Abstract
Laforin is the only phosphatase in the animal kingdom that contains a carbohydrate-binding module (CBM). Mutations in the gene encoding laforin result in Lafora disease (LD), a fatal autosomal recessive neurodegenerative disorder, which is diagnosed by the presence of intracellular deposits of insoluble complex carbohydrates known as Lafora bodies (LB). We demonstrate that laforin interacts with proteins known to be involved in glycogen metabolism and rule out several of these proteins as potential substrates. Surprisingly, we find that laforin displays robust phosphatase activity against a phosphorylated complex carbohydrate. Furthermore, this activity is unique to laforin as several other phosphatases are unable to dephosphorylate polysaccharides. Finally, fusing the CBM of laforin to the dual specific phosphatase VHR does not result in the ability of this phosphatase to dephosphorylate polysaccharides. Therefore, we hypothesize that laforin is unique in its ability to utilize a phosphorylated complex carbohydrate as a substrate and that this function may be necessary for the maintenance of normal cellular glycogen.
Introduction
Lafora disease (LD, OMIM 254780) is an autosomal recessive neurodegenerative disorder that falls into the broad category of progressive myoclonus epilepsies (1–3). These diseases include Unverricht-Lundborg disease, myoclonic epilepsy with ragged red fibers, neuronal ceroid lipofuscinosis and type I sialidosis, all of which manifest myoclonic seizures, tonic-clonic seizures and progressive neurological dysfunction (4). In each case, the causal gene mutations are known and mouse models have been generated, but despite these advances the molecular mechanisms of the diseases remain unknown.
Two genes have been identified that are mutated in Lafora disease. The first is EPM2A (epilepsy of progressive myoclonus type 2 gene A), which encodes laforin and is responsible for approximately 48% of LD cases (5,6). Laforin is a dual specificity phosphatase that contains an NH2-terminal carbohydrate-binding module (CBM) and a COOH-terminal phosphatase active site motif, HCXXGXXRS/T (CX5R). Accordingly, recombinant laforin displays two functions in that it can bind complex polysaccharides as well as hydrolyze phosphotyrosine and phosphoserine/threonine substrates (7,8). Disease mutations found in the gene encoding laforin include several missense mutations that disrupt the phosphatase activity as well as several that abrogate the ability of the carbohydrate-binding domain to bind complex polysaccharides (7,9–11). A point mutation also exists that reduces the interaction of laforin with a glycogen scaffolding protein, protein targeted to glycogen (PTG) (12). Furthermore, the CBM targets laforin to sites of glycogen metabolism (7), a cellular process historically known to be regulated by phosphorylation. Collectively, these data suggests that both the phosphatase activity and the carbohydrate binding functions are critical for laforin’s function in glycogen metabolism.
The second gene involved in Lafora disease, EPM2B, encodes an E3 ubiquitin ligase, called malin, and is responsible for approximately 40% of LD cases (13,14). Malin is a multidomain protein containing a RING-HC and six NHL domains. RING domains are indicative of a class of E3 ubiquitin ligases while NHL domains form a six-bladed β-propeller involved in protein-protein interactions (15–18). We previously identified laforin as a binding partner of malin and provided evidence that malin binds laforin and polyubiquitinates it both in vitro and in vivo (13). Furthermore, this polyuibiquitination leads to laforin’s degradation in tissue culture cells (13). Lending support to this surprising finding, Chan et al. (19), reported that while laforin cannot be detected in wild type tissues, it could be detected in EPM2B null tissues.
One of the clinical manifestations of LD is the appearance of insoluble carbohydrate deposits called Lafora bodies (LB) in the cytoplasm of nearly all cell types (2,20–24). Because of this and the fact that laforin contains a CBM, it is hypothesized that laforin is involved in glycogen metabolism, either its synthesis or degradation. Normal cells store carbohydrates in the form of glycogen, a polymer of glucose residues linked together by α-1,4-glycosidic linkages with branches occurring every 8–12 residues via α-1,6-glycosidic linkages. This level of branching makes glycogen a homogenous water-soluble polymer. In contrast, while LBs are composed of the same backbone structure as glycogen, there are fewer α-1,6-glycosidic branches (25). This decreased branching gives LBs a crystalline structure and renders them insoluble (25). Additionally, LBs are significantly more phosphorylated than glycogen (26). Surprisingly, while LBs and glycogen differ in multiple structural aspects, LBs and amylopectin appear to be very similar.
