Abstract
A method for the design and rapid manufacture of a patient specific tissue slicing device based on in vivo images in order to facilitate the process of correlating the images with histopathology is presented. The method is applied to radical prostatectomy specimens where the customized mold is designed using magnetic resonance (MR) images of each patient obtained prior to surgery. In this case, the mold holds the prostate in place while a knife with a single blade or multiple blades is inserted in slots which are positioned to obtain tissue blocks corresponding to the slices in the MR images. The resulting histological specimens demonstrate good anatomical correlation with the MR images.
INTRODUCTION
Prostate cancer (PCa) is the most commonly diagnosed cancer in American men and the second leading cause of cancer-related death. In 2008, it is estimated that 28 660 American men will die of PCa (American Cancer Society, 2008). Improved diagnosis with screening prostate specific antigen (PSA) and higher prevalence in an aging population contribute to the increased incidence of PCa. The cancers detected today are smaller and less advanced than those detected before the PSA era and therefore more curable.1, 2, 3 The standard methods of diagnosis are the digital rectal exam (DRE) and serum PSA levels followed by transrectal ultrasound (TRUS) guided needle biopsy. While TRUS guided biopsy is adequate for diagnosis, it does not provide the spatial resolution needed to visualize the extent of the tumor within and outside the gland. Currently, such information is available only from whole-mount histopathology sections of the prostate gland.2, 4, 5, 6, 7, 8, 9
Recent studies have shown that magnetic resonance imaging (MRI) can help to detect and localize PCa.1, 2, 10, 11, 12, 13 The MRI techniques that have been shown to be effective include T2-weighted imaging (T2W), diffusion-weighted imaging (DWI), dynamic contrast-enhanced MRI (DCE-MRI), and three-dimensional spectroscopic imaging (3DSI). However, because the orientation of the specimen and its sections may be different from the imaging, mismatches between imaging and histopathology can occur making it difficult to assess the true accuracy of MRI.
The correlation between in vivo MR images and histopathology is challenging. The prostate is an easily deformable organ, hence, it often deforms during and after prostatectomy. Once the anatomic orientation in the body is lost, it may be difficult to section the prostate in the same plane as the images are obtained. Additionally, in order to improve image quality, prostate MRI is often performed using an endorectal coil, which further deforms the gland.14 A number of methods have been reported to solve this problem. One approach uses a generic cradle to hold the specimen while the tissue is sliced with a series of evenly spaced blades.5 The generic cradle does not account for the deformation of the gland or variations in prostate size, thus the slices obtained may not be in the same plane with the MR images. In another approach, the fresh prostate specimen is embedded in 2% agar (30 mM NaCl) at 50 °C and cooled to 4 °C to solidify agar in a small Plexiglas box.8 MR images are obtained and parallel histological sections are generated using a rotary knife. However, this approach cannot be applied to functional MRI sequences such as DCE-MRI that require a living subject. Thus, there is a need to improve the method of processing the specimens in order to improve correlation of MR images with histopathology.
Techniques for three-dimensional (3D) modeling and rapid prototyping have been known for several years. Rapid prototyping15 technology can be used to automatically convert a computer generated 3D model into a physical model. Application of these technologies is not limited to building a prototype model for visualization but is actually revolutionizing the medical industry.16, 17 A literature survey shows that these technologies have not been used to process histopathology specimens. This paper presents a new method of processing a prostatectomy specimen using a customized mold which is designed for each patient using image processing and computer-aided design, and is fabricated using rapid prototyping technology. The customized mold holds the prostate in the same position and shape as the in vivo images and guides the cutting knife to obtain tissue blocks that corresponds to the image slices.
MATERIALS AND METHODS
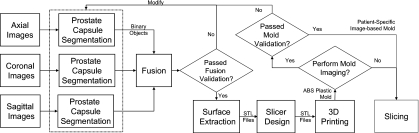
The general block diagram for developing the image-based mold that includes in vivo imaging of the specimen, creation of the virtual 3D model, and fabrication and verification of the physical mold are shown in Fig. 1. Each section of the block diagram is discussed in Secs. 2A, 2B, 2C, 2D, 2E, 2F, 2G.
Figure 1.
Block diagram for the method of generating the individualized image-based mold.
