Abstract
The microsporidian Encephalitozoon cuniculi is an intracellular eukaryotic parasite considered to be an emerging opportunistic human pathogen. The infectious stage of this parasite is a unicellular spore that is surrounded by a chitin containing endospore layer and an external proteinaceous exospore. A putative chitin deacetylase (ECU11_0510) localizes to the interface between the plasma membrane and the endospore. Chitin deacetylases are family 4 carbohydrate esterases in the CAZY classification, and several bacterial members of this family are involved in evading lysis by host glycosidases, through partial de-N-acetylation of cell wall peptidoglycan. Similarly, ECU11_0510 could be important for E. cuniculi survival in the host, by protecting the chitin layer from hydrolysis by human chitinases. Here, we describe the biochemical, structural, and glycan binding properties of the protein. Enzymatic analyses showed that the putative deacetylase is unable to deacetylate chitooligosaccharides or crystalline β-chitin. Furthermore, carbohydrate microarray analysis revealed that the protein bound neither chitooligosaccharides nor any of a wide range of other glycans or chitin. The high resolution crystal structure revealed dramatic rearrangements in the positions of catalytic and substrate binding residues, which explain the loss of deacetylase activity, adding to the unusual structural plasticity observed in other members of this esterase family. Thus, it appears that the ECU11_0510 protein is not a carbohydrate deacetylase and may fulfill an as yet undiscovered role in the E. cuniculi parasite.
Keywords: cell wall, chitin, peptidoglycan, deacetylase, protein structure, glycobiology, carbohydrates
Introduction
The Microsporidia Phylum, comprising over 1200 spore-forming species, encompasses unicellular eukaryotes, which are all obligate intracellular parasites. These fungi-related1–3 parasites are considered opportunistic pathogens4 and infect a wide range of invertebrate and vertebrate hosts.5 These emerging human pathogens are responsible for various digestive and nervous clinical syndromes in immunocompromised AIDS and organ transplant patients.6–8
Microsporidia display an unusual invasion mechanism, involving the polar tube, a cylindrical and very long structure that is coiled within the spore. Microsporidian spores can also be internalized after interactions between the parasite spore wall and the host plasma membrane, followed by phagocytosis.9–11 The microsporidian spore is surrounded by a spore wall, which plays a major role in host cell invasion12 and in the protection against environmental stresses, allowing long-term survival of the parasite after its release from the host cell.13,14 This cell wall usually comprises a proteinaceous outer electron-dense layer (exospore) and an inner electron-transparent layer (endospore) which consists mainly of chitin, a homopolymer of β(1-4) linked N-acetylglucosamine frequently found in fungi, which is of a fibrillar nature and connected to the plasma membrane.15–17
Recent postgenomic studies18,19 allowed us to identify the ECU11_0510 protein of the mammalian parasite Encephalitozoon cuniculi as a spore wall protein located at the sporont surface then in the inner part of the mature spore endospore. This 254 amino acid protein is predicted to contain an N-terminal signal peptide with a cleavage site between 15 and 16 or 17 and 18. Residues 234–253 are predicted to be a transmembrane helix in the outside-inside orientation, suggesting that this protein is anchored to the plasma membrane with a large extracellular domain. ECU11_0510 shows significant homology to the carbohydrate esterase 4 (CE4) family (CAZY database http://www.cazy.org). CE4 esterases are metal dependent enzymes that de-N-acetylate or de-O-acetylate saccharides, such as peptidoglycan, chitin in fungal cell walls, and acetyl xylan in plants.20–28 The crystal structures and reaction mechanism of several members of this family have been determined.21–23,25 In some bacteria, these enzymes are important virulence factors and act by de-N-acetylating sugar residues on cell wall peptidoglycan.28,29 The positively charged de-N-acetylated peptidoglycan is no longer a substrate for mammalian lysozyme and helps the bacteria evade the host immune system.28 It is possible that the ECU11_0510 protein, like other fungal CE4 esterases, is able to deacetylate chitin in the microsporidian spore.21,30,31 This process may help the parasite evade the host immune response by conferring resistance against host glycoside hydrolases (e.g. chitinases) present in bacteria, plants and animals, including humans.
Here, we report on the cloning, recombinant expression, characterization, and structure of this protein. Through a combination of enzymology, carbohydrate binding studies, and high-resolution X-ray crystallography, we show that this is an unusual member of the CE4 esterase family, which appears to have lost its catalytic activity and ability to bind carbohydrates.
Results
E. cuniculi ECU11_0510 is a putative chitin deacetylase
A recent report investigating cell wall proteins of the E. cuniculi spore identified a protein, ECU11_0510, that exhibited sequence similarity with carbohydrate esterase 4 family members. The protein sequence of the putative E. cuniculi chitin deacetylase was aligned with several fungal and bacterial CE4 esterases of known activity [Fig. 1(A)]. The alignment shows that sequence identity within the CE4 esterase family is low (8%; 16/202 residues in Colletotrichum lindemuthianum chitin deacetylase—(ClCDA). However, throughout the CE4 family, there are five common structural motifs, which contain important residues involved in metal binding and general acid-base catalysis [Fig. 1(A)].21–23
Figure 1.
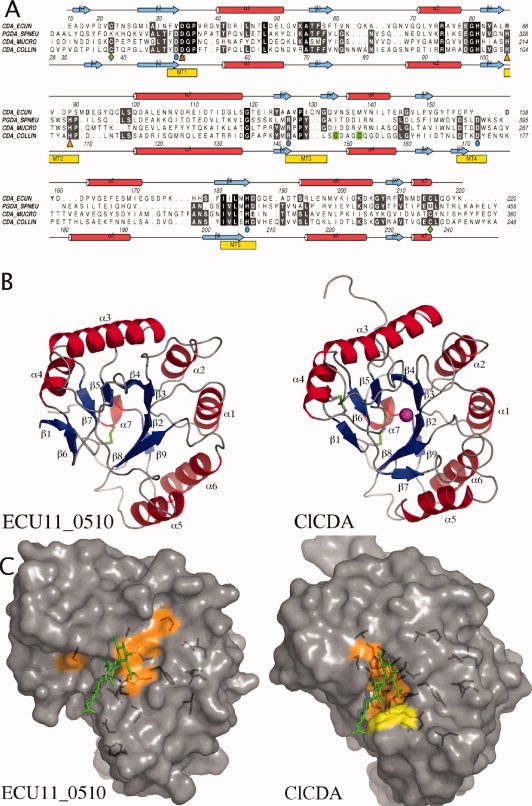
(A) A structure-based sequence alignment. Secondary structure of the ClCDA and the ECU11_0510 proteins are shown; α helixes are colored red and β strands in blue. Metal-coordinating residues are highlighted with orange triangles. Catalytic residues are highlighted with cyan circles. Conserved disulphide bridges are highlighted with green diamonds. A second nonconserved disulphide bridge in ClCDA is highlighted in green. CDA-ECCUN = ECU11_0510, PGDA_SPNEU = Streptococcus pneumoniae PgdA, CDA_MUCRO = Mucor Rouxii Chitin Deacetylase and CDA_COLLIN = Colletotrichum lindemuthianum Chitin Deacetylase. (B) Overall structure comparison of Encephalitozoon cuniculi putative chitin deacetylase, ECU11_0510 (left panel), and ClCDA (right panel). α helixes are colored red and β strands in blue. Conserved disulphide bridges are highlighted in green. Zinc in the active site of the ClCDA is shown as a magenta sphere. (C) ECU11_0510 and ClCDA are shown in a surface representation. A chitotrioside carrying the previously proposed oxyanion reaction intermediate was docked into the ClCDA active site as described previously21 and then superposed onto the active sites of the ECU11_0510. The consensus sequence was generated by aligning 12 CE4 esterase sequences using T-coffee. Residues that are identical or show conservative substitutions and are present in 11/12 sequences are represented on the surface in orange and yellow, respectively.