Amylopectin is the major component of plant starch and is composed of the same backbone structure as glycogen but with branches occurring every 24–30 glucose residues. This decreased amount of branching also renders amylopectin crystalline and insoluble. Additionally, the glucose monomers of amylopectin are phosphorylated on approximately 1 in every 300 residues at either the C3 or C6 position (27). Strikingly, the definitive biochemical studies on the structure of LBs revealed that LBs are more similar to amylopectin than to any other naturally occurring or synthetic compound, including mammalian glycogen (25,28,29).
In order to understand laforin’s molecular role in glycogen metabolism, we analyzed laforin’s protein-protein interactions in the cell. We further tested interacting proteins for their ability to act as substrates for laforin’s phosphatase activity. Since none of the proteinaceous substrates we tested appeared to be substrates for laforin, we questioned whether laforin could act on a non-proteinaceous substrate. Since LBs are similar to amylopectin, we tested amylopectin as a substrate and demonstrate that laforin effectively removes phosphate from this carbohydrate. We further demonstrate that this activity is specific for the laforin phosphatase and that replacing laforin’s phosphatase domain with that of VHR, an active dual specificity phosphatase, does not confer activity towards amylopectin. Finally, we speculate on the consequences this unexpected activity could have on glycogen metabolism.
Materials and Methods
Plasmids and Proteins
Wild type and C/S FLAG-tagged laforin for use in mammalian expression studies and bacterially expressed laforin in pET21a (Novagen, San Diego, CA) were described previously (7). PTG family members were amplified from ESTs and inserted into the pcDNA3.1/myc-His eukaryotic expression vector (Invitrogen, Carlsbad, CA). HA-tagged GSK3β was a kind gift from David Pagliarini (Harvard, Cambrigde, MA).
Recombinant His-tagged VHR expressed in Escherichia coli BL21 (DE3) CodonPlus RIL cells (Stratagene, La Jolla, CA) was purified using Ni2+-agarose (Qiagen, Germany) as described previously (30). PTPMT1 was a kind gift from David Pagliarini and Ji Zhou (University of CA at San Diego, La Jolla, CA) and dullard was a kind gift from Youngjun Kim (University of CA at San Diego, La Jolla, CA). TCPTP and PP1 were purchased from New England Biolabs (Beverly, MA) and alkaline phosphatase (AlkP) was purchased from Roche Applied Science (Indianapolis, IN).
Potato amylopectin and glycogen were purchased from Sigma (St. Louis, MO).
Cell Culture and Transfection
Adenovirus-transformed human embryonic kidney (HEK) 293T cells were maintained at 37°C with 5% CO2 in DMEM (Invitrogen) supplemented with 10% FBS and 50 units/ml penicillin/streptomycin. CHO cells stably transformed with the insulin receptor, (CHO-IR), were maintained at 37°C with 5% CO2 in Eagle Minimal Essential Media (Invitrogen) containing 10% FBS, 50 units/ml penicillin/streptomycin, and 50 μg/ml Geneticin (Invitrogen).
Subconfluent cultures of HEK293T or CHO-IR cells (1–2 × 106 cells per 100-mm dish) were transfected with FuGENE transfection reagent (Roche Applied Sciences) according to the manufacturer’s protocol. Transfected cells recovered 24 – 48h prior to harvest to allow for protein expression.
Immunoprecipitations (IPs)
Twenty four to 48h after transfection, cells were washed once with ice-cold PBS, drained, and harvested in ice-cold lysis buffer consisting of 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Nonidet P-40 (NP-40), 1mM dithiothreitol (DTT) and Complete protease inhibitor cocktail (Roche Applied Sciences). The cells were lysed by tituration and cleared by centrifugation at 8,000 × g for 10 min. The supernatants were mixed with anti-FLAG M2 affinity resin (Sigma) or anti-myc agarose (Sigma) for 2–4 h at 4°C with constant agitation. The resins were pelleted by centrifugation at 500 × g for 1 min and washed three times with 1 ml lysis buffer. The beads were resuspended in 30 μl of 4X NuPage sample buffer (Invitrogen) and subjected to Western analyses. Western blots were probed with the following antibodies; α-FLAG HRP (Sigma), α-GS (Chemicon, Temecula, CA), α-myc HRP (Roche Applied Sciences), α-HA HRP (Roche applied Sciences), GSK3β α-PSer9 (Biosource, Camarilo, CA) and α-PTyr 4G10 (Upstate, Charlottesville, VA). Goat α-mouse-HRP was used as needed. The HRP signal was detected by using SuperSignal West Pico (Pierce, Rockford, IL).
