | photocycle τ (s) |
|---|---|---|
| AppA1–126 | 446 nm (458 nm) | 948.5 ±6.0 |
| AppA17–133 | 446 nm (458 nm) | 690.9 ±3.1 |
| AppA-full | 443 nm (457 nm) | 795.0 ±2.3 |
| AppA W302F | 445 nm (458 nm) | 786.2 ±5.1 |
| AppA1–126 W64F | 446 nm (458 nm) | 776.3 ±5.4 |
| AppA17–133 W64F | 446 nm (458 nm) | 477.3 ±2.0 |
| AppA1–126 W104A | 443 nm (450 nm) | 6.0 ±0.16 (58%), 18.0 ±0.7 (42%) |
| AppA-full W104A | 443 nm (443 nm) | nda |
| AppA1–126 M106A | 447 nm (458 nm) | 643.2 ±6.0 |
| AppA-full M106A | 444.5 nm (458 nm) | 473.9 ±4.6 |
| W104M-M106W in AppA1–126 | 445 nm (nd)a | 11.6 ±0.1 (20%), 50.8 ±0.1 (80%) |
nd = not determined.
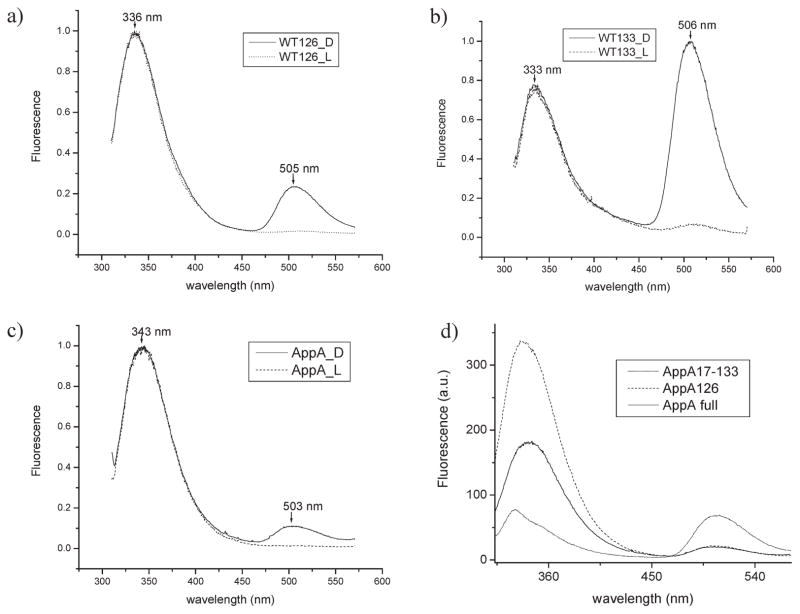
The Trp emission spectrum (λex = 295 nm) of AppA1–126 shows an emission maximum of 336 nm (Figure 3a) that is slightly red shifted relative to AppA17–133 that has a maximum at 333 nm with a slight shoulder around 355 nm (Figure 3b, d). The observation that Trp emission of AppA1–126 is slightly red shifted over that of AppA17–133 is consistent with the crystal structures in which Trp104 is partially solvent exposed in AppA1–126 but not in AppA17–133 where it is buried near the flavin. The emission maximum of full-length AppA (Figure 3c) is further red shifted by 10 nm to 343 nm nm relative to the BLUF domains AppA17–133 and AppA1–126.
Figure 3.
Normalized fluorescence emission spectra of AppA and its various clones: (a) AppA126, (b) AppA17–133, and (c) full-length AppA. D represents dark-adapted protein (solid lines), and L represents light-excited protein (dashed lines). (d) Nonnormalized fluorescence spectra of dark-adapted full-length AppA, AppA133wt, and AppA126wt using protein of the same concentration (A280= 0.2).