In previously studied active CE4 esterases, the second aspartic acid of motif 1 (Asp50 in ClCDA) and two histidine residues in motif 2 (His104 and His 108 in ClCDA) coordinate a divalent metal cation that is essential for activity.21,22 ECU11_0510 retains the metal-coordinating aspartic acid (Asp34); however, sequence alignments show that in ECU11_0510, the two histidines are nonconservatively substituted. Although this could suggest loss of activity, the structure of an active CE4 esterase with only two metal coordinating amino acids was recently reported.25 This structure, of the Clostridium thermocellum acetyl xylan esterase, showed that only one of the two metal binding histidines in motif 2 is required for de-O-acetylation of acetyl xylan. ECU11_0510 contains a single residue, Asp92, that is, in theory, able to coordinate a divalent metal cation, perhaps with assistance of side chains from regions not directly identifiable by sequence alignment alone.
CE4 esterases employ a general acid base catalysis (GABC) mechanism to deacetylate sugar substrates. In the ClCDA enzyme, Asp49 in motif 1 acts as the catalytic base. Hydrogen bonded interactions between the catalytic base and a highly conserved arginine residue (Arg142) present in motif 3 are also essential, as site-directed mutation of either of these residues resulted in complete loss of activity in the Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) protein.22 However, in the E. cuniculi protein, these residues align with valine (Val33) and an alanine (Ala126), which would be unable to participate in GABC catalysis. It has been proposed that an aspartic acid (Asp172 in ClCDA) interacts with a histidine (His206) to form an activated histidinium ion, which is proposed to act as the catalytic acid, protonating the reaction intermediate.22 In the E. cuniculi protein, these amino acids are retained at the sequence level in motifs 4 (Asp158) and 5 (His206). This partial conservation of the catalytic machinery prompted an investigation of the ability of the ECU11_0510 protein to function as an active CE4 esterase.
ECU11_0510 does not display chitin deacetylase activity
A region of the ECU11_0510 gene that encoded amino acids 17–233 was cloned into the Pichia pastoris secretory plasmid, pPICZαA. This region of the mature protein lacked a predicted signal peptide sequence (amino acids 1–16) and a predicted C-terminal transmembrane helix (amino acids 234–254), but retained a conserved N-terminal Cys22 [Fig. 1(A)]. This truncated gene was inserted into the genome of the Pichia pastoris X-33 strain, yielding over 100 mg of secreted recombinant ECU11_0510 per liter of culture. Contaminating proteins were successfully removed by anion exchange chromatography, and a subsequent gel filtration step yielded large amounts of pure protein suitable for biochemical characterization and crystallization trials.
Chitin deacetylase activity of recombinant ECU11_0510 was investigated using chitotriose, chitotetraose, and chitohexaose as substrates. As positive control, 500 nM ClCDA was tested in a 60 min assay containing 2 mM chitotriose. The ClCDA reaction that was subsequently incubated with fluorescamine generated relative fluorescence units corresponding to conversion of >90% of available substrate (data not shown).21 In assays using chitotriose, chitotetraose, and chitohexaose, no chitin deacetylase activity was observed when 4 μM ECU11_0510 protein was incubated with chitotriose, chitotetraose, and chitohexaose substrates for 60 min in the absence of additional divalent metal cations. No enzyme activity was observed when 5 μM ZnCl2 or 5 μM CoCl2 were added to these assays. When the protein was incubated with 2 mM of all the aforementioned chitooligosaccharides for up to 16 h, no activity was observed under any of the conditions (Fig. 2). This result suggests that the ECU11_0510 protein is unable to deacetylate short chitin oligomers but does not necessarily exclude its role in deacetylation of chitin polymers, other polysaccharides, or a carbohydrate binding function.
Figure 2.
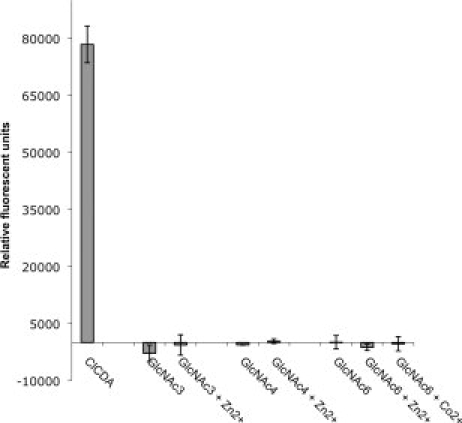
Activity of ECU11_0510 against oligomeric N-acetyl glucosamine (NAG) in the presence or absence of metal ions. ECU11_0510 (4 μM) was assayed using a fluorescamine-based method for 16 h at 37°C with 2 mM of NAG substrates and 5 μM. Colletotrichum lindemuthianum chitin deacetylase (ClCDA, 500 nM) was used as a positive control and the assayed contained 2 mM NAG3 and 5 μM ZnCl. Experiments were performed in triplicate and standard deviations from the mean are shown as error bars.
For analysis of activity toward β-chitin, a new assay was developed that exploits the fact that conversion of partially deacetylated chitin by family 18 chitinases will yield longer products than their conversion of chitin. This is because of the fact that family 18 chitinases cannot cleave polymer chains with certain deacetylation patterns (DP) (Sørbotten et al., 2005; Horn et al., 2006). After a variety of enzymatic treatments, reaction products were analyzed by MALDI-TOF-MS (see Fig. 3). The results show that the chitinase alone led to production of primarily dimers [and minor amounts of trimers; Fig. 3(A), as expected (Horn et al., 2006)]. A similar result was obtained after pretreatment of the β-chitin with ECU11_0510, suggesting that this protein has no deacetylating activity on β-chitin (see Fig. 3). As a positive control, reactions where β-chitin was incubated with Aspergillus nidulans CDA (AnCDA) and an endo-acting family 18 chitinase from Serratia marcescens chitinase C (ChiC) (Horn et al., 2006; Synstad et al., 2008) successively or simultaneously both yielded short partially deacetylated oligomers (D1A1 and D2A1, D being glucosamine) as major products [Fig. 3(A)]. When β-chitin was first incubated with AnCDA and then treated with ChiC, all observed oligomers had the composition DnA1, which was to be expected because AnCDA is known to deacetylate all but one of the sugars in chitooligosaccharides [Control reactions in which chitohexaose (A6) was incubated with AnCDA yielded D5A1, whereas incubation with ECU11_0510 did not have any effect (not shown)]. Most importantly, the mass spectrum of this AnCDA -> ChiC two-step reaction showed the presence of longer reaction products with a DP up to 10 [Fig. 3(B)]. These longer products were not observed when β-chitin was incubated with AnCDA and chitinase C simultaneously. In the latter case, only trace amounts of oligomers with a DP of up to six were observed, as was the case for the reaction with only ChiC. Thus, under the conditions of the assay, ChiC works faster than AnCDA, meaning that we do not see an effect of deacetylation on the DP of the product when both enzymes are added simultaneously. All in all, these observations clearly show that AnCDA is capable of deacetylating chitin, whereas EcCDA is not.