Clinical MR imaging
The patients, who later underwent radical prostatectomy, were enrolled in a single institution study that has been approved by the local institutional review board. Prior to surgery, each patient was scanned on a 3.0 T whole-body MRI system (Achieva, Philips Healthcare, Best, NL) using an endorectal coil (BPX-30, Medrad, Pittsburgh, PA) combined with a manufacturer supplied six-channel cardiac coil. After performing a DRE, the endorectal coil was placed into the rectum and the balloon around the coil was distended using a liquid perfluorocarbon (Fluorinert FC-77, 3M, St. Paul, MN) to a volume of approximately 50 ml to avoid susceptibility artifacts caused by air in the balloon. Three-plane scout views of the prostate were obtained to confirm correct positioning of the endorectal coil. Multislice T2-weighted turbo spin-echo (TSE) images of the entire prostate were obtained in three orthogonal planes (sagittal, axial, and coronal) at a scan resolution of 0.461×0.598×3.0 mm3 field of view (FOV), 140 mm; acquisition matrix, 234×304; TR∕TE, 8869∕120 ms; flip angle, 90°; slice thickness, 3 mm without gaps; image reconstruction, 512×512). To reduce scan time, a lower refocusing pulse of 100° was used for the sagittal and coronal images. The axial images with the phase encode direction going left to right were positioned perpendicular to the rectal wall guided by the sagittal TSE images. The clinical study also included DWI, DCE-MRI, and 3DSI after the T2W images.
Generation of high-resolution 3D model of the organ
Virtual 3D models of each prostate were created using the ANALYZE software (Mayo Clinics, AnalyzeDirect, Inc., Overland Park, KS). The 3D model generation included segmentation of the prostate capsule on in vivo images, fusion of the binary objects, and the surface extraction of high-resolution 3D surface from the binary object.
A radiologist helped to trace the boundary of the prostate capsule on the three orthogonal TSE images using the trace boundary tool of the software (Fig. 2). The region of interests (ROIs) of all three views is converted into binary objects that have the same nonisotropic resolution and FOV as the MR images. A shape-based interpolation algorithm was used to process these binary objects to have an isotropic resolution. The images were then cropped to create the same FOV for all the binary objects. The resulting binary object was obtained by fusing these three cropped isotropic resolution binary objects and applying a majority rule. The resultant binary object was validated by overlaying it on the MR images. The ROIs in different views were modified or manual registration was performed on the binary objects if the fused binary object was not satisfactory.
Figure 2.
Segmentation of the prostate boundary in the MR images at three different views; axial (a), coronal (b), and sagittal (c).
An adaptive deformation algorithm based on an adaptive mesh model was used for the surface extraction from the binary object. The accuracy of the final model depended on the number of iterations and the cube edge size. Heuristically, it was determined that setting the number of iterations to 1000 yielded an acceptable model. The cube edge size determined the number of triangles generated to create the final surface; eight were selected experimentally. These selected parameters provided a good compromise between the number of triangles and the appropriate virtual surface model. The extracted surface was saved as a stereolithography (STL) or virtual reality modeling language (VRML) file.
Solid models marking the location of the urethera at the apex and base were generated using the same method. In the apex, the ROIs were drawn on the axial slices, while in the base, the ROIs were drawn on the sagittal slices.
Mold design
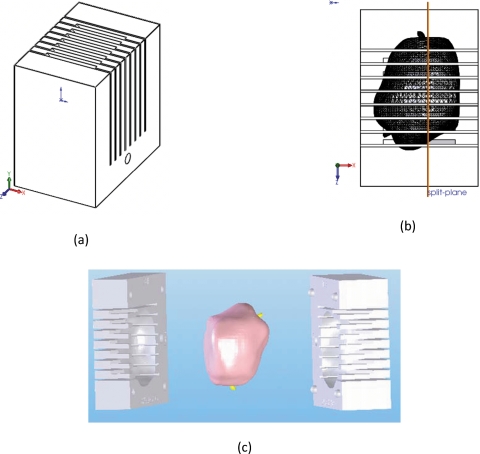
Each mold was designed using a commercially available 3D computer-aided design software (Solidworks, Dassault Systèmes SolidWorks Corp., Concord, MA) starting with the same solid rectangular part with parallel cutting slots spaced 6 mm apart, as shown in Fig. 3a. The customized cavity for the prostate was created by importing the 3D models of the prostate with the urethra markers from the STL or VRML file created in Sec. 2C, and subtracting them from the solid mold block using the “features combine” tool. Since the size of the gland varied between patients, the outside dimensions of the block and the number of slots were adjusted for each patient. The resulting mold was split perpendicular to the cutting slots to create two–halves, as shown in Fig. 3b. Alignment posts and holes were added to help line up the two-halves and labels were applied to help orient the prostate specimen. The cutting slots in the mold were parallel to the slices of the axial MR images and positioned such that the each resulting tissue block corresponded to two axial TSE MRI slices. Two possible configurations were used: one where the prostate cavity was oriented such that the split in the mold was along the sagittal plane, and another where the split in the mold was along the coronal plane [Fig. 3c].
