Abstract
In multiple myeloma (MM), malignant plasma cells produce large amounts of antibodies and have highly active protein translational machinery. It is not known whether regulation of the abundance and aminoacylation (charging) of transfer RNA (tRNA) takes place in myeloma cells to accommodate for the increased amount of protein translation. Using tRNA-specific microarrays, we demonstrate that tRNA levels are significantly elevated in MM cell lines compared to normal bone marrow cells. We furthermore show that the addition of the proteasome inhibitor, Bortezomib (Velcade™, PS-341) results in decreased charging levels of tRNAs, in particular those coding for hydrophobic amino acids. These results suggest that tRNA properties are altered in MM to accommodate for its increased need for protein translation, and that proteasome inhibition directly impacts protein synthesis in MM through effects on tRNA charging.
Keywords: Myeloma, tRNA, Bortezomib
Introduction
tRNAs are molecular carriers of amino acids and translate genes into proteins according to the genetic code. Cancer cells are often associated with over-expression of TFIII factor components which can lead to over-expression of tRNA [1]. However, how over-expression of tRNA plays a role in cancer development and maintenance is not well understood. Recent work showed that a less than 2-fold increase in the abundance of tRNAMet used in translational initiation was sufficient to transform a non-cancerous cell, whereas a similar increase in tRNAMet used in translational elongation had little effect [2]. This remarkable discovery suggests a delicate tRNA balance exists in normal and transformed cells. A tipped balance in tRNA abundance may be characteristic for cancer. In normal human tissues, however, tRNA abundance can vary by up to 4-fold [3]. It is therefore not yet possible to predict how this balance is perturbed in any particular type of cancer or what effect perturbation will have.
The malignant plasma cells in multiple myeloma (MM) are characterized by a high level of translation of monoclonal antibodies [4; 5; 6]. To accommodate the higher rate of protein synthesis, myeloma cells might have a higher abundance of tRNA molecules compared to normal cells. Myeloma cells are very sensitive to inhibition of the 26S proteasome complex by bortezomib which results in the accumulation of misfolded proteins and leads to apoptosis [7]. Proteasome inhibition also perturbs amino acid recycling [8; 9] which is crucial to maintain high levels of translation. Only tRNAs aminoacylated (charged) with a 3' amino acid are used in translation. We hypothesize therefore that tRNA charging levels are altered when myeloma cells are treated with bortezomib. In this study, we used recently developed tRNA-specific microarrays [3; 10] to test these hypotheses on the abundance and charging levels of tRNA in myeloma cells (Fig. 1).
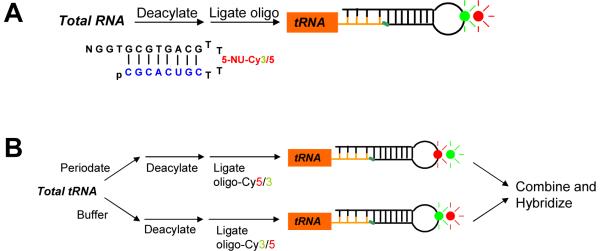
Fig. 1. Schemes for measuring tRNA abundance (A) and aminoacylation (charging, B).
(A) Total RNA was deacylated to remove 3' amino acids. An RNA-DNA hybrid oligo containing Cy3 or Cy5 was ligated to the 3' end of tRNA using T4 DNA ligase. Fluorescent-labeled tRNAs were directly hybridized to microarrays containing tRNA-specific probes. (B) Total charged tRNA sample was split in two halves. One half was treated with periodate which oxidized only uncharged tRNAs and represents the amount of charged tRNA; the other half was not treated with periodate but otherwise treated identically and represented the amount of total tRNA. After removal of periodate, tRNAs were deacylated and labeled with the Cy3/Cy5 containing RNA-DNA hybrid oligos.
Fluorescent labeling and array hybridization of tRNA was applied previously to demonstrate the existence of tissue-specific expression of tRNA species in humans, implicating a role for tRNA heterogeneity in regulating translation [3]. To measure relative tRNA charging levels, we used selective oxidation of uncharged tRNA followed by fluorescent labeling of charged tRNA. A similar method was applied previously to demonstrate differential tRNA charging upon amino acid starvation in E. coli [10]. Here, we adopted this method for the first time in human cells. We demonstrate that tRNA abundance is increased in myeloma cells and that the proteasome inhibitor bortezomib selectively down-regulates charging of tRNAs coding for hydrophobic amino acids.