Excitation at 295 nm also leads to emission of the flavin between 503 and 506 nm. Specifically, excitation of dark-adapted AppA1–126 at 295 nm leads to high Trp emission at 336 nm and measurable, but low flavin emission at 505 nm (Figure 3a, solid spectrum). When AppA1–126 is first photoconverted into a signaling (lit) state prior to excitation at 295 nm, the flavin emission peak becomes nearly fully quenched (Figure 3a, dotted spectrum). This is contrasted by AppA17–133 where dark-adapted protein exhibits a flavin emission peak that is dramatically enhanced over that of light-excited protein and actually exceeds the level of the Trp emission peak (Figure 3b, solid spectrum). Upon light illumination, the fluorescence of flavin is also strongly quenched in this BLUF domain construct (Figure 3b, dotted spectrum). We also compared the Trp and flavin emission intensities of these BLUF domains with comparable amounts of protein. Figure 3d shows that AppA17–133 has a Trp emission peak that is blue shifted and significantly reduced and a flavin peak that is significantly elevated, relative to that of AppA1–126. Interestingly, the flavin emission pattern of full-length wild-type AppA (Figure 3d, solid line) closely resembles that of the shorter AppA1–126 in respect to low intensity of flavin emission relative to Trp emission in the dark state as well as by quenching of flavin emission when exposed to white light prior to excitation (Figure 3c, dotted spectrum). The total Trp emission is also lower in full-length AppA compared to AppA1–126, indicating that the tryptophan fluorescence is more quenched in the full-length protein than in AppA1–126, probably due to the higher exposure of tryptophan residues to solvent in full-length protein that is monomeric (see below).
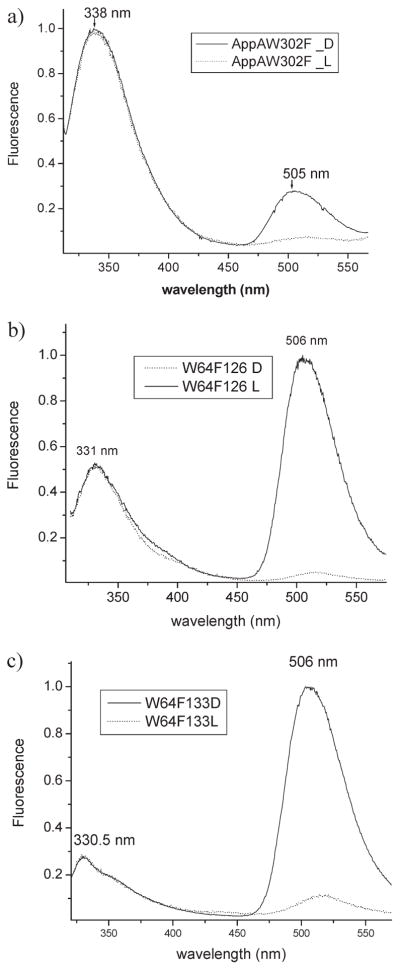
Full-length AppA actually contains three tryptophans with two located in the BLUF domain at positions 64 and 104 and one at the C-terminus at position 302. We therefore assessed whether the red-shifted Trp emission spectrum in full-length AppA is a result of the third tryptophan by constructing a Trp to Phe mutation at this position (AppAW302F). The full-length AppA W302F mutant thus has the same number of Trp (W64 and W104) as is present in AppA1–126 and AppA17–133. Spectral analysis of full-length AppA W302F shows that it has an absorption spectrum more like that of the shorter BLUF proteins (Table 1) and a Trp emission maximum at 338 nm that is close to AppA1–126 tryptophan emission of 336 nm (Figures 4a and 3a, respectively). This suggests that Trp302 in full-length AppA is likely solvent exposed. As is the case of AppA1–126, excitation of dark-adapted full-lengthAppAW302F at 295 nm leads to high Trp emission at 338 nm and measurable, but low flavin emission at 505 nm (Figure 4a, solid spectrum). When full-length AppA W302F is photoconverted into a light-excited state prior to excitation at 295 nm, the flavin emission peak becomes nearly fully quenched as is the case with AppA1–126 (Figure 4a, dotted spectra).
Figure 4.
Normalized fluorescence emission spectra of AppA mutants: (a) W302A, (b) W64F 1–126, and (c) W64F 17–133. D represents dark-adapted protein (solid lines), and L represents light-excited protein (dashed lines).
The BLUF crystal structures show that Trp104 is very near the flavin in AppA17–133 but not in AppA1–126 so it is possible that the reduced Trp fluorescence and increased flavin fluorescence in AppA17–133 may be a result of excitation energy transfer from Trp104 to the flavin. To test this hypothesis, we constructed W64F mutations in both AppA1–126 and AppA17–133 so that Trp104 is the only tryptophan present in these BLUF domains. The W64F mutation does not influence absorption maxima (Table 1); however, both AppA1–126 and AppA17–133 W64F mutant proteins have Trp emission slightly blue shifted, indicating that Trp104 is in more hydrophobic environment than Trp64 (Figure 4b, c). There is also an interesting ~20–30% increase in the rate of the photocycle for both the AppA1–126 and AppA17–133 W64F mutants (Table 1). Importantly, the Trp fluorescence emission of dark state AppA17–133 W64F is lower and remains slightly blue shifted over that of dark state AppA1–126 W64F (panels c and b of Figure 4, respectively). The AppA17–133 W64F emission spectrum retains a shoulder around 355 nm meaning that Trp104 in AppA17–133 is likely in heterogeneous environment. Collectively, these results suggest that Trp104 is closer to the flavin in the dark state in AppA17–133 than in dark state AppA1–126.