Figure 3.
The individualized mold showing the starting rectangular solid part having parallel cutting slots (a), after subtraction of the 3D prostate model and showing the location of the split plane (b), and after being split in two halves with the 3D model of the prostate with urethra markers in yellow. The mold shown is split along the coronal plane of the prostate that is shown with the base toward the top and the anterior toward the right.
Fabricating the mold
A 3D printer (Dimension Elite 3D printer, Stratasys, Inc., Eden Prairie, MN) was used to fabricate each mold. This printer creates functional parts via fused deposition of acrylonitrile butadiene styreneplastic (ABS) plastic. The ABS plastic comes in the form of a wire spooled within a cartridge and is extruded through a heated print-head tip. The extruded 0.010×0.020 in.2 (height×width) filament is deposited as the print head moves in the printer bed. The split mold parts were simultaneously built layer by layer. Depending on the mold geometry, some areas of the prostate cavity required the automatic printing of non-ABS plastic support material to provide a rigid substrate for the subsequent layers. This support material was either mechanically broken off or dissolved in a solvent bath after printing. Depending on the amount of ABS plastic required to build the prostate mold, print times ranged from 8 to 24 h. The CATALYST EX 3D printer driver software was used to preselect the mold orientation during printing. It was advantageous to orient the parts with the prostate cavity up so that minimal or no support material was used during the printing process; therefore, avoiding the time consuming step of dissolving the support material away. To aid in orienting the prostate specimen in the mold, the cavity surface was color coded to match the color of the paint on the specimen (vide infra).
Validation of the mold
In some cases, the fabricated mold was validated using MRI by placing it in a box filled with water and scanning it with the same TSE protocol used to scan the patients. These MR images of the mold were overlaid on the drawn ROI and the patient’s MR images for comparison.
Preparation of human specimens for histopathology
The radical prostatectomy specimen obtained from each patient was painted so that the anterior was black, the right posterior was blue, the left posterior was red, and the urethra was yellow. Afterward, the specimen was fixed in formalin for 2–24 h at room temperature. The seminal vesicles were removed before placing the specimen in the mold. A pair of clamps was used to press the two halves together. The specimen was sliced using a 10 in. long autopsy knife (Scientific Supplies, order No. MII-5000), as shown in Fig. 4a. To complete the fixation process, the resulting slices were kept in formalin for an additional 48–72 h. The formalin was removed by wax addition and graded alcohol was added to dehydrate the specimen. The alcohol was cleared by adding xyline and paraffin to the media. The histology slides were prepared after this process was repeated in reverse order. The slices were expected to shrink by approximately 10%–15% after all of these procedures.
Figure 4.
Knives with a single blade (a) and with multiple blades (b) used for slicing the specimens.