Materials and Methods
Myeloma cell lines MM.1S, MM.1R, NCI-H929, U266 and RPMI-8266 are maintained in RPMI-1640 containing 10% FBS. Cells were grown in fresh medium overnight prior to the addition of 10–20nM bortezomib. Cells were harvested for RNA isolation using TRIzol reagent (Invitrogen, CA). Apoptosis of bortezomide-treated cells were monitored by TMRM apoptosis asaay [11]. Total RNA isolated from bone marrows of healthy donors was purchased from BioChain Insititute, Inc (Hayward, CA).
To isolate RNA for tRNA charging measurements, cells were resuspended in 500μl 0.3M sodium acetate/acetic acid, 10mM EDTA (pH 4.8), and homogenized by adding 0.6g 0.5mm diameter glass beads, 500μl acetate (pH 4.8)-saturated phenol-chloroform, 50μl 10% SDS, and vortexed at 4°C for 30 min. After centrifugation, the aqueous layer was extracted with 1 volume of acetate-saturated phenol-chloroform. RNA was precipitated with 1 volume of isopropanol, washed with 75% EtOH/50mM NaOAc/HOAc (pH 4.8), and resuspended in 10mM NaOAc/HOAc/1mM EDTA (pH 4.8).
tRNA microarray experiments were conducted as described previously [3; 10]. A brief description follows. The array contains 42 probes for human chromosomal-encoded tRNAs and 21 probes for human mitochondrial-encoded tRNAs, plus 84 probes from non-human tRNAs as hybridization and normalization controls. Each probe is represented at least 20 times on the same array.
To measure tRNA charging, total tRNA isolated at pH 4.8 was first supplemented with 0.66 pmole/μg each of charged E. coli tRNALys, E. coli tRNATyr and yeast tRNAPhe and then split in two halves. One half was subjected to periodate oxidation at 100 mM KOAc/HOAc, pH 4.8, 50 mM NaIO4, 0.1 μg/μl total RNA; the other half was treated in the same way, but NaIO4 was substituted with NaCl. The oxidation proceeded at 22°C for 30 min and the reaction was stopped by the addition of 100 mM glucose. Excess periodate was removed using a G25 spin column followed by precipitating twice with ethanol mixed with 100 mM NaOAc/HOAc, pH 4.8, 100 mM NaCl. tRNAs were deacylated in 0.1 M Tris-HCl, pH 9.0 at 37°C for 30–60 min, followed by ethanol precipitation. Ligation reaction to the tagging oligonucleotides (Fig. 1A) proceeded at 0.2 μg/μl RNA in 66 mM trisHCl, pH 7.6, 6.6 mM MgCl2, 10 mM DTT, 66 μM ATP, 25% (v/v) DMSO, 4 μM tagging oligonucleotides and 0.5 U/μl T4 DNA ligase (US Biochemicals, Inc.) overnight (>16h). The ligated samples were extracted with phenol/CHCl3 and precipitated with ethanol.
Array hybridization was carried out with ~0.5 μg each of Cy3 or Cy5-labeled total tRNAs at 60 °C overnight on a Hyb4 station from Genomic Solutions (Ann Arbor, Michigan).