Probing Exposure of Trp104 by Acrylamide Quenching
Acrylamide quenching of Trp fluorescence has previously been used to reveal the extent of solvent exposure of tryptophans in proteins (25, 27, 28). A quenching constant Ksv obtained from quenching plots that is close to 0 indicates complete Trp burial whereas a Ksv above 6 indicates complete solvent exposure. The plot curves upward in proteins that contain two or more fluorophores with similar Ksv constants or when static quenching is present. If the two Ksv constants differ by a factor of ~2, the plot appears linear, while the plot curves downward when the difference in Ksv is larger than 2 (29).
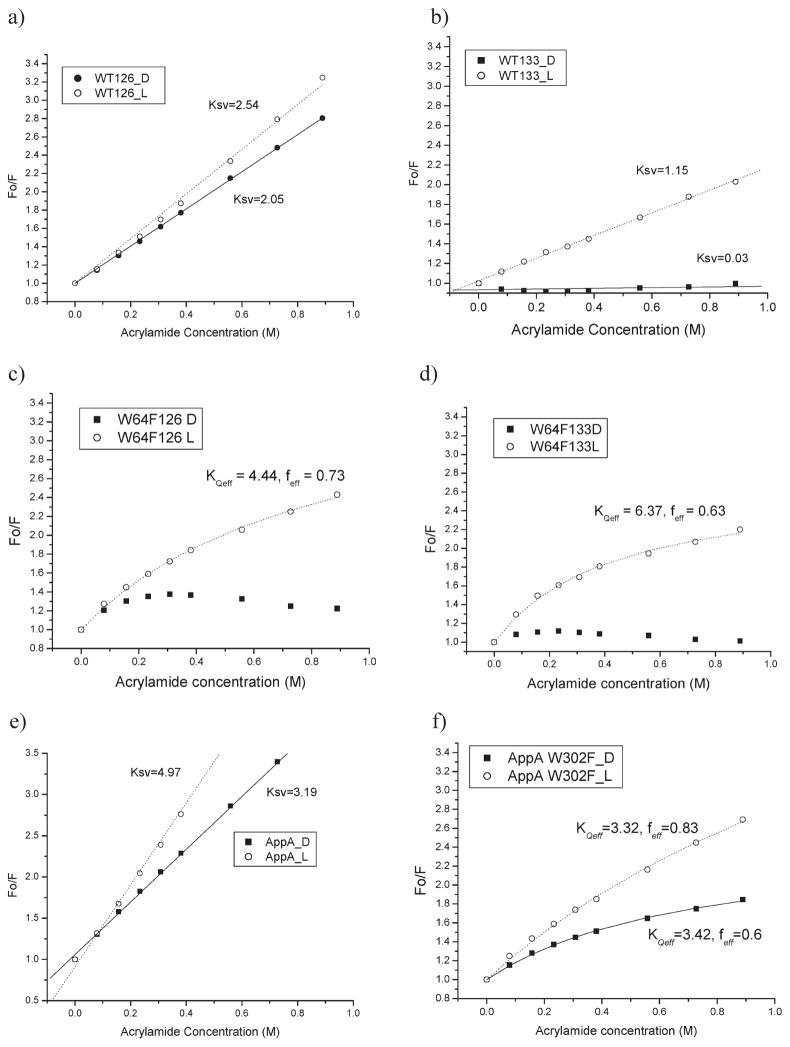
AppA1–126 and AppA17–133 both exhibit linear acrylamide quenching curves indicating that the two tryptophans (W64 and W104) present in this BLUF domain differ in a degree of exposure by a factor of ~2 (panels a and b of Figure 5, respectively). A comparison of Ksv values of dark- and light-adapted AppA1–126 reveals that tryptophan residues are partially solvent exposed in both the dark- and light-adapted samples with Ksv values of 2.05 ± 0.01 M−1 and 2.54 ± 0.04 M−1, respectively. Similar analysis of AppA17–133 is significantly different with a Ksv = 0.03 ± 0.01 M−1 under dark conditions and Ksv = 1.15 ± 0.02 M−1 under light-excited conditions (Figure 5b). This indicates that AppA17–133 has both Trp residues deeply buried and inaccessible to acrylamide under dark conditions, which is in agreement with the crystal structure, and that light excitation allows at least one of the Trp to become more solvent exposed upon illumination. However, the light-excited Ksv value of 1.15 for AppA17–133 remains significantly lower than that of Ksv 2.05 for AppA1–126 in the dark, indicating that the Trp in AppA17–133 never reaches a degree of exposure as observed with AppA1–126. Together with the different crystallography structures, these results indicate that AppA1–126 and AppA17–133 have different conformations in the dark with Trp64/Trp104 partially exposed in AppA1–126 and buried in AppA17–133. Upon light illumination Trp residues become only slightly more solvent exposed in AppA1–126 while in AppA17–133 there is movement from a deeply buried state to a partially solvent exposed state that never reaches the level of solvent exposure observed with AppA1–126.