Figure 3.
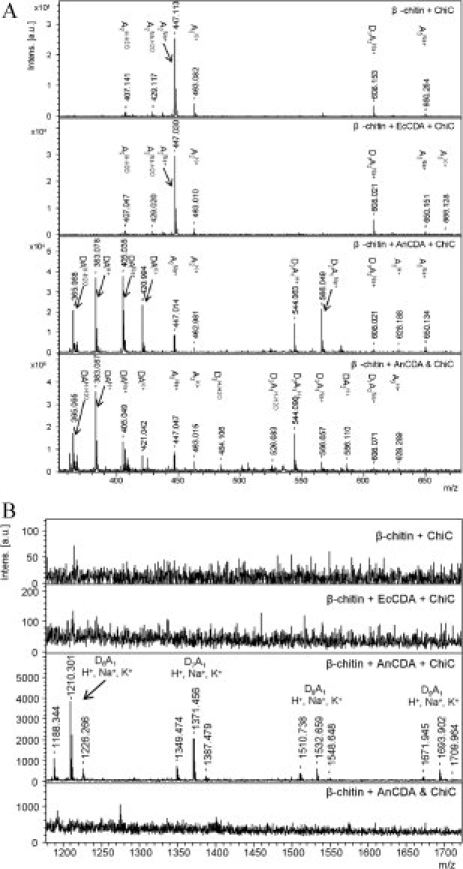
MALDI-TOF-MS analysis of reaction products obtained after treating β-chitin with various enzymes. Sample codes: ChiC, treated with ChiC only; EcCDA + ChiC and AnCDA + ChiC, treated first with the CDA and then with ChiC; AnCDA and ChiC, treated with both enzymes simultaneously. (A) m/z 350–670 region. Note that the partially deacetylated D1A2 species are not a result of enzymatic deacetylation, because they are also observed in the sample treated with ChiC only. (B) m/z 1180–1720 region. Note the difference in scale on the Y-axis. Also note that all the longer species observed in the AnCDA+ChiC sample have the composition DnA1, because of the fact that the AnCDA will deacetylate any DxAy oligosaccharide emerging during the chitinase reaction to DnA1. Oligosaccharides such as D9A1 are not likely to be produced directly from deacetylated chitin because the chitinase has little or no ability to cleave highly acetylated substrates (Sørbotten et al., 2005; Horn et al., 2006).
The ECU11_0510 reveals structural plasticity in the CE4 esterase family
To understand why, despite showing significant homology to the CE4 esterase family, ECU11_0510 does not deacetylate chitooligosaccharides, the crystal structure of the enzyme was determined. The protein was crystallized in the presence of 10 mM DTT and 10 mM ZnCl2. Synchrotron diffraction data were collected to 1.5 Å, the structure solved by molecular replacement and refined to a final model with an R-factor of 0.197 (Rfree = 0.214), and good geometry [RMSD (root mean square deviation) from ideal bond lengths and angles is 0.01 Å and 1.4°, respectively]. Two residues, Glu15 and Ala16, were introduced at the N-terminus as a result of the nucleotide sequence of the pPICZαA expression plasmid. Thirteen residues (221–233) at the C-terminus did not have well-defined density and were not modeled in the structure. The corresponding amino acids at the C-terminus of the structures of SpPgdA and ClCDA also do not form any secondary structure.21,22
The ECU11_0510 structure revealed a compact single domain similar to the deformed (β/α)8 fold adopted by other CE4 family members [Fig. 1(B)]. Superposition on the structure of ClCDA gives an RMSD of 2.2 Å on 194 equivalenced Cα atoms. However, there are a few notable differences between the ECU11_0510 and ClCDA structures [Fig. 1(B)]. Firstly, the β1 strand of ECU11_0510 is displaced by 6 Å compared with the β1 strand of the ClCDA protein. One of the two N-terminal residues introduced from the expression vector sequence, Ala16 form part of the ECU11_0510 β1 strand [Fig. 1(A,B)]. Ala16 forms two hydrogen bonds from its backbone to the backbone of Val134. There is no structural equivalent of Val134 in the ClCDA, as the aligned residue, Cys148, is part of an intramolecular disulphide bridge. In ClCDA, two hydrogen bonds between the first five amino acids and its β6 strand, anchor the N-terminus into the alternative position. Secondly, the insertion of two amino acids in Motif 3 creates an additional strand (β6) between the β5 strand and the α4 helix in ECU11_0510 (see Fig. 1). This causes significant shifts in the backbone of the β5-α4 loop. Thirdly, in the area between the α4 helix and the α5 helix, ECU11_0510 is missing the β7 strand present in ClCDA. This strand is positioned at the start of motif 4, which contains a conserved aspartic acid essential for catalysis.
In addition, one of the two disulfide bridges present in ClCDA is also observed in the ECU11_0510 protein, linking the N- and C-termini [Fig. 1(B)]. Two further cysteines, Cys97 and Cys130, are found on the surface of the protein and participate in the formation of crystal contacts by coordinating zinc.
The ECU11_0510 active site is not compatible with amidase activity
The sequence conservation generated from aligning 12 CE4 esterases was mapped on to the surface of the ECU11_0510 and ClCDA structures [Fig. 1(C)]. The results showed that ClCDA contains a small patch of highly conserved residues covering the catalytic residues and the putative oligosaccharide binding site, formed by a well defined groove. In contrast that the ECU11_0510 protein contains only a minority of conserved residues within the active site. Furthermore, the surface in this region is very smooth and there is no active site groove that would be conducive to substrate binding.
A superposition of the ECU11_0510 protein onto the ClCDA structure shows significant differences in the active site region (see Fig. 4). The first and most obvious difference was that despite the growth of ECU11_0510 crystals in the presence of 10 mM ZnCl2, no electron density was observed for an octahedrally coordinated divalent metal cation within the active site. Analysis of the residues in motifs 1 and 2 involved in metal binding in other CE4 esterases show that only Asp34 of motif 1 is conserved, and maintains a position similar to the metal coordinating residues of other CE4 esterases (see Fig. 4). Two residues in motif 2 (Arg86 and Asp92) that align with the zinc coordinating residues of ClCDA, form a salt bridge with each other and therefore in the ECU11_0510 protein they are not positioned correctly to form a metal binding site with Asp34 (see Fig. 4). Surprisingly, a zinc ion is present close to the active site region of ECU11_0510, ∼5.4 Å away from the metal ions observed in other CE4 esterases. The zinc atom coordinates to His185, which aligns with the histidine that in other CE4 esterases is thought to act as a catalytic acid within the GABC mechanism, and Cys97′ from a symmetry related molecule (see Fig. 4).
Figure 4.