Isolation of phosphorylated GSK3β
CHO-IR cells were transfected with HA-tagged GSK3β and allowed to recover 24h. Immediately before harvesting, the cells were treated with 50 nM insulin for 5 min. Extracts were prepared as described above and α-HA affinity resin (Roche Applied Sciences) was used to IP GSK3β. The α-HA affinity resin was washed 3X with lysis buffer, 1X with lysis buffer containing 1 M NaCl, 1X with lysis buffer, and 2X with phosphatase buffer. The final product was resuspended in 150 μl of phosphatase buffer (1X phosphatase reaction buffer; 0.1 M sodium acetate, 0.05 M bis-Tris, 0.05 M Tris-HCL, 2 mM DTT, pH 6.5) and 20 μl was used in the phosphatase reaction (30 μl total) containing 500 ng of laforin. Tungstate (1 mM) was added prior to the addition of laforin.
Phosphatase Activity Assays
Hydrolysis of para-nitrophenylphosphate (pNPP) was performed in 50 μl reactions containing 1X phosphatase buffer (above), 50 mM pNPP, and 100 – 500 ng of enzyme at 37°C for 1–5 min. The reaction for dullard also contained 10 mM MgCl2 and the PP1 reaction mix contained 1 mM MnCl2. The reaction was stopped by addition of 200 μl of 0.25 N NaOH. Absorbance was measured at 410nm. Malachite green assays containing 1X phosphatase buffer (MgCl2 or MnCl2 when appropriate), 100–500 ng of enzyme and approximately 45 μg of amylopectin or glycogen were performed in a final volume of 20 μl. Reactions were terminated by addition of 20 μl of 0.1 M N-ethylmaliemide and 80 μl of malachite green reagent. Absorbance was measured at 620nm.
Results and Discussion
GSK3β is not a substrate of laforin
Laforin is unique among phosphatases found in the animal kingdom in that it contains an NH2-terminal starch-binding domain of the subtype CBM20 (31). Accordingly, we previously demonstrated that laforin binds to glycogen in vitro (7). In order to elucidate laforin’s role in cellular signaling, we sought to evaluate which proteins involved in glycogen metabolism would co-immunoprecipitate with laforin, with the idea that co-immunoprecipitating proteins could be potential substrates for the phosphatase.
During the course of this study, it was reported that GSK3β co-immunoprecipitated with laforin and that laforin dephosphorylated Ser9 of GSK3β (32). To test these findings, we transfected HA-tagged GSK3β into HEK293 and CHO-IR cells along with FLAG-tagged laforin and immunoprecipitated laforin using anti-FLAG. Despite robust expression of both laforin and GSK3β, GSK3β did not co-immunoprecipitate with laforin from HEK293 or CHO-IR cells (Fig. 1A, data not shown).