Figure 5.
Quenching of tryptophan fluorescence with acrylamide: (a) AppA1–126, (b) AppA17–133, (c) AppA126W64F, (d) AppA17–133 W64F, (e) full-length AppA WT, and (f) AppA W302F. Solid lines, dark state; dashed lines, light excited state.
Analysis of acrylamide quenching of the AppA BLUF domains that contain a W64F mutation allows measurement of solvent exposure of only Trp104. The acrylamide quenching curves indicate that Trp104 is mostly buried in dark-adapted AppA17–133 W64F (Figure 5d) and slightly exposed in AppA1–126 W64F (Figure 5c). Interestingly for both proteins, the dark-adapted curve cannot be fitted using either a linear or heterogeneous quenching model reflecting the conclusions of Grinstead et al. (4) that the positioning of Trp104 is quite dynamic in solution. Very reproducibly, we obtained higher exposure of Trp104 to acrylamide when the protein is light excited in both AppA17–133 W64F and AppA1–126 W64F. The light-excited quenching curve is also not linear, indicating that Trp104 is present in various conformations and the light state occupancy never reaches 100%. We also cannot exclude that a mutation of the nonconserved Trp64 residue does not somewhat destabilize the AppA BLUF domain as the photocycle rate increases by 20–30% in this mutant. The light-excited quenching curves have been fitted with the heterogeneous model (eq 5) and provided values of effective (or “observed”) quenching constant: KQeff = 4.44 ± 0.21 M−1, feff = 0.73 ± 0.01 for AppA1–126W64F and KQeff =6.37 ± 0.42 M−1, feff=0.63 ± 0.01 for AppA17–133 W64F. Both results indicate a partial movement of Trp104/uncovering of Trp104 residue upon light excitation in both mutant BLUF domains. Finally, we also constructed a W104A mutant and examined the accessibility of W64 alone (see below). W64 does not get more exposed upon light excitation but becomes slightly more hidden (Figure 6a).
Figure 6.
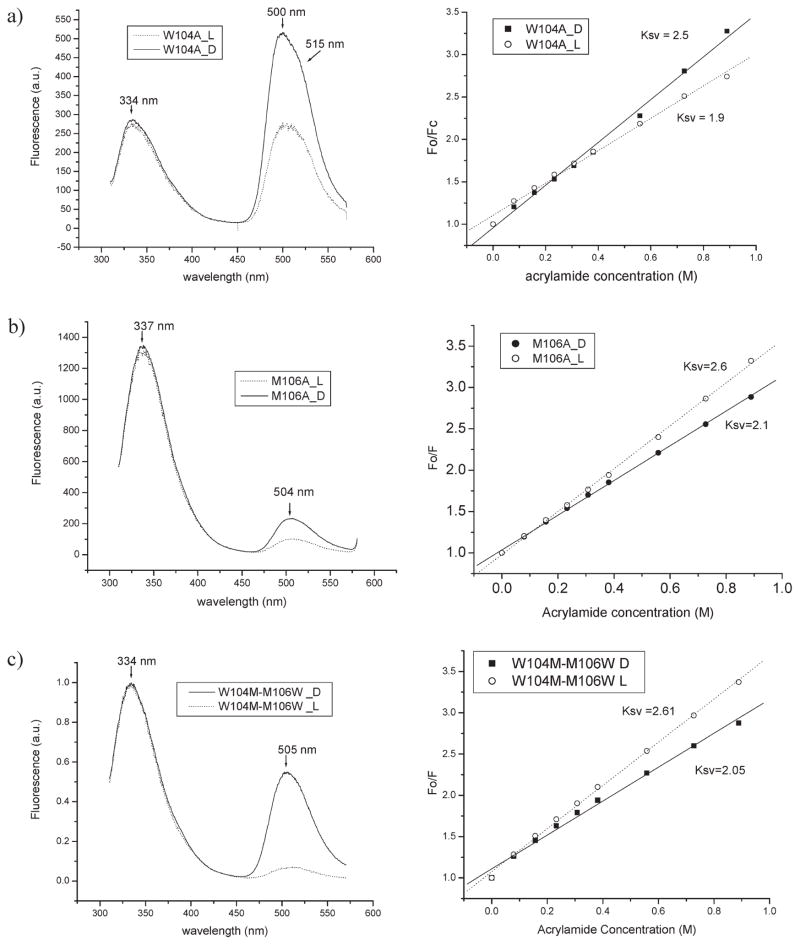
Fluorescence emission spectra and acrylamide quenching of W104A(a), M106A (b), andW104M-M106W(c) mutants in AppA126 clone.