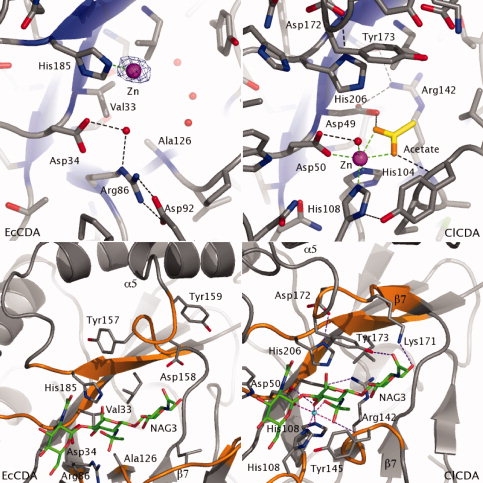
Active site differences between the E. cuniculi and C. lindemuthianum chitin deacetylases. Top Panel: Comparison of the active sites of ECU11_0510 and ClCDA proteins. β strands are shown in blue and cartoon representation. Zinc ions are shown as magenta spheres and waters as red spheres. An anomalous difference electron density map contoured at 5σ identifies the zinc ion at the surface of the ECU11_0510 protein. An acetate present in the ClCDA structure is shown with carbons colored yellow. Catalytically important and metal binding residues are labeled. Metal coordinating interactions are shown as green dotted lines, all other interactions are shown as black dotted lines. Bottom panel: Conformational change of Tyr159. The secondary structure of ECU11_0510 and ClCDA is shown as a grey ribbon, with residues conserved in the CE4 family colored orange. The chitotrioside reaction intermediate model is shown as sticks with green carbons. Residues lining the putative substrate binding cleft are shown as sticks with grey carbons.
Another major difference between the two structures is that the position of the catalytic base of ClCDA (Asp49) is occupied by Val33 in ECU11_0510 (see Fig. 4). No other residue capable of acting as the general base in catalysis is observed near the active site of ECU11_0510. Furthermore, a previous study has shown that the catalytic acid of CE4 esterases is activated by a conserved arginine residue (Arg142 in ClCDA) and hydrogen bonding interactions between its side chain and the catalytic base are observed (see Fig. 4).22 Ala126 replaces this residue in the E. cuniculi protein and this amino acid is not able to perform a similar role (see Fig. 4). In the ECU11_0510 structure three water molecules (226, 254, and 266) lie in similar positions to the Arg142 side chain of ClCDA, filling a pocket within the centre of the protein.
Interestingly, both the catalytic acid (His185) and Asp158, which interacts with the catalytic acid, are conserved in the E. cuniculi protein. The Nɛ atoms of the catalytic histidines in ECU11_0510 and ClCDA occupy similar positions. However, the activating Asp158 has shifted significantly, away from the active site [Fig. 4(B)], with a 7.1 Å shift in the position of the Cα atoms of the aspartate residues in ECU11_0510 and ClCDA proteins. The difference in the position of this Asp residue is due to a movement of the β7-α5 loop, most likely caused by a region which is eight amino acids shorter in this protein (see Fig. 1). The different conformation of the β7-α5 loop also affects the position of the neighboring conserved aromatic residue, Tyr159, which is thought to contribute to hydrophobic stacking interactions with the sugar rings of the polysaccharide substrate in ClCDA (Tyr173, Fig. 4).21 In the ECU11_0510 protein, Tyr159 is positioned 11 Å away from the active site, and no other similarly exposed aromatic residue is present to form such interactions with the substrate.
Does ECU11_0510 bind other carbohydrate ligands?
Although the putative active site of ECU11_0510 does not appear to be compatible with chitooligosaccharide deacetylation, it is possible that the protein has evolved from a carbohydrate active enzyme to a lectin, binding carbohydrate structures within the E. cuniculi cell wall. It has been shown recently that the microsporidian spore wall contains glycosylated proteins, decorated by O-mannosylation with up to eight α1,2 linked mannoses.32 To test this hypothesis, the recombinant ECU11_0510 protein produced in Pichia pastoris was analyzed for its ability to bind over 250 glycans on carbohydrate microarrays by the Functional Glycomics Consortium. The top four hits for ECU11_0510 binding on the microarray were all human serum glycoproteins [Fig. 5(B)]. Typically, each of these glycoproteins has a different assortment of carbohydrate chains. This may suggest that either ECU11_0510 is able to bind to a wide range of sugars present on the surface of these proteins or that it binds nonspecifically to these proteins.
Figure 5.
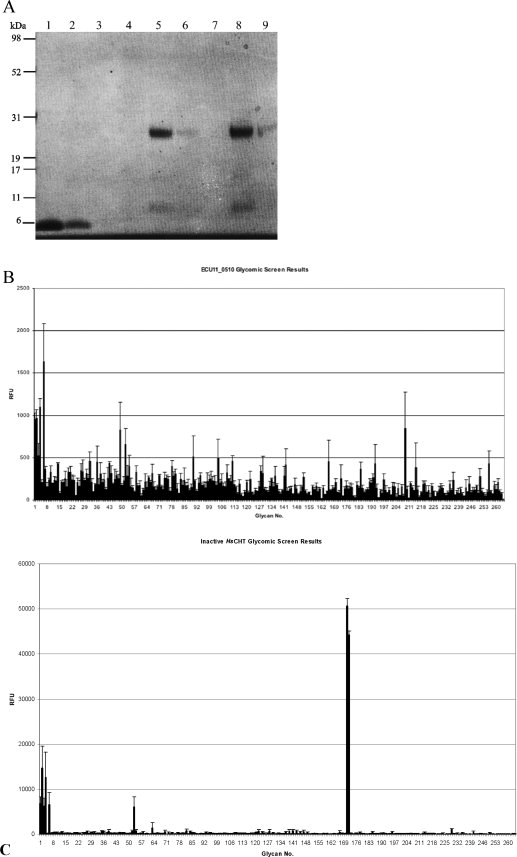
(A) SDS-PAGE analysis of the fourth domain of the Vibrio cholerae GbpA protein and Encephalitozoon cuniculi ECU11_0510 binding to chitin beads. Lanes 1–3, 8 kDa VcGbpA fourth domain (incubated for 16 h at 4°C) and lanes 4–6, 28 kDa ECU11_0510 (incubated for 16 h at 4°C) and lanes 7–9, 28 kDa ECU11_0510 (incubated for 4 h at 37°C). Lanes 1, 4, and 7, washed chitin beads; lanes 2, 5, and 8, supernatant after incubation and lanes 3, 6, and 9, supernatant after first wash of beads. The majority of the Vibrio cholerae GbpA domain is bound to the chitin beads with a smaller amount present in the supernatant. None of the ECU11_0510 protein has bound to the chitin beads at either temperature. (B/C) Histogram of glycomic screen hits. The mean relative fluorescence units (rfu) values observed when Alexa Fluor labelled ECU11_0510 (B) or inactive HsCHT (D138V/E140I, panel C) bound to the corresponding carbohydrates on printed Array Version 2, containing 264 carbohydrate ligands. Each ligand was tested in six replicates. The mean rfu values shown here are calculated after removal of the maximum and minimum replicate rfu values. Selected glycan numbers are as follows; No. 1 = Alpha1-acid glycoprotein (AGP), No. 2 = AGP-A (AGP ConA flowthrough), No. 4 = Ceruloplasmin, No. 6 = Transferrin, No. 171 = (GlcNAcb1-4)6β-Sp8 and No. 172 = (GlcNAcβ1-4)5β-Sp8.