Results and Discussion
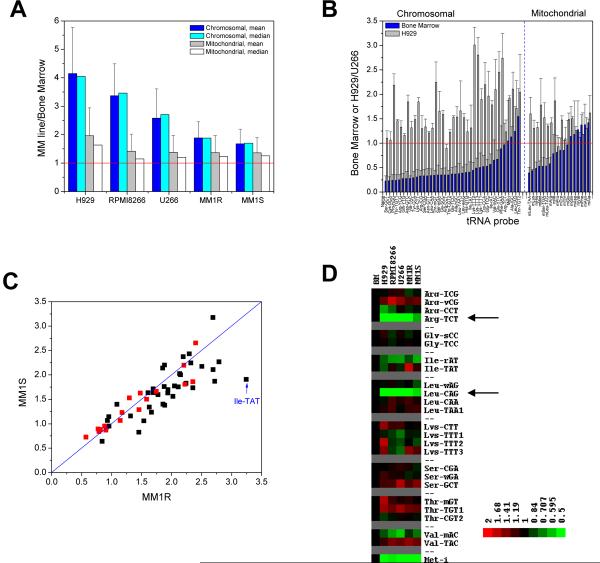
We compared tRNA abundance in MM cell lines MM.1S, MM.1R, NCI-H929, U266 and RPMI-8266 to that of bone marrow cells from healthy donors. tRNAs in total RNA were specifically fluorescent-labeled and then directly hybridized onto microarrays printed with complementary oligonucleotide probes (Fig. 1A). The overall tRNA abundance in each cell line compared to bone marrow cells from a healthy donor is presented as the median and mean values of individual tRNA probes after each tRNA in MM line is normalized to those from bone marrow (Fig. 2A). For all five MM lines, the levels of tRNAs derived from chromosomal-encoded genes were elevated by 2–4 fold, whereas the levels of tRNAs derived from mitochondrial-encoded genes increased by 1.2–1.7-fold. These results demonstrate that tRNA levels are elevated in MM cells which accommodate for their aberrantly high translation activities. Mitochondrial translation may be less affected in MM as seen by the smaller increase in abundance of mitochondrial tRNA.
Fig. 2. Comparing tRNA abundance in MM cell lines and normal bone marrow.
(A) Mean and Median values of chromosomal-encoded and mitochondrial-encoded tRNAs in MM lines compared to bone marrow cells. (B) Ratio of each probe for the sample with lowest (bone marrow) and highest (H929) relative tRNA abundance, plotted against the sample with intermediate tRNA abundance (U266). Left: chromosomal-encoded tRNAs; right: mitochondrial-encoded tRNAs. (C) Relative abundance of MM.1R (dexamethasone-resistent) versus MM.1S (dexamethasone-sensitive) showing similar tRNA levels in these two lines derived from the same parental line. Chromosomal-encoded tRNAs are in black and mitochondrial-encoded tRNAs are in red. tRNAIle(TAT) shows highest deviation between these two lines (1.7-fold). (D) Relative abundance of tRNA isoacceptor families after normalization to the Median value of all chromosomal-encoded tRNAs in the respective samples. Only isoacceptor families are shown that can be distinguished by our tRNA microarray and have the potential for translational regulation. The significant difference among arginine, leucine isoacceptor families (arrows) suggests the possibility for adjusting translational efficiency in genes with distinct codon usages. The relative abundance of initiator-tRNAMet (tRNA-i) after normalization to the Median value is also shown.
The relative abundance of individual tRNA species was more cell line specific (Fig. 2B). For example, some tRNAs in U266 showed a slight decrease and others an increase up to 4-fold. In H929, no tRNA showed a decrease and others an increase up to 6-fold. These differences are likely due to biological and genetic differences between U266 and H929 lines. For instance, U266 and H929 cells have different proliferative indexes and secrete immunoglobulin type IgE and IgA, respectively [12; 13]. In contrast, the relative tRNA levels in MM.1R and MM.1S lines were very similar (Fig. 2C) which was expected as they were derived from the same parental line [14].
Because various tRNA isoacceptors read codons for the same amino acid, differences among tRNA isoacceptor levels can predict the translational efficiency of specific genes according to their codon frequencies [15; 16; 17]. The tRNAArg and tRNALeu isoacceptor families show more than a 2-fold variation among myeloma cells compared to bone marrow (Fig. 2D). Changes in tRNAArg(TCT) which reads the codon AGA are much lower than the three other tRNAArg isoacceptors. Similarly, changes in tRNALeu(CAG) which reads the codon CTG are much lower than the three other tRNALeu isoacceptors. These results suggest that AGA (Arg) and CTG (Leu) codons are translated less efficiently than other codons for arginine and leucine in MM. Alternatively, these two tRNAs may be more tightly regulated in MM for other reasons. Additionally, a notable exception to the overall similarity of the MM.1 lines was the difference in tRNAIle(TAT), which had a 1.7-fold higher level in MM.1R. tRNAIle(TAT) specifically reads one of the three isoleucine codons (ATA), suggesting that MM.1R is better able to translate the ATA codon.