Fixed dog prostate experiment
Prior to processing human specimens, the method was tested on two formalin-fixed dog prostate glands. High resolution multislice T2W TSE MR images were obtained in three orthogonal orientations through a dog prostate specimen, using a homebuilt saddle-shaped receiver coil (28 mm i.d.×70 mm length) with the following parameters: TR∕TE, ∼4000∕35 ms; flip angle, 90°; FOV, 40 mm; acquisition matrix, 200×200; slice thickness, 0.5 mm without gaps; reconstruction, 256×256. Since the images were close to having isotropic resolution, only the axial images were used to generate the 3D prostate model. Similar to the clinical study, the molds were equipped with slots spaced 6 mm apart. The single blade knife shown in Fig. 4a was used on one prostate specimen. A knife comprised of multiple parallel steel blades (Catalog No. 6727-C18 Tissue Slicer Blades, Thomas Scientific, USA) mounted to a plastic handle created using the 3D printer, as shown in Fig. 4b, was used on the other prostate specimen. Each prostate required four cuts to generate five tissue blocks. In the case of the multiblade knife, only four of the detachable blades were mounted on the handle. The resulting tissue blocks were removed from the mold and arranged in the same sequence in a plastic holder fabricated with the 3D printer. The plastic holder had 6 mm slots for the tissue blocks separated by 1 mm spacers. MR images were taken in the same axial orientation using the same protocol with additional slices to account for the spacers and longer scan time to reduce noise. In planning the MRI scans of the tissue blocks, the location and angulation of the slices were set to be parallel and coincident with the tissue blocks. The MR images of the specimens before and after slicing were then compared by locating the middle slice through each tissue block and visually matching them to the MR slices of the whole specimens.
RESULTS
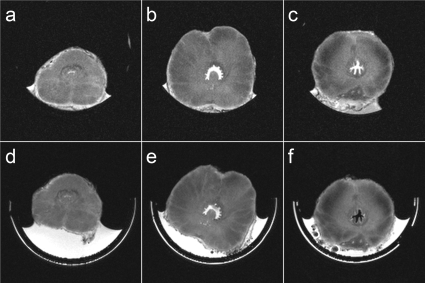
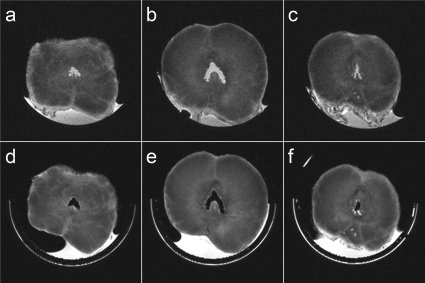
Comparisons of MR slices from two whole fixed dog prostate specimens and the corresponding tissue blocks obtained using the customized molds with the single blade and the multiblade knives are shown in Figs. 56, respectively. Figures 5a, 5b, 5c, 6a, 6b, 6c show the MR image slices from the whole specimens, which best matched the MR image slices positioned around the middle of the second, third, and fourth tissue blocks shown in Figs. 5d, 5e, 5f, 6d, 6e, 6f. Note the in-plane rotation of the images of the individual tissue blocks relative to the images of the whole specimens due to rotation of each tissue block in the plastic holder. Accounting for the 1 mm spacer between tissue blocks, the slice positions for the specimen sliced with the multiblade knife were exactly as expected, while the fourth tissue block sliced with the single blade was off by one slice (0.5 mm).
Figure 5.
Comparison of the MR slices from a fixed dog prostate specimen [(a)–(c)] that matched the MR slices through the middle of the second (d), third (e), and fourth (f) of five tissue blocks obtained from that specimen using a knife with a single blade.
Figure 6.
Comparison of the MR slices from a fixed dog prostate specimen [(a)–(c)] that matched the MR slices through the middle of the second (d), third (e), and fourth (f) of five tissue blocks obtained from that specimen using a knife with multiple blades.
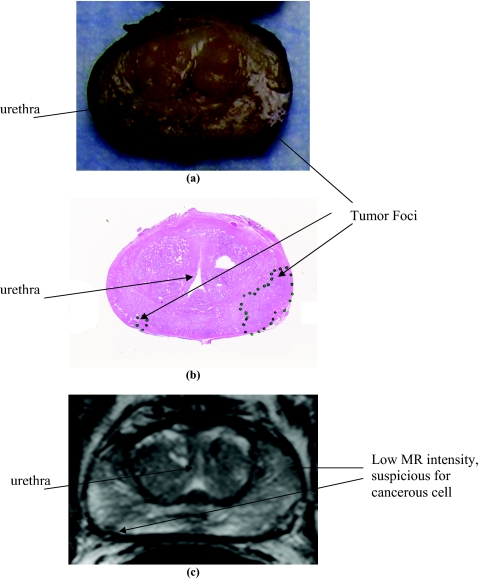
Prostatectomy specimens from patients were processed using individually generated MRI-based molds. Figure 7 shows a picture of the individualized mold and the resulting the tissue blocks (shown from the base to the apex) from a specimen sliced after fixation for 2 h. A photograph of one of the tissue blocks (third from the apex side), the histopathology section from the tissue block, and the corresponding T2W image are compared in Fig. 8. There was good anatomical correlation between the MR image and the histology section. The urethra served as a guide for correlating the images. A suspected tumor was present in the left posterior region, which was confirmed on the slice-matched histological section. The smaller tumor area as seen on the right-posterior region was also mapped to the low signal intensity in the T2W images.