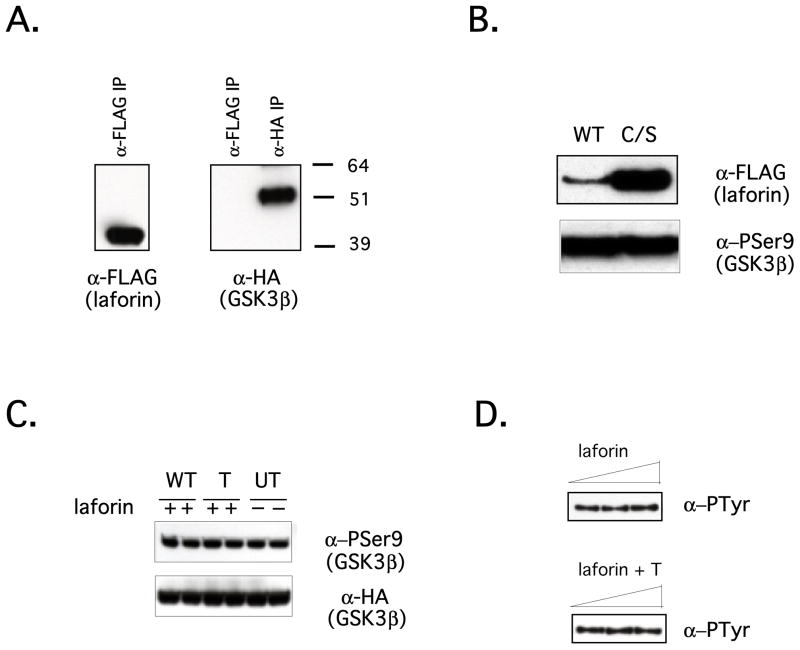
Figure 1.
GSK3β is not a substrate of laforin in vivo or in vitro. (A) HEK293 cells were cotransfected with FLAG-tagged laforin and HA-tagged GSK3β. Western analysis probed with α-FLAG HRP of the FLAG IP is shown in the left panel, while Western analysis probed with α-HA HRP of the FLAG and HA IPs is shown in the right panel. (B) WT and C/S FLAG-tagged laforin were co-transfected along with HA-tagged GSK3β into CHO-IR cells. Western analysis of WCL probed with α-FLAG demonstrates the expression level of laforin (top panel). Western analysis of α-HA immunoprecipitates using an antibody directed against PSer9 of GSK3β is shown in the bottom panel. C/S laforin is consistently expressed at a higher level than WT laforin in all cell types analyzed. (C) WT His-tagged laforin was expressed in and purified from bacteria. HA-tagged GSK3β was immunoprecipitated from CHO-IR cells treated with insulin as described in Materials and methods. Laforin and GSK3β were allowed to react in the presence or absence of tungstate (T) in standard phosphatase assays followed by Western analysis of the samples using α-PSer9 (top panel) or α-HA to assess equal loading (bottom panel). Samples were run in duplicate. (D) HA-tagged GSK3β was immunoprecipitated from transiently transfected HEK239 cells. Increasing amounts of bacterially expressed laforin were allowed to react with immunoprecipitated GSK3β in the absence (top panel) or presence (bottom panel) of tungstate. Western analysis of the samples was performed using α-PTyr (4G10).
Despite this lack of interaction, we went on to determine if GSK3β was a substrate of laforin. For these experiments, we took advantage of the finding that laforinC266S (C/S laforin) acts as a dominant negative in the mouse model (19), potentially “trapping” the substrate in the phosphorylated form. Thus, we hypothesized that overexpression of C/S laforin in tissue culture cells might “trap” laforin’s substrate in the phosphorylated form. The major regulatory site of phosphorylation on GSK3β is Ser9, and this was the site previously reported to be dephosphorylated by laforin (32). This is a particularly attractive hypothesis to explain the molecular mechanism of LD since phosphorylation of Ser 9 by an upstream kinase such as Akt results in inactivation of GSK3β (33,34). Inactive GSK3β is not able to phosphorylate glycogen synthase (GS), resulting in a more active form of GS and leading to increased glycogen synthesis. WT or C/S FLAG-tagged laforin along with HA-tagged GSK3β were transiently introduced into HEK293 cells. GSK3β was immunoprecipitated using anti-HA resin and a Western analysis using anti-P Ser9 antibody was performed to determine the phosphorylation level of Ser9 in vivo (Fig. 1B). There was no change in the phosphorylation status of this residue upon expression of WT versus C/S laforin. Nonetheless, we pursued the claim that GSK3β is a substrate of laforin and tested whether laforin could dephosphorylate GSK3β in vitro. Cells transiently overexpressing HA-tagged GSK3β were treated with insulin or PDGF to maximally phosphorylate GSK3β on Ser9. GSK3β was immunoprecipitated from cells and subjected to treatment with laforin in the presence or absence of tungstate, a potent phosphatase inhibitor. The ability of laforin to remove the phosphate from Ser9 was assessed by Western analysis using anti-PSer9 antibodies. Consistent with our previous results, laforin was unable to dephosphorylate GSK3β in vitro (Fig. 1C). Since GSK3β activity is also thought to be regulated by Tyr phosphorylation (35), we tested to see if laforin could dephosphorylate GSK3β on Tyr residues using an anti-phosphotyrosine antibody. As shown in Figure 1D, laforin did not dephosphorylate Tyr residues on GSK3β. In an effort to be fully confident that GSK3β is not a substrate of laforin, we monitored dephosphorylation of GSK3β by radiolabeling cells and checking for changes in the phosphate content of immunoprecipitated GSK3β in the presence of WT versus C/S laforin. These results were also negative (data not shown). Therefore, we conclude that contrary to a published report GSK3β is not a substrate of laforin.