To address which BLUF domain structure best approximates the BLUF domain structure in full-length AppA, we measured Trp exposure in a full-length AppA W302F mutant. Interestingly, the acrylamide quenching curve from full-length wild-type AppA is linear (Figure 5e) while that of the full-length W302F mutant is not linear and instead curves downward (Figure 5f). The downward slopping curve could be best fitted using the heterogeneous fluorophore model (eq 5) providing only “effective” quenching constant and suggesting that the exposure of the Trp residues to acrylamide in the BLUF domain is heterogeneous in the W302 mutant of full-length AppA. We interpret this as evidence that the output domain may be influencing the accessibility of Trps in the BLUF domain. The two Trps in AppA W302F are certainly not buried in the dark as they have KQeff= 3.42 ± 0.11 M−1, feff = 0.6 ± 0.01. At light, overall higher exposure of Trps to acrylamide quencher is still seen with KQeff= 3.32 ± 0.16 M−1 and feff = 0.82 ± 0.01 (bigger fraction of Trps becomes exposed). These results are more consistent with the results of acrylamide quenching of AppA1–126 rather than AppA17–133, indicating that the Jung et al. (7) structure better approximates the BLUF structure in full-length AppA.
Met106 and Trp104 Mutants Exhibit Different Spectral Characteristics
As discussed previously, AppA1–126 and AppA17–133 not only have different positions of Trp104 but also have different positioning of Met106. In AppA1–126, Trp104 is swung away from the flavin and partially solvent exposed while Met106 is deeply buried near the flavin. Conversely, the crystal structure of AppA17–133 has Trp104 buried near the flavin whileMet106 is solvent exposed. To further probe this dichotomy, we constructed single Ala substitution mutations of Trp104 (W104A) and Met106 (M106A). Since AppA1–126 represents the more native conformation of full-length AppA (based on the above spectroscopy data), these mutations were constructed in both AppA1–126 and full-length AppA. The first observation is that mutations ofW104 andM106 inAppA1–126 do not affect the level of expression or solubility; however, the same mutations in the full-length AppA severely decreased both yield and solubility. Spectroscopic analysis of W104A mutation in AppA1–126 yielded a biphasic photocycle that is ~50–100 times faster than wild-type AppA1–126with the light-excited red shift only extending to 450 nm(Table 1). TheW104Amutant also exhibits an overall decrease in Trp fluorescence as would be expected with the loss of one of the two Trp present in the BLUF domain with the flavin emission maximum blue shifted to 500 nm with a shoulder at ~515 nm (Figure 6a). An acrylamide quenching curve of Trp fluorescence with AppA1–126 W104A shows that the remaining Trp (W64) has Ksv=2.5 in the dark-adapted protein and actually becomes slightly more hidden in the light-adapted protein with Ksv =1.9 (Figure 6a). Interestingly, the same W104A mutation in full-length AppA significantly reduces expression and solubility impeding similar spectral analyses.
A M106A mutation in AppA1–126 and in full-length AppA modestly speeds up the photocycle about 1.5–2-fold and only slightly affects the flavin absorption maximum (Table 1). M106A mutant inAppA1–126 exhibits a fluorescence emission spectrum similar to wild-type AppA1–126 with a small red shift to 337 nm of the tryptophan fluorescence (Figure 6b). Acrylamide quenching of this mutant protein is also similar to the wild-type AppA1–126 quenching curve. As is the case of the full-length W104A mutation, theM106A mutation is also poorly expressed and has low solubility impeding spectral analyses.