Figure 7.
Image-based mold designed from the MR images of the patient acquired prior to surgery and fabricated with a 3D printer (a), and the tissue blocks obtained after slicing the prostatectomy specimen that have been fixed in formalin for approximately 2 h. The tissue blocks are arranged from the base to the apex.
Figure 8.
The third tissue block (a) from the apex end of the prostatectomy specimen in Fig. 7, a histopathology slide (b) taken from that tissue block, and the corresponding slice from a multislice T2W image (c) of the patient.
DISCUSSION
A number of factors affected the performance of the image-based mold in correlating radiology and histology: the fit of the mold to the specimen, the knife and slicing technique used, and the thickness of image slices. The fit of the mold depended on the accuracy of the image segmentation and mold creation process, change in gland size between image acquisition and surgery, and tissue deformation and shrinkage that occurred after surgery and tissue fixation. The knife and slicing technique were critical in order to cut through the tissue without deforming the prostate especially when processing fresh or partially fresh specimens. The almost three orders of magnitude difference in the thickness of the image slice and the histology slides made it more difficult to perform the correlation.
In the dog prostate experiment, nearly isotropic in vitro 3D images of the fixed prostate glands were used to generate the mold. There were no changes due to shrinkage and no noticeable deformation in the specimens. Only extra fatty tissues, which were removed with the specimen, needed to be removed before processing the specimens in the mold. As a result, the location of the axial MR images acquired of the whole prostate and those acquired of the tissue blocks matched regardless whether they were cut with a single or a multiple knife. This gave an indication of the accuracy of the mold generation process. As in Ref. 8, the image-based mold process can be used to process prostatectomy specimens using ex vivo images after surgery. The power of the in vivo mage-based mold, however, lies in the ability to directly correlate in vivo images with histology by personalizing the mold to each patient.
In the clinical study, the acquisition of high-resolution isotropic 3D in vivo images was not practical due to time constraints. Thus, nonisotropic 3D images with thicker slices were acquired in three different orthogonal orientations and were used to design the 3D prostate models. The ROIs were drawn separately on the MR images obtained along the axial, coronal, and sagittal orientations, as shown in Fig. 2. Additionally, the ROIs included any additional tissues that were removed during the surgery. Extra tissues surrounding the prostate usually resulted when extra-capsular extension was suspected. The majority rule method used to combine the ROIs from each of the orthogonal images was degraded by movement of the patient. This resulted in time consuming iterative coregistration to make the combined ROIs consistent with the individual orthogonal ROIs. We are currently working on more efficient methods of combining the orthogonal ROIs as well as implementing automatic segmentation of prostate capsule to accelerate the model generation process.
The fit of the mold to the specimen was also affected by changes in the shape and size of the prostate. The prostate is not only deformable, but studies have shown that the size can change over time due to benign prostatic hyperplasia (BPH).18, 19, 20 If the time between the acquisition of the in vivo images and the prostatectomy is too long, the customized mold may not fit the specimen. For this study, the time interval between the MRI scan and prostatectomy varied from two days to approximately 4 months. In most cases, the prostate fits well, except for two cases where the MR images were obtained 4 months prior to surgery resulting in a mold that was too small to fit the prostate. Hence, if possible there should be minimum time gap between MRI and surgery.
Another factor that can significantly affect the fit of the mold is the shrinkage and toughening of tissues after fixation.21, 22 This depends on the formalin concentration and the duration of the fixation. Our fixation procedure includes the immersion of the prostate specimen in formalin for approximately 2–24 h before slicing. In most cases, the prostate gland fitted well in the mold even after 20 h of fixation, indicating that shrinkage can be negligible. However, the fixative made the gland more rigid over time. Thus, it was difficult to deform when placing it in the mold. In addition, the alignment of the MR slices and the histology can be off due to shrinkage in the fixed specimens. The fit and image coregistration would improve if fresh specimens were processed with the mold. In addition, this would facilitate procurement of the tissues directly from the tissue blocks for “-omics” analysis (e.g., genomics, proteomics, etc.).