The relative fluorescent units (rfu) observed when ECU11_0510 binds to these glycoproteins is much lower than that observed when a protein with known carbohydrate binding capacity binds to a bonafide ligand on the microarray. For example, an inactive mutant (D138V/E140I) of human chitotriosidase (HsCHT) (Ref.35) was analyzed under the same conditions and the rfu value observed when it binds chitopentaose is one order of magnitude higher than those seen when ECU11_0510 binds to transferrin [Fig. 5(C)]. Additionally, relatively low rfu values are observed for the top hits of the ECU11_0510 protein and this may indicate that binding to these glycoproteins is not significant. Indeed, HsCHT exhibited similar or up to 14 times greater rfu values against the same human serum glycoproteins than those observed with ECU11_0510. Despite these generally higher levels of background rfu values, known chitooligosaccharide ligands of HsCHT exhibited much greater rfu values on the microarray.
To test the hypothesis that ECU11_0510 may bind to high molecular weight chitin, not present on this microarray, both the ECU11_0510 and the chitin binding domain of the VcGbpA protein were analyzed for their capacity to bind to polystyrene beads coated in chitin (average size 750 kDa) (NEB). Both proteins were added to 50% of the binding capacity of the beads, incubated at 4°C for 16 h and 37°C for 1 h before washing. Analysis of binding to chitin beads by SDS-PAGE [Fig. 5(A)] showed that the majority of the 8 kDa VcGbpA chitin binding domain remained bound to the chitin beads and a small amount of this protein was observed in the supernatant. No protein was observed in the first wash of the beads suggesting it was tightly bound to the beads. In contrast, when the 24 kDa ECU11_0510 protein was incubated at either 4 or 37°C, no binding to the beads was observed [Fig. 5(A)]. The vast majority is observed within the supernatant fractions suggesting that this protein is unable to bind to the high molecular weight chitin present on the surface of the beads.
Discussion
Here, we have reported the biochemical and structural characterization of the only E. cuniculi carbohydrate esterase family 4 (ECU11_0510) annotated within the CAZY database (www.cazy.org). We previously reported the production of a recombinant form of the ECU11_0510 protein.19 However, the original construct containing residue 32–329 expressed poorly and produced insufficient quantities for protein crystallography studies. This low level of expression can be explained by analysis of the structure (see Fig. 1). This shows that the β2 strand (residues 27–33) that contributes to motif 1 would have been disrupted within the original construct. Furthermore, the original construct lacks highly conserved cysteine (Cys22) of fungal chitin deacetylases and the intramolecular disulphide bridge with Cys237 that we observe in the structure could not have formed. Expression of the region encoding amino acids 17–233 in a Pichia pastoris expression system described in this article produced extremely high yields and enabled further characterization.
The deacetylase activity of the protein toward chitooligosaccharides was investigated using a fluorescamine-based assay that measured the production of free amines.22 Although the chitin deacetylase from the fungi Colletotrichum lindemuthianum showed significant deacetylase activity against chitooligosaccharides, the E. cuniculi protein exhibited no de-N-acetylase activity. Lack of activity toward β-chitin was confirmed using an MS-based assay. Apart from confirming that ECU11_0510 does not work on chitohexaose, this assay showed that while AnCDA could deacetylate crystalline β-chitin, ECU11_0510 could not. Structural analysis revealed that several known CE4 esterase catalytic and metal binding residues are missing from active site of the E. cuniculi protein, suggesting that the ECU11_0510 protein is not an active chitin deacetylase. Because of the mutated catalytic machinery it is improbable that this enzyme would be able to catalyze the deacetylation of any substrate, unless a second separate active site is present.
The lack of CE4 esterase activity does not rule out the possibility that the ECU11_0510 protein has evolved into a lectin, as reported, for instance, for inactive homologues of chitinases in mammals.33–36 There is already evidence to suggest that the protein may be involved in development of the endospore layer in E. cuniculi. After infection of a host cell, the parasite proliferates to form meronts that differentiate into sporonts (sporogony) then sporoblasts. The latter ultimately transform into mature spores where the deposition of chitin in the cell wall is first observed. Immunocytochemical experiments revealed that during sporogony the ECU11_0510 protein is regularly distributed at the interface between the plasma membrane and the chitinous endospore.19 Therefore, it is attractive to hypothesize that ECU11_0510 may be involved in the deposition and modeling of chitin within the endospore during sporogony within the human host cell or in the spore wall structure maintenance by binding to spore wall glycoproteins. To investigate this hypothesis the binding of ECU11_0510 to polymeric chitin and oligomeric chitin on carbohydrate microarrays was investigated. These studies suggested that ECU11_0510 lacks the ability to bind short chitooligosaccharides and polymeric chitin.
Screening of 256 carbohydrate ligands, predominantly present in humans, was performed in an attempt to identify any such ligands. No significant binding was observed to any of the carbohydrates on these microarrays. The results of all of the binding studies were corroborated by the analysis of the crystal structure. Three aromatic residues (Trp79, Tyr145, and Tyr173) thought to be involved in forming stacking interactions between the sugar substrate and the ClCDA are missing from the active site of ECU11_0510. We cannot exclude that this lack of binding to carbohydrates of human origin is linked to the nature of the glycosylation. Recently, in E. cuniculi we detected only O-linked glycans which consist of fungi-like linear mano-oligosaccharides containing up to eight α1,2-linked mannose residues.32 The glycomic screen contained several carbohydrates that contained two α1,2-linked mannose residues (Glycan numbers 189, 194, and 196) but longer chains were not present. No significant binding was observed to these short mannose chains [Fig. 5(B)].
Analysis of chitin deacetylase protein sequence of other fungi and a second, distantly related, Microsporidian parasite, Antonospora locustae, shows that the putative E. cuniculi enzyme has one striking feature that distinguishes it from all of the other CE4 esterases. ECU11_0510 is the only member of the entire CE4 family (>1700 sequences in the current CAZY database) where the aspartic acid thought to act as the catalytic base is mutated. As a result of its parasitism, E. cuniculi has one of the smallest and highly condensed eukaryotic genomes.37 Moreover, comparison of E. cuniculi and A. locustae genome sequences has suggested that Microsporidia paradoxically possess rapidly evolving genes in slowly evolving genomes.7 Therefore, it is tempting to suggest that the ECU11_0510 protein was once an active CE4 esterase, essential to the ancestor of the parasite. Interestingly, studies of yeast and fungal CDA knockouts have revealed that they are essential for spore germination and/or the separation of mother and daughter cells.20,30,31 The product of chitin deacetylation, chitosan, is a much more flexible polymer than chitin. Indeed, chitosan synthesis is regulated and increases during the infection of plant leaves by the maize fungal pathogen Colletotrichum graminicola.38 However, in E. cuniculi, the development of the unique polar tube invasion mechanism negates any need for a more motile cell wall during invasion. Furthermore, the rigidity of the infectious spore is enhanced by the presence of chitin in the endospore, and this important property for spore survival would be disrupted by chitosan production. As the parasite evolved, the CE4 esterase may have become functionally obsolete. It is tempting to speculate that the protein we have studied currently is evolving/has evolved toward a new and as yet unidentified function, which should be important enough for the parasite to maintain it. Studies of a knockout of the ECU11_0510 gene would help to elucidate the function of this enigmatic protein. We are currently developing methods with the aim to producing knockouts in E. cuniculi, which would be of considerable use in further studies of this emerging human pathogen.