Laforin interacts with proteins involved in glycogen metabolism
In an effort to widen our search for laforin’s substrate, we turned our attention to proteins that co-immunoprecipitate with laforin. We previously demonstrated that laforin co-localizes with glycogen synthase (GS) in cells overexpressing both GS and laforin (7). In addition, transgenic mice overexpressing GS in muscle manifest an aberrant form of glycogen that resembles LBs (36). To ascertain if GS co-immunoprecipitates with laforin, wild type (WT) or catalytically inactive (C/S) FLAG-tagged laforin expression vectors were transfected into CHO-IR cells followed by immunoprecipitation using anti-FLAG. Endogenous GS immunoprecipitated with both WT and C/S laforin (Fig. 2A, left panels). Similarly, both WT and C/S laforin were immunoprecipitated with endogenous GS using antibodies directed against GS (Fig. 2A, right panels). However, efforts utilizing both antibodies directed against phosphorylated GS and radiolabeling of cells overexpressing WT or C/S laforin followed by analysis of the radioactive labeling of GS, failed to support the hypothesis that GS was a substrate of laforin (data not shown).
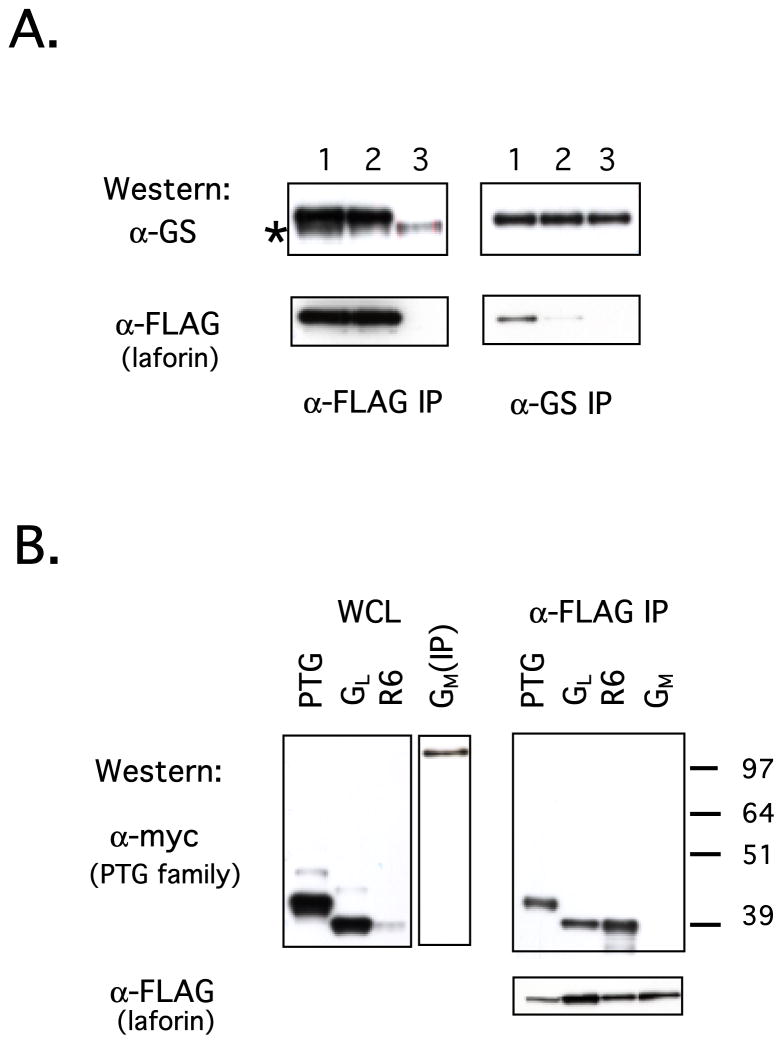
Figure 2.