We also constructed a double mutant where Trp104 and Met106 are swapped (W104M-M106W) in AppA1–126 (Figure 6c). Interestingly, the absorption maximum of the flavin (445 nm) of the double mutant is similar to that of wild-type AppA1–126 (an absorption maximum of 446 nm). The double mutant actually exhibits a biphasic decay from light-excited to dark state. Approximately 20% of the protein decays fast with τ1 = 11 s that is close to the AppA1–126 W104A mutant. The second decay phase (~80% of the sample) is much longer with τ2 = 50 s. This second phase is still >10-fold faster than wild-type AppA1–126 that has τ = 948 s.
We also accessed the location of the swapped Trp by analyzing the fluorescence emission profile as well as an acrylamide quenching curve. We would expect Trp at position 106 to be close to the flavin and possibly transfer some of its energy to the flavin, thus increasing the flavin emission as in AppA17–133wt. However, the emission spectrum of the double mutant has only a slight increase in flavin emission and an emission maximum that is identical to wild-type AppA1–126 (Table 1, Figure 6c). This indicates that Trp at position 106 is likely not close enough to the flavin to promote excitation transfer as is occurs with wild-type AppA17–133. The acrylamide quenching profile of the double mutant is virtually the same as wild-type AppA1–126 under light and dark conditions, leading us to a conclusion that Trp at location 106 is partially solvent exposed under both dark and light conditions. This result indicates that there is enough flexibility in the fifth β-strand that swapping the two residues still allows the protein to obtain nearly native conformation and function with the exception of the speed of the photocycle which is biphasic and faster.
DISCUSSION
Previous models proposed that light excitation of the flavin induces movement of Trp104. One model proposes that light absorption causes movement of Trp104 from a buried position near the flavin to a more solvent exposed position, and the other model proposes that Trp104 moves in the opposite direction, from a solvent-exposed position to a buried position. These models were based on several different crystal structures that had Trp104 in AppA, or its equivalent in other BLUF domains, located at different positions (5, 7, 9). Even though movement of Trp104 has been adopted by many groups, the dark- and light-state positions, and actual light-induced movement of this residue, have never been directly addressed.
Using NMR spectroscopy, we addressed whether AppA1– 126 and AppA17–133 domains are different in solution with buffers devoid of detergent and DTT (DTT was covalently attached to Cys19 in the Jung et al. structure (7)). The 2D NMR spectra showed small but distinct differences. Since several residues neighboring the truncated N-terminus in AppA17–133 are affected in the 1H–15N HSQC spectra (Figure 2b), we conclude that the N-terminal truncation in AppA17–133 may be responsible for some of the differences observed in the crystallographic structures. Chemical shift changes were observed at the end of the β4 strand and in the highly dynamic flexible loop between β4 and β5 that contains Trp104, suggesting that this flexible loop may occupy slightly different positions as observed in the crystal structures.
Since the side chain protons of Trp104 and Met106 were not assigned in the NMR spectra, we used a combination of fluorescence emission spectroscopy and acrylamide quenching analyses to probe the position of Trp104 in AppA1–126 vs AppA17–133 in solution in the dark as well as when light excited. The acrylamide quenching data indicate that there is only partial change in exposure of Trp104 to acrylamide upon light excitation of the flavin in the AppA BLUF domain. While we cannot rule out that there may be other interpretations of these results, such as opening and/or closing light-induced solvent-exposed channels, the most straightforward interpretation is that AppA1–126 and AppA17–133 indeed have different fixed conformations of Trp104 in solution. Specifically, AppA17–133 BLUF domain appears to have Trp104 buried in the dark state and slightly more solvent exposed in the light state. This is contrasted by AppA1–126 where Trp104 appears partially solvent exposed in both dark and light-excited states. Thus the crystallization structures of AppA17–133 and AppA124 C20S that have Trp104 in different positions also appear to represent similar different positions of Trp104 in AppA17–133 and AppA1–126 in solution. There is, however, no evidence that AppA1–126 and full-length AppA undergo light-induced movement of Trp104 from a deeply buried state to a solvent-exposed state as proposed to occur in the Trp-in to Trp-out model. The involvement, if any, of Trp104 as an output signal remains in question.
Fluorescence emission and acrylamide quenching data also demonstrated that AppA1–126 more closely resembles the BLUF domain present in full-length wild-type AppA and is therefore the more biologically relevant structure. AppA17–133 with its Trp-in conformation in the dark must therefore represent a different fold likely due to a different energy minimum of this N-terminally truncated protein. Collectively, our results do not support movement of Trp104 from a deeply buried state to a solvent-exposed state (or vice versa) in full-length AppA, suggesting that the above models are likely incorrect. Our results do indicate that Trp104 is dynamic and capable of occupying several different positions in the dark and light-induced states, which supports previous NMR data (4, 20).