The abundance of tRNAMet used in translational initiation (tRNA-i, Fig. 2D) was significantly below most tRNAs after normalization to the median values. Without median normalization, only the two MM lines with the highest increase in tRNA abundance (H929 and RPMI8266) showed ~1.8-fold increase in tRNAiMet, whereas the three other lines (MM.1R/1S and U266) showed a similar level of tRNAiMet as Bone Marrow. This result contrasts with previous results using fibroblast cells [2], suggesting that increased abundance of initiator-tRNA is not a mechanism for all cancer. Alternatively, MM cells may suppress the elevation of tRNAiMet.
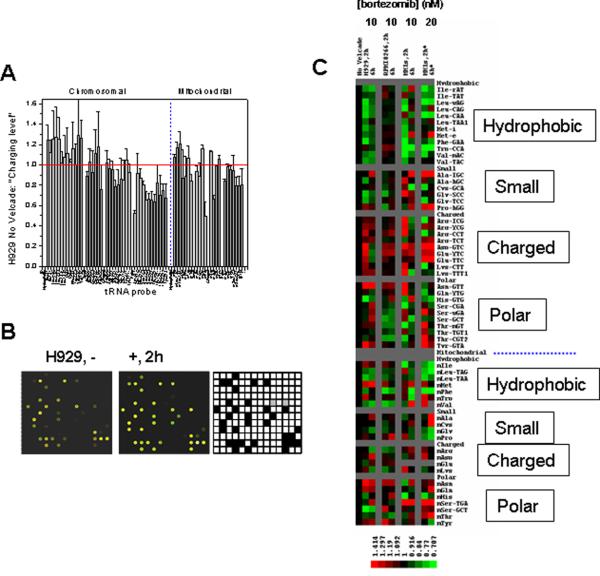
Although bortezomib is FDA-approved for the treatment of relapsed/refractory MM, its effects beyond being a proteasome inhibitor are unclear [18; 19]. Since bortezomib inhibits proteasome activity and thereby reduces amino acid recycling, we would expect a change in charged tRNA levels after treatment with this drug. To test this idea, H929, MM.1S and RPMI8266 lines were treated with 10 nM bortezomib. At this concentration, H929 and MM.1S undergo apoptosis at 6 hours, whereas RPMI8266 does not. Because charged tRNA turns over very rapidly, we also measured tRNA charging prior to induction of apoptosis at 2 hours after bortezomib treatment. The tRNA charging level was determined by the fluorescent ratios between periodate oxidized total tRNA (representing charged tRNA) and the control total tRNA (Fig. 1B). The fluorescent ratios of most tRNAs for H929 line prior to treatment with Bortezomib were approximately one suggesting that most tRNAs are nearly fully charged before Bortezomib addition (Fig. 3A). Addition of Bortezomib resulted in decreased charging levels for some tRNAs, lending support to our hypothesis that proteasome inhibition affects tRNA charging (Figs. 3B, 3C).
Fig. 3. Comparing tRNA charging levels in MM cell lines upon Bortezomib treatment.
(A) H929 line before Bortezomib addition (0h). “Charging level” represents fluorescent ratios of periodate-treated tRNA/total tRNA. (B) H929 line at 0h and 2h after Bortezomib addition. A small, identical section of the microarray image is shown. Total tRNA is in green and charged tRNA is in red. More green spots are present in the 2h sample, indicating decreased tRNA charging levels. Filled squares in the grid indicate human tRNA probes and are expected to show fluorescent signals. Open squares in the grid indicate non-human tRNA probes and are expected to be devoid of fluorescent signals. (C) TreeView presentation of changes in tRNA charging levels, grouped by amino acid properties. Chromosomal-encoded tRNAs are above and mitochondrial-encoded tRNAs are below the dashed line. Gray columns divide the cell lines and gray rows divide tRNAs grouped by amino acid properties. Charging level changes are normalized to the 0h sample of each respective cell line. The dominant feature is the decrease in charging of chromosomal-encoded tRNAs coding for hydrophobic amino acids, only in the Bortezomib sensitive lines at 2h (H929 and MM.1S), but not in the resistant line (RPMI8266). This feature is maintained at higher concentration of Bortezomib for MM1S line (MM.1S*).