A slight change in the design of the mold was necessary to improve the fit. Initially, the molds were designed such that the prostate mold was split along the midsagittal plane. Since the endorectal coil compresses the prostate in the anterior-posterior direction, the molds generated from the MR images were also compressed in the same way. Although it was possible to compress half the gland by hand before inserting it into the mold, it was very difficult to compress the other half while placing the two-halves together. This became more difficult with the larger specimens due to BPH and with more rigid specimens after longer fixing time. Rotating the prostate cavity, as shown in Fig. 3c, such that the mold was split parallel to the coronal plane proved more effective since the specimen was easily compressed between the two-halves of the mold.
The slicing tool and technique were critical in obtaining evenly spaced tissue blocks with uniform thickness. The single blade and multiblade knives shown in Fig. 4 were used in this study. Although both knives performed equally well in the experiment with the fixed dog prostate, as shown in Figs. 56, the multiblade was not as effective with the clinical specimens because the blades were not long enough to enable a smooth slicing motion for the larger human prostatectomy specimens. In general, we consistently obtained uniformly sliced tissue blocks with longer fixation times which made the specimens more rigid. With less fixation time, the result was not as consistent. This was probably due to variations in the cutting technique as the fresh tissue was subject to deformation with too much downward force. This can be improved with the design of an automatic knife to eliminate variations in the manual cutting technique. A multiblade design, which would keep the tissue blocks from shifting within the mold, should also perform better on fresh specimens. Future work will investigate the use of manual and automatic tissue cutters made of wires and rotary blades. Improvements in the cutting method would allow us to reduce the thickness of the tissue blocks from 6 mm, which was considered thick for histopathology, to a more desirable thickness of 3 or 4 mm.
Lastly, ideal correlation between MR images and histology slides was complicated by the difference in the slice thickness of the MR images (3 and 6 mm in this study) and the very thin histology sections (5 μm in this study) required for light microscopy. The MR image slices may have significant anatomical variation along the slice direction. Hence, the histology sections, taken from a tissue block that exactly matches the MR slice, may not look like the thick MR slice. The correlation process can be simplified by matching significant structural landmarks (nodules, cysts, urethra, etc.). Since the histology slides were taken from tissue blocks that match the spacing and angulations of the MR slices, they could be easily matched in sequence.
CONCLUSIONS
We have successfully designed and applied patient specific image-based molds to process radical prostatectomy specimens to improve the coregistration of whole-mount histology sections to in vivo MR images. The method presented here can be easily adapted for use with other tissues such as kidney, breast, and uterus, and with other imaging modalities such as computed tomography and ultrasonography. This technique will be crucial in validating the results of imaging studies with histology. This will help to improve the efficacy of imaging modalities in detecting cancer. Future work will focus on developing the technique to slice fresh prostate specimens to improve coregistration and to aid in the procurement of tissues for spatially resolved “-omics” analysis that can be correlated with radiology and histology.