Our spectral analyses demonstrate that AppA1–126 and AppA17–133 have similar absorption maxima, similar light-induced spectral shifts, and only slight changes in the rate of photocycle. Thus, the initial position of Trp104 in the dark state has no significant influence on the ability of the BLUF domain to undergo a photocycle, indicating that the H-bonding pattern includingGln63-Trp104 or Gln63-Met106 is not essential. This is further confirmed byM106Amutation that behaves in all aspects as the wild-type protein. Furthermore, the fact that light excitation of AppA1–126 does not promote movement of Trp104 to a deeply buried “Trp-in” position clearly refutes previous models invoking a light-induced exchange of Trp104 with Met106. Despite this conclusion, Trp104 does have an important role in controlling the rate of the photocycle in AppA given thatW104A mutation in AppA1–126 increases the speed of the photocycle 50–100-fold. We propose that the bulky and perhaps aromatic character ofW104 stabilizes light-induced structural changes that occur to the fifth β-strand and to the flexible loop between β4 and β5. In support of this conclusion is a recent report revealing that substitution of Trp104 in AppA5–125 to a more conservative phenylalanine only enhances the speed of the photocycle 4- fold (30). Thus, introduction of Ala substitution into the flexible loop appears to uncouple the flavin photocycle from a stable output signaling state. This result also suggests that Trp104 and Met106 may have roles in “tethering” this loop in the dark into a conformation that allows AppA to interact with PpsR and provides further evidence that the loop between the fourth and fifth β-strand and the β5 strand itself are involved in the generation of an output signal. This is consistent with hydrogen– deuterium exchange measurement on AppA5–125, which showed that amide protons of the β5 edge strand are poorly protonated from exchange with solvent in the dark state (4). Clearly, many questions remain as to what stabilizes this region of the molecule in the dark state and how light absorption by the flavin causes destabilization of this region.
Acknowledgments
Our thanks go to Dr. Shinji Masuda for construction of the clones used in this report.
Footnotes
Abbreviations: BLUF, blue-light sensing using FAD; DTT, dithiothreitol.
This study was supported by National Institutes of Health Grants GM40941 to C.E.B. and GM042569 to D.P.G.
References
- 1.Gomelsky M, Kaplan S. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavo-protein. Identification of a novel fad binding domain. J Biol Chem. 1998;273:35319–35325. doi: 10.1074/jbc.273.52.35319. [DOI] [PubMed] [Google Scholar]
- 2.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 3.Okajima K, Yoshihara S, Fukushima Y, Geng X, Katayama M, Higashi S, Watanabe M, Sato S, Tabata S, Shibata Y, Itoh S, Ikeuchi M. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J Biochem (Tokyo) 2005;137:741–750. doi: 10.1093/jb/mvi089. [DOI] [PubMed] [Google Scholar]
- 4.Grinstead JS, Hsu ST, Laan W, Bonvin AM, Hellingwerf KJ, Boelens R, Kaptein R. The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling. Chem BioChem. 2006;7:187–193. doi: 10.1002/cbic.200500270. [DOI] [PubMed] [Google Scholar]
- 5.Anderson S, Dragnea V, Masuda S, Ybe J, Moffat K, Bauer C. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44:7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung A, Domratcheva T, Tarutina M, Wu Q, Ko WH, Shoeman RL, Gomelsky M, Gardner KH, Schlichting I. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc Natl Acad Sci USA. 2005;102:12350–12355. doi: 10.1073/pnas.0500722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung A, Reinstein J, Domratcheva T, Shoeman RL, Schlichting I. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J Mol Biol. 2006;362:717–732. doi: 10.1016/j.jmb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005;349:1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 9.Yuan H, Anderson S, Masuda S, Dragnea V, Moffat K, Bauer C. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 2006;45:12687–12694. doi: 10.1021/bi061435n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragnea V, Waegele M, Balascuta S, Bauer C, Dragnea B. Time-resolved spectroscopic studies of the AppA blue-light receptor BLUF domain from Rhodobacter sphaeroides. Biochemistry. 2005;44:15978–15985. doi: 10.1021/bi050839x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauden M, van Stokkum IH, Key JM, Luhrs D, van Grondelle R, Hegemann P, Kennis JT. Hydrogen- bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc Natl Acad Sci USA. 2006;103:10895– 10900. doi: 10.1073/pnas.0600720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zirak P, Penzkofer A, Schiereis T, Hegemann P, Jung A, Schlichting I. Absorption and fluorescence spectroscopic characterization of BLUF domain of AppA from Rhodobacter sphaeroides. Chem Phys. 2005;315:142–154. [Google Scholar]