Specifically, bortezomib selectively decreased charging of chromosomal-encoded tRNAs coding for hydrophobic amino acids, but not the charging of tRNA coding for charged amino acids (Fig. 3C). This selectivity suggests that bortezomib differentially affects the availability of amino acids for protein synthesis. This effect is primarily found in cells that were sensitive to a low bortezomib concentration (H929 and MM.1S). This effect is much more pronounced at 2h than at 6h, which may be explained by a decreased demand for amino acids at the later time when cellular translation rates are low due to apoptosis. At a higher bortezomib concentration (20nM = MM.1S*), decreased charging of tRNAs coding for hydrophobic amino acids were apparent at both 2h and 6h in MM.1S (Fig. 3C).
In summary, we demonstrate that the abnormally high translation activity in MM cells is accommodated by over-expression of tRNAs and tRNA charging becomes increasingly sensitive to amino acid supply. These results suggest that regulatory pathways exist in MM cells that link translation activity to tRNA properties. Further studies of tRNA in cancer biology may lead to therapeutic applications that specifically target translational regulation.
Acknowledgments
This work was in part supported by grants from the National Institutes of Health RO1CA98550 (AW), leukemia Lymphoma Society SCOR (AW) and CDMRP (T.P.). J.G. was supported by a genetics training grant from NIH.
Abbreviations
- MM
multiple myeloma
Footnotes
The authors declare no competing financial interests.
References
- [1].White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- [2].Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- [3].Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104:607–18. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- [5].van Rhee F, Bolejack V, Hollmig K, Pineda-Roman M, Anaissie E, Epstein J, Shaughnessy JD, Jr., Zangari M, Tricot G, Mohiuddin A, Alsayed Y, Woods G, Crowley J, Barlogie B. High serum-free light chain levels and their rapid reduction in response to therapy define an aggressive multiple myeloma subtype with poor prognosis. Blood. 2007;110:827–32. doi: 10.1182/blood-2007-01-067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kyle RA, Rajkumar SV. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. Hematol Oncol Clin North Am. 2007;21:1093–113. ix. doi: 10.1016/j.hoc.2007.08.005. [DOI] [PubMed] [Google Scholar]
- [7].Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr., Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kadowaki M, Kanazawa T. Amino acids as regulators of proteolysis. J Nutr. 2003;133:2052S–2056S. doi: 10.1093/jn/133.6.2052S. [DOI] [PubMed] [Google Scholar]
- [9].Bechet D, Tassa A, Combaret L, Taillandier D, Attaix D. Regulation of skeletal muscle proteolysis by amino acids. J Ren Nutr. 2005;15:18–22. doi: 10.1053/j.jrn.2004.09.005. [DOI] [PubMed] [Google Scholar]
- [10].Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–7. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Galluzzi L, Zamzami N, de La Motte Rouge T, Lemaire C, Brenner C, Kroemer G. Methods for the assessment of mitochondrial membrane permeabilization in apoptosis. Apoptosis. 2007;12:803–13. doi: 10.1007/s10495-007-0720-1. [DOI] [PubMed] [Google Scholar]
- [12].Nilsson K, Bennich H, Johansson SG, Ponten J. Established immunoglobulin producing myeloma (IgE) and lymphoblastoid (IgG) cell lines from an IgE myeloma patient. Clin Exp Immunol. 1970;7:477–89. [PMC free article] [PubMed] [Google Scholar]
- [13].Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood. 1986;67:1542–9. [PubMed] [Google Scholar]
- [14].Goldman-Leikin RE, Salwen HR, Herst CV, Variakojis D, Bian ML, Le Beau MM, Selvanayagan P, Marder R, Anderson R, Weitzman S, et al. Characterization of a novel myeloma cell line, MM.1. J Lab Clin Med. 1989;113:335–45. [PubMed] [Google Scholar]
- [15].Santos MA, Moura G, Massey SE, Tuite MF. Driving change: the evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- [16].Kreitman M, Comeron JM. Coding sequence evolution. Curr Opin Genet Dev. 1999;9:637–41. doi: 10.1016/s0959-437x(99)00034-9. [DOI] [PubMed] [Google Scholar]
- [17].Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- [18].Guedat P, Colland F. Patented small molecule inhibitors in the ubiquitin proteasome system. BMC Biochem. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2091-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kanagasabaphy P, Morgan GJ, Davies FE. Proteasome inhibition and multiple myeloma. Curr Opin Investig Drugs. 2007;8:447–51. [PubMed] [Google Scholar]