Materials and Methods
Cloning of the ECU11_0510 gene
E. cuniculi was grown in vitro on MDCK (Madin-Darby canine kidney) cells, in minimum essential medium (MEM) supplemented with 5% fetal calf serum and 2 mM glutamine at 37°C in a 5% CO2 atmosphere. Spores were harvested from supernatants (18,000 g, 2 min), washed and stored in phosphate buffered saline (PBS) at 4°C. E. cuniculi genomic DNA was purified using the Elu-Quik Glaspulver Kit (Schleider and Schuell) and resuspended in water and used to amplify the entire ORF of the ECU11_0510 protein using the following primers:
11-0510D 5′-AAAAAGCAGGCTCCGCCATGTTAC TTTGCCTACTATAC-3′ and 11-0510R 5′-AGAAAGCTG GGTTTACAGAAGCCTTACTAAC-3′.
A second PCR was performed using the first PCR product as template with the aim of extending the region homologous to the plasmid pDONR221. We used the following primers:
attB1 5′-GGGGACAAGTTTGTACAAAAAAGCAGG CT-3′ and attB2 5′-GGGGACCACTTTGTACAAGAAA GCTGGGT-3′.
The second PCR product was inserted in the pDONR221 vector (Invitrogen) by recombination (BP reaction). The pDONR221 ECU11_0510 plasmid was used as template to amplify a region corresponding to amino acids 17–233, using the following primers: ECU11_051017_For 5′-CTCGAGAAAAGAGAGGCTGAA GCTGATGTCCCTGACGTGTGCAC-3′ and ECU11_0510 233_Rev 5′-CCGCGGTCATCACTTTCCTCTCAGGCT GAG-3′. A 5′ XhoI and 3′ SacII site are shown in italics and two stop codons in the reverse primer are shown in bold. A secretory peptide cleavage sequence of amino acids was encoded in the forward primer (underlined) for cloning into the P. pastoris secretory expression vector, pPICZαA (Invitrogen).
The nucleotide sequence of the PCR fragment was determined by automated nucleotide sequencing on an automatic sequencer (ABI 377, BioRad). An insert was excised from the pDONR221 ECU11_0510 plasmid by digestion with XhoI and SacII and ligated into the pPICZαA plasmid digested with the same enzymes.
Expression and purification
Competent X-33 Pichia cells were produced using the LiCl method described by the supplier (Invitrogen). A total of 5 μg of the pPICZαA ECU11_0510 Δ17–233 plasmid was linearized with PmeI at 37°C for 2 h and transformed into the competent X33 cells according to the manufacturer's instructions. Transformed cells were incubated in 1 mL of YPD at 30°C for 1 h. Subsequently, 100 μL was removed and plated onto YPD plates containing zeocin (100 μg/mL) and incubated at 30°C. Colonies appeared after 3 days.
A single colony was used to inoculate 2 mL × 100 mL of BMGY media, and the cultures were grown overnight at 30°C. Cells were collected by centrifugation at 3500 g, 30 min, 4°C, resuspended and used to inoculate 2 mL × 500 mL of BMMY media in 5 L flasks. Cultures were incubated at room temperature at 150 rpm, with the addition of 5 mL (1% v/v) methanol every 24 h. After 4 days, cells were removed by centrifugation as above, and the supernatant was passed through 0.8 μm and 0.2 μm filter units. The filtrate was concentrated using a Vivaflow 50 mL 10,000 Mw cutoff concentrator (Sartorius) and dialyzed twice against 5 L of 25 mM Tris, pH 8.0, at 4°C. A final dialysis step into 5 L of 25 mM Hepes, pH 8.0, 25 mM NaCl, 4°C for 4 h was performed.
Approximately 40 mg (20 mL) of the dialyzed sample was loaded on to a Hi-trap QC anion exchange column, previously equilibrated with five column volumes of 25 mM Hepes, pH 8.0, 25 mM NaCl. Proteins were eluted using a gradient of NaCl from 25 to 500 mM over 20 column volumes and 3 mL fractions were collected. The major contaminating proteins were eluted before 150 mM NaCl had passed through the column. Recombinantly produced ECU11_0510 was present in the fractions containing 190–480 mM NaCl. The corresponding fractions were pooled and concentrated to a 5 mL volume using a Vivaspin 10,000 Mw cutoff filter and loaded onto a Superdex 75 26/60 gel filtration column equilibrated with 25 mM Hepes, 150 mM NaCl. Two peaks at OD280 were observed and a calibration curve suggested these were close to the theoretical molecular weights of monomeric and dimeric forms of ECU11_0510. Fractions containing the ECU11_0510 monomer were pooled and concentrated to 19.4 mg/mL as above.
Fluorescamine-based assay of activity toward chitooligosaccharides
Both dimeric and monomeric forms of ECU11_0510 were tested for chitin deacetylase activity using a 96-well plate assay previously reported by Blair et al.22 Standard reactions consisted of 4 μM chitin deacetylase, 5 μM CoCl2 or ZnCl2, 50 μM Bis-Tris (pH 7.0) and 2 mM chitooligosaccharide (Sigma)in a total volume of 50 μL, incubated for 1 h or 16 h at 37°C. The reactions were stopped by the addition of 0.4M borate buffer, pH 9.0. Free amines were labeled by the addition of 20 μL of 2 mg/mL fluorescamine in dimethylformamide (DMF) for 10 min at room temperature. The addition of 150 μL H2O/DMF (1:1) stopped the labeling reaction. Fluorescence was quantified using a FLX 800 Microplate Fluorescence Reader (Bio-Tek, Burlington, USA), with excitation and emission wavelengths of 360 and 460 nm, respectively. A total of 500 nM of recombinant ClCDA purified as described previously was tested under identical conditions as a positive control.39 All measurements were performed in triplicate.
Activity assay against β-chitin
Squid pen β-chitin (France Chitin, Marseille, France) was incubated with recombinant ECU11_0510 or AnCDA, a purified recombinant chitin deacetylase from Aspergillus nidulans (gene accession code AN9380.2, details to be published elsewhere), at 37°C overnight, and subsequently incubated with ChiC, an endo-acting family 18 chitinase from Serratia marcescens that is capable of cleaving partially deacetylated chitin (Horn et al., 2006; Synstad et al., 2008), for another 24 h. Standard reactions in a final volume of 50 μL consisted of 5 mg β-chitin, 50 μM CoCl2 and 500 nmol of deacetylase in 20 mM Tris, pH 7.0. Addition of the chitinase (500 nmol) increased the reaction volume by 3 μL. Two control reactions were run in parallel: β-chitin incubated with ChiC solely, and β-chitin incubated with AnCDA and ChiC simultaneously. The reactions were stopped by boiling for 3 min. After centrifugation, oligosaccharide products in the liquid phase were analyzed using MALDI-TOF-MS, using an Ultraflex instrument (Bruker Daltonics) with a Nitrogen 337 nm laser beam operated in positive acquisition mode, using 2,5-dihydrobenzoic acid as matrix.