- 13.Unno M, Masuda S, Ono TA, Yamauchi S. Orientation of a key glutamine residue in the BLUF domain from AppA revealed by mutagenesis, spectroscopy, and quantum chemical calculations. J Am Chem Soc. 2006;128:5638–5639. doi: 10.1021/ja060633z. [DOI] [PubMed] [Google Scholar]
- 14.Grinstead JS, Avila-Perez M, Hellingwerf KJ, Boelens R, Kaptein R. Light-induced flipping of a conserved glutamine sidechain and its orientation in the AppA BLUF domain. J Am Chem Soc. 2006;128:15066–15067. doi: 10.1021/ja0660103. [DOI] [PubMed] [Google Scholar]
- 15.Domratcheva T, Grigorenko BL, Schlichting I, Nemukhin AV. Molecular models predict light-induced glutamine tautomerization in BLUF photoreceptors. Biophys J. 2008;94:3872–3879. doi: 10.1529/biophysj.107.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadeghian K, Bocola M, Schutz M. A conclusive mechanism of the photoinduced reaction cascade in blue light using flavin photoreceptors. J Am Chem Soc. 2008;130:12501– 12513. doi: 10.1021/ja803726a. [DOI] [PubMed] [Google Scholar]
- 17.Masuda S, Hasegawa K, Ono TA. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44:1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- 18.Masuda S, Hasegawa K, Ono TA. Tryptophan at position 104 is involved in transforming light signal into changes of beta-sheet structure for the signaling state in the BLUF domain of AppA. Plant Cell Physiol. 2005;46:1894–1901. doi: 10.1093/pcp/pci208. [DOI] [PubMed] [Google Scholar]
- 19.Obanayama K, Kobayashi H, Fukushima K, Sakurai M. Structures of the chromophore binding sites in BLUF domains as studied by molecular dynamics and quantum chemical calculations. Photochem Photobiol. 2008;84:1003–1010. doi: 10.1111/j.1751-1097.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu Q, Ko WH, Gardner KH. Structural requirements for key residues and auxiliary portions of a BLUF domain. Biochemistry. 2008;47:10271–10280. doi: 10.1021/bi8011687. [DOI] [PubMed] [Google Scholar]
- 21.Laan W, Bednarz T, Heberle J, Hellingwerf KJ. Chromophore composition of a heterologously expressed BLUF-domain. Photochem Photobiol Sci. 2004;3:1011–1016. doi: 10.1039/b410923f. [DOI] [PubMed] [Google Scholar]
- 22.Ghlron MREaCA. Fluorescence Quenching of Indole and Model Micelle Systems. J Phys Chem. 1976;80:486–493. [Google Scholar]
- 23.Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 24.Prieto ACaM. Ribonuclease T1 and alchohol dehydrogenase fluorescence quenchign by acrylamide. J Chem Educ. 1993;70:425. [Google Scholar]
- 25.Stern O, Volmer M. On the quenching time of fluorescence. Phys Z. 1919;20:183–188. [Google Scholar]
- 26.Eftink MR, Selvidge LA. Fluorescence quenching of liver alcohol dehydrogenase by acrylamide. Biochemistry. 1982;21:117–125. doi: 10.1021/bi00530a021. [DOI] [PubMed] [Google Scholar]
- 27.Lehrer SS. Solute perturbation of protein fluorescence—Quenching of tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 1971;10:3254. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- 28.Eftink MR, Selvidge LA. Fluorescence quenching of liver alcohol-dehydrogenase by acrylamide. Biochemistry. 1982;21:117–125. doi: 10.1021/bi00530a021. [DOI] [PubMed] [Google Scholar]
- 29.Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins—Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 30.Laan W, Gauden M, Yeremenko S, van Grondelle R, Kennis JT, Hellingwerf KJ. On the mechanism of activation of the BLUF domain of AppA. Biochemistry. 2006;45:51–60. doi: 10.1021/bi051367p. [DOI] [PubMed] [Google Scholar]
- 31.Arunkumar AI, Klimovich V, Jiang X, Ott RD, Mizoue L, Fanning E, Chazin WJ. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat Struct Mol Biol. 2005;12:332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]