ACKNOWLEDGMENTS
This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Choi Y. J., Kim J. K., Kim N., Kim K. W., Choi E. K., and Cho K. S., Radiographics 27, 63 (2007). 10.1148/rg.271065078 [DOI] [PubMed] [Google Scholar]
- Ocak I., Bernardo M., Metzger G., Barrett T., Pinto P., Albert P. S., and Choyke P. L., AJR, Am. J. Roentgenol. 189, W192 (2007). 10.2214/AJR.06.1329 [DOI] [PubMed] [Google Scholar]
- Rogerson P. A., Sinha G., and Han D., Am. J. Prev. Med. 30, S50 (2006). 10.1016/j.amepre.2005.09.006 [DOI] [PubMed] [Google Scholar]
- Graser A., Heuck A., Sommer B., Massmann J., Scheidler J., Reiser M., and Mueller-Lisse U., AJR, Am. J. Roentgenol. 188, 84 (2007). 10.2214/AJR.06.0401 [DOI] [PubMed] [Google Scholar]
- Jhavar S. G., Fisher C., Jackson A., Reinsberg S. A., Dennis N., Falconer A., Dearnaley D., Edwards S. E., Edwards S. M., Leach M. O., Cummings C., Christmas T., Thompson A., Woodhouse C., Sandhu S., Cooper C. S., and Eeles R. A., J. Clin. Pathol. 58, 504 (2005). 10.1136/jcp.2004.021808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling F., Le-Huu M., Kunert T., Thorn M., Vosseler S., Schmidt K., Hoffend J., Meinzer H. P., Fusenig N. E., and Semmler W., Eur. Radiol. 15, 1079 (2005). 10.1007/s00330-005-2701-5 [DOI] [PubMed] [Google Scholar]
- Kozlowski P., Chang S. D., Jones E. C., Berean K. W., Chen H., and Goldenberg S. L., J. Magn. Reson Imaging 24, 108 (2006). 10.1002/jmri.20626 [DOI] [PubMed] [Google Scholar]
- Madabhushi A., Feldman M. D., Metaxas D. N., Tomaszeweski J., and Chute D., IEEE Trans. Med. Imaging 24, 1611 (2005). 10.1109/TMI.2005.859208 [DOI] [PubMed] [Google Scholar]
- Van As N., Charles-Edwards E., Jackson A., Jhavar S., Reinsberg S., Desouza N., Dearnaley D., Bailey M., Thompson A., Christmas T., Fisher C., Corbishley C., and Sohaib S., Br. J. Radiol. 81, 456 (2008). 10.1259/bjr/29869950 [DOI] [PubMed] [Google Scholar]
- Futterer J. J., Heijmink S. W., Scheenen T. W., Veltman J., Huisman H. J., Vos P., Hulsbergen-Van de Kaa C. A., Witjes J. A., Krabbe P. F., Heerschap A., and Barentsz J. O., Radiology 241, 449 (2006). 10.1148/radiol.2412051866 [DOI] [PubMed] [Google Scholar]
- Haider M. A., van der Kwast T. H., Tanguay J., Evans A. J., Hashmi A. T., Lockwood G., and Trachtenberg J., AJR, Am. J. Roentgenol. 189, 323 (2007). 10.2214/AJR.07.2211 [DOI] [PubMed] [Google Scholar]
- Miao H., Fukatsu H., and Ishigaki T., Eur. J. Radiol. 61, 297 (2007). 10.1016/j.ejrad.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Noworolski S. M., Henry R. G., Vigneron D. B., and Kurhanewicz J., Magn. Reson. Med. 53, 249 (2005). 10.1002/mrm.20374 [DOI] [PubMed] [Google Scholar]
- Heijmink S. W., Scheenen T. W., van Lin E. N., Visser A. G., Kiemeney L. A., Witjes J. A., and Barentsz J. O., Int. J. Radiat. Oncol., Biol., Phys. 73, 1446 (2009). 10.1016/j.ijrobp.2008.06.1491 [DOI] [PubMed] [Google Scholar]
- Ashley S., Mech. Eng. 117, 62 (1995). [Google Scholar]
- Gibson I., Advanced Manufacturing Technology for Medical Applications-Reverse Engineering, Software Conversion and Rapid Prototyping (Wiley, New York, 2006). [Google Scholar]
- Martin Masters T. V. and McBagonluri F., in Rapid Manufacturing Technologies: The Next Industrial Revolution, edited by Hopkinson N., Haque R., and Dickens P. (Wiley, New York, 2006), pp. 195–209. [Google Scholar]
- Nickel J. C., Urol. Clin. North Am. 35, 109 (2008). 10.1016/j.ucl.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C. R., Crowe R., Gilpin S. A., Gosling J., and Burnstock G., J. Urol. 146, 1637 (1991). [DOI] [PubMed] [Google Scholar]
- Orth M., Pharm. Unserer Zeit 37, 315 (2008). 10.1002/pauz.200700275 [DOI] [PubMed] [Google Scholar]
- Jonmarker S., Valdman A., Lindberg A., Hellstrom M., and Egevad L., Virchows Arch. 449, 297 (2006). 10.1007/s00428-006-0259-5 [DOI] [PubMed] [Google Scholar]
- Schned A. R., Wheeler K. J., Hodorowski C. A., Heaney J. A., Ernstoff M. S., Amdur R. J., and Harris R. D., Am. J. Surg. Pathol. 20, 1501 (1996). 10.1097/00000478-199612000-00009 [DOI] [PubMed] [Google Scholar]