Carbohydrate microarray analysis
Recombinant ECU11_0510 protein and an inactive mutant of full length Homo sapiens HsCHT D138V/E140I mutant protein was generated by site-directed mutagenesis (Ref.35) and analyzed for glycan binding by Core H of the Consortium of Functional Glycomics (http://www.functionalglycomics.org) using the Printed Array Version 2. This microarray contains a library of 264 natural and synthetic glycans with amino linkers, printed onto chemically-modified glass microscope slides. Glycans were spotted at 100 μM concentrations on the array. The rECU11_0510 and HsCHT proteins were labeled with Alexa Fluor 488 using the Invitrogen Alexa Fluor 488 Protein labeling kit. Both labeled proteins were incubated on the glycan slides for 1 h under a cover slip at room temperature in a humidified chamber and the fluorescence of bound protein was analyzed after several washing steps. Samples were washed four times each in the following three solutions: (1) 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween-20, (2) Buffer 1 without Tween-20 and (3) deionized water. Slides were spun dry for 30 s and then scanned on the ProScanArray Express from Perkin Elmer using fluorophore Alexa488 (Excitation at 495 nm and Emission measured at 520 nm).
Detection of chitin binding
Recombinant ECU11_0510 protein was incubated with chitin beads (NEB) for either 16 h at 4°C or for 1 h at 37°C. The chitin binding domain of the Vibrio cholerae GbpA protein (kindly provided by Dr. Edmond Wong, Dundee University) (Kirn, Jude et al., 2005) was used as a positive control for chitin bead binding. 100 μL of chitin beads, with a predicted binding capacity of 200 μg protein were incubated with 100 μg of either recombinant ECU11_0510 or VcGbpA protein. This suspension was made to a final volume of 400 μL with 25 mM Tris, pH 8.0, 150 mM NaCl and placed on a slowly rotating platform. After incubation with the protein the beads were collected by centrifugation at 400 g for 3 min and washed three times in the above buffer. The supernatant from the centrifuged suspension and from the first wash of the beads was retained. 15 μL of beads, supernatant and first wash were analyzed on a 10% acrylamide SDS-PAGE gel in MES buffer. The expected size of the ECU11_0510 protein is 24 kDa, whilst the GbpA protein domain is 8 kDa. A small amount of degradation had occurred in the recombinant ECU11_0510 sample and a second band of 10 kDa was observed on the gel.
Crystallization and structure solution
Before crystallization, 10 mM dithiothreitol and 1 mM ZnCl2 were added to both the concentrated ECU11_0510 protin and the reservoir solutions of all crystallization conditions. Crystals were produced with the sitting drop vapor diffusion method using 0.5 μL of protein solution and an equal volume of mother liquor consisting of 10% PEG 8000, 100 mM imidazole and 200 mM calcium acetate. Better quality diffracting crystals formed following the addition of ZnCl2 to a final concentration of 10 mM, producing small rod-like crystals after 2 days. Crystals were cryoprotected by transferring to mother liquor containing 20% PEG 400 and were then frozen directly in a nitrogen cryostream for data collection. Data were collected on beamline ID14-2 at the European Synchrotron Radiation Facility (Grenoble, France) and processed with the HKL suite of programs.40 The data between 20 and 1.50 Å were scaled in P21212 (a = 64.98, b = 81.92, c = 39.20 Å; one molecule per asymmetric unit) with an overall Rmerge of 0.050 (0.374 for the last shell), 99.5% completeness (97.8% for the last shell) and 3.4-fold redundancy (3.1-fold for the last shell). The structure was solved by molecular replacement with AMoRe,41 using the catalytic domain of the SpPgdA structure (PDB entry 2C1G) as a search model against 12–3.5 Å data. The model phases from the top solution (R-factor of 0.523) were used as input for warpNtrace,42 which built 201 of 217 residues. Refinement was performed by REFMAC43 interspersed with model building in COOT.44 Figures were generated using the PyMOL Molecular Graphics System, DeLano Scientific (http://www.pymol.org).
Acknowledgments
The authors thank David Smith and his colleagues at Core H, Functional Glycomics Consortium for the data generated by their microarray analysis. They thank Sharon Shepherd for assistance in protein purification, David Blair for guidance on the fluorescamine assay, Edmond Wong for his kind gift of the VcGbpA protein and Marianne Schimpl for the use of inactive chitotriosidase data. They also thank the European Synchrotron Radiation Facility, Grenoble, for the time at beamline ID14-EH2. The structure has been deposited in the Protein Data Bank (2VYO).
References
- 1.Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeling PJ, Luke MA, Palmer JD. Evidence from β-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 3.Thomarat F, Vivares CP, Gouy M. Phylogenetic analysis of the complete genome sequence of Encephalitozoon cuniculi supports the fungal origin of microsporidia and reveals a high frequency of fast-evolving genes. J Mol Evol. 2004;59:780–791. doi: 10.1007/s00239-004-2673-0. [DOI] [PubMed] [Google Scholar]
- 4.Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 2005;94:61–76. doi: 10.1016/j.actatropica.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Keeling PJ, Fast NM. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 6.Dascomb K, Clark R, Aberg J, Pulvirenti J, Hewitt RG, Kissinger P, Didier ES. Natural history of intestinal microsporidiosis among patients infected with human immunodeficiency virus. J Clin Microbiol. 1999;37:3421–3422. doi: 10.1128/jcm.37.10.3421-3422.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira FM, Bezerra L, Santos MB, Bernardes RM, Avelino I, Silva ML. Intestinal microsporidiosis: a current infection in HIV-seropositive patients in Portugal. Microbes Infect. 2001;3:1015–1019. doi: 10.1016/s1286-4579(01)01465-4. [DOI] [PubMed] [Google Scholar]
- 8.Kottler DP, Orentstein JM. Clinical syndromes associated with microsporidiosis. In: Wittner M, editor. The microsporidia and microsporidiosis. Washington, DC: ASM Press; 1999. pp. 258–292. [Google Scholar]
- 9.Couzinet S, Cejas E, Schittny J, Deplazes P, Weber R, Zimmerli S. Phagocytic uptake of Encephalitozoon cuniculi by nonprofessional phagocytes. Infect Immun. 2000;68:6939–6945. doi: 10.1128/iai.68.12.6939-6945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franzen C, Muller A, Hartmann P, Salzberger B. Cell invasion and intracellular fate of Encephalitozoon cuniculi (Microsporidia) Parasitology. 2005;130:285–292. doi: 10.1017/s003118200400633x. [DOI] [PubMed] [Google Scholar]
- 11.Leitch GJ, Ward TL, Shaw AP, Newman G. Apical spore phagocytosis is not a significant route of infection of differentiated enterocytes by Encephalitozoon intestinalis. Infect Immun. 2005;73:7697–7704. doi: 10.1128/IAI.73.11.7697-7704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzen C. Microsporidia: how can they invade other cells? Trends Parasitol. 2004;20:275–279. doi: 10.1016/j.pt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Koudela B, Kucerova S, Hudcovic T. Effect of low and high temperatures on infectivity of Encephalitozoon cuniculi spores suspended in water. Folia Parasitol (Praha) 1999;46:171–174. [PubMed] [Google Scholar]
- 14.Shadduck JA, Polley MB. Some factors influencing the in vitro infectivity and replication of Encephalitozoon cuniculi. J Protozool. 1978;25:491–496. doi: 10.1111/j.1550-7408.1978.tb04174.x. [DOI] [PubMed] [Google Scholar]
- 15.Bigliardi E, Selmi MG, Lupetti P, Corona S, Gatti S, Scaglia M, Sacchi L. Microsporidian spore wall: ultrastructural findings on Encephalitozoon hellem exospore. J Eukaryot Microbiol. 1996;43:181–186. doi: 10.1111/j.1550-7408.1996.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 16.Vavra J. Structure of the microsporidia. In: Bulla LA Jr, Cheng TC, editors. Comparative pathology, Vol. 1. Biology of the microsporidia. London: Plemum Press; 1976. pp. 1–86. [Google Scholar]
- 17.Vavra J, Larson J. Structure of the microsporidia. In: Wittner M, editor. The microsporidia and microsporidiosis. Washington, DC: ASM Press; 1999. pp. 7–84. [Google Scholar]
- 18.Brosson D, Kuhn L, Delbac F, Garin JCPV, Texier C. Proteomic analysis of the eukaryotic parasite Encephalitozoon cuniculi (microsporidia): a reference map for proteins expressed in late sporogonial stages. Proteomics. 2006;6:3625–3635. doi: 10.1002/pmic.200500796. [DOI] [PubMed] [Google Scholar]
- 19.Brosson D, Kuhn L, Prensier G, Vivares CP, Texier C. The putative chitin deacetylase of Encephalitozoon cuniculi: a surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol Lett. 2005;247:81–90. doi: 10.1016/j.femsle.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blair DE, Hekmat O, Schuttelkopf AW, Shrestha B, Tokuyasu K, Withers SG, van Aalten DM. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry. 2006;45:9416–9426. doi: 10.1021/bi0606694. [DOI] [PubMed] [Google Scholar]
- 22.Blair DE, Schuttelkopf AW, MacRae JI, van Aalten DM. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc Natl Acad Sci USA. 2005;102:15429–15434. doi: 10.1073/pnas.0504339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blair DE, van Aalten DM. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Lett. 2004;570:13–19. doi: 10.1016/j.febslet.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Kafetzopoulos D, Martinou A, Bouriotis V. Bioconversion of chitin to chitosan: purification and characterization of chitin deacetylase from Mucor rouxii. Proc Natl Acad Sci USA. 1993;90:2564–2568. doi: 10.1073/pnas.90.7.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor EJ, Gloster TM, Turkenburg JP, Vincent F, Brzozowski AM, Dupont C, Shareck F, Centeno MS, Prates JA, Puchart V, Ferreira LM, Fontes CM, Biely P, Davies GJ. Structure and activity of two metal-ion dependent acetyl xylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. J Biol Chem. 2006;281:10968–10975. doi: 10.1074/jbc.M513066200. [DOI] [PubMed] [Google Scholar]
- 26.Tsigos I, Zydowicz N, Martinou A, Domard A, Bouriotis V. Mode of action of chitin deacetylase from Mucor rouxii on N-acetyl chitooligosaccharides. Eur J Biochem. 1999;261:698–705. doi: 10.1046/j.1432-1327.1999.00311.x. [DOI] [PubMed] [Google Scholar]
- 27.Vollmer W, Tomasz A. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J Biol Chem. 2000;275:20496–204501. doi: 10.1074/jbc.M910189199. [DOI] [PubMed] [Google Scholar]
- 28.Vollmer W, Tomasz A. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect Immun. 2002;70:7176–7178. doi: 10.1128/IAI.70.12.7176-7178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prevost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christodoulidou A, Bouriotis V, Thireos G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J Biol Chem. 1996;271:31420–31425. doi: 10.1074/jbc.271.49.31420. [DOI] [PubMed] [Google Scholar]
- 31.Christodoulidou A, Briza P, Ellinger A, Bouriotis V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999;460:275–279. doi: 10.1016/s0014-5793(99)01334-4. [DOI] [PubMed] [Google Scholar]
- 32.Taupin V, Garenaux E, Mazet M, Maes E, Denise H, Prensier G, Vivares CP, Guerardel Y, Metenier G. Major O-glycans in the spores of two microsporidian parasites are represented by unbranched manno-oligosaccharides containing alpha-1,2 linkages. Glycobiology. 2007;17:56–67. doi: 10.1093/glycob/cwl050. [DOI] [PubMed] [Google Scholar]
- 33.Harbord M, Novelli M, Canas B, Power D, Davis C, Godovac-Zimmermann J, Roes J, Segal AW. Ym1 is a neutrophil granule protein that crystallizes in p47phox-deficient mice. J Biol Chem. 2002;277:5468–5475. doi: 10.1074/jbc.M110635200. [DOI] [PubMed] [Google Scholar]
- 34.Bigg HF, Wait R, Rowan AD, Cawston TE. The mammalian chitinase-like lectin, YKL-40, binds specifically to type I collagen and modulates the rate of type I collagen fibril formation. J Biol Chem. 2006;281:21082–21095. doi: 10.1074/jbc.M601153200. [DOI] [PubMed] [Google Scholar]
- 35.Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, Aerts JM, van Aalten DM. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J Biol Chem. 2002;277:25537–25544. doi: 10.1074/jbc.M201636200. [DOI] [PubMed] [Google Scholar]
- 36.Houston DR, Recklies AD, Krupa JC, van Aalten DM. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J Biol Chem. 2003;278:30206–30212. doi: 10.1074/jbc.M303371200. [DOI] [PubMed] [Google Scholar]
- 37.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 38.El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 2002;156:103–112. [Google Scholar]
- 39.Shrestha B, Blondeau K, Stevens WF, Hegarat FL. Expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris: purification and characterization. Protein Expr Purif. 2004;38:196–204. doi: 10.1016/j.pep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Navaza J, Saludjian P. AMoRe: an automated molecular replacement program package. Macromol Crystallogr A. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 42.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Horn SJ, Sørbotten A, Sikorski P, Sørlie M. Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia Marcescens. FEBS J. 2006;273:491–503. doi: 10.1111/j.1742-4658.2005.05079.x. [DOI] [PubMed] [Google Scholar]
- 46.Sørbotten A, Varum KM. Degradation of chitosans with chitinase B from Serratia marcescens. Production of chito-oligosaccharides and insight into enzyme processivity. FEBS J. 2005;272:538–549. doi: 10.1111/j.1742-4658.2004.04495.x. [DOI] [PubMed] [Google Scholar]
- 47.Synstad B, Vaaje-Kolstad G, Cederkvist FH, Saua SF, Horn SJ, Eijsink VGH, Sørlie M. Expression and Characterization of Endochitinase C from Serratia Marcescens BJL200 and Its Purification by One-Step General Chitinase Purification Method. Biosci Biotechnol Biochem. 2008;72:715–723. doi: 10.1271/bbb.70594. [DOI] [PubMed] [Google Scholar]
- 48.Kirn TJ, Jude BA, Taylor RK. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature. 2005;438(7069):863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]