Laforin interacts with proteins involved in glycogen metabolism. (A) CHO-IR cells were transfected with WT FLAG-tagged laforin (lane 1), C/S FLAG-tagged laforin (lane 2) or empty vector (lane 3). Laforin was immunoprecipitated using α-FLAG resin and endogenous GS was immunoprecipitated using α-GS. Western analyses of the FLAG IPs using α-GS and α-FLAG are shown in the left panels (* denotes a nonspecific band) while Western analyses of the GS IPs are shown in the right panels. (B) HEK293 cells were co-transfected with WT FLAG-tagged laforin and myc-tagged PTG family members. Whole cell lysates (WCLs) were immunoblotted with α-myc (PTG, GL, R6) or immunoprecipitated using α-myc (GM) followed by immunoblotting with α-myc (left panels) to ascertain the expression levels of the PTG family members. The remainder of the WCLs was immunoprecipitated using α-FLAG resin and immunoblotted with α-myc or α-FLAG (right panels).
Since PTG had previously been shown by two-hybrid analysis to interact with laforin, we next turned our attention to the members of the PTG family (12). PTG (R5) and related family members GL, GM, R6 serve as scaffolds to assemble proteins involved in glycogen metabolism. Although the binding partners of all the family members have not yet been defined, PTG interacts with enzymes that regulate glycogen metabolism including protein phosphatase 1 (PP1), glycogen synthase, phosphorylase, phosphorylase kinase, and laforin (37–39). The PTG family members display differential expression patterns in that PTG is expressed in all insulin-sensitive tissues while GL is expressed mainly in the liver and GM is expressed in the muscle (40,41). R6 displays a more ubiquitous expression pattern (42). Each of the PTG family members was expressed as a myc-tagged fusion protein in CHO-IR cells along with FLAG-tagged laforin. Laforin was immunoprecipitated from these cells and analyzed for the association of PTG family members using antibodies directed towards the myc epitope. All of the PTG family members were detectable in CHO-IR cell extracts except GM which was expressed at such low levels that the fusion protein could only be detected after immunoprecipitation (Fig. 2B, left panel). PTG, GL and R6 all co-immunoprecipitated with laforin, with R6 being the most robust (Fig. 2B, right panel). GM could not be detected in the co-immunoprecipitate possibly due to its low expression level (Fig. 2B, left panel). PTG was further evaluated as a substrate for laforin as described above for GS and similar negative results were obtained (data not shown).
We utilized similar strategies to test the ability of laforin to dephosphorylate other enzymes involved in glycogen metabolism including malin (13,14), glycogen branching enzyme (43), PP1 inhibitor 2 (44), β-catenin (45), and the AMPKα/β subunits (46) (data not shown). Our conclusion is that although laforin is found in a complex with many proteins involved in glycogen metabolism, it does not dephosphorylate any of the other proteins associated with glycogen metabolism that were tested. These results are in agreement with multiple studies that have failed to find any changes in the activities of enzymes associated with glycogen metabolism in LD patients (47–49).
Laforin dephosphorylates a complex polysaccharide
CBM20 domains are commonly found in a variety of glycosylhydrolases in plants, fungi and bacteria. The vast majority of enzymes that contain a CBM20 domain, such as α-amylase or glucoamylase, use this domain to bind directly to the carbohydrate, and then enzymatically act on the sugar itself (31). As previously mentioned, LBs are structurally amylopectin-like in nature and are phosphorylated. Therefore, we hypothesized that laforin might dephosphorylate the LB itself. Since we have been unable to obtain enough pure LB material to test as a substrate, we turned to its closest equivalent, plant starch (25,28,29). In particular, potato amylopectin, is phosphorylated on approximately one glucose residue in 300 (27). Phosphorylation occurs on either the C-3 (30–40% of the time) or the C-6 (60–70% of the time) position of the glucose residue and is important in starch metabolism (50–52). Recently, a putative laforin functional homologue has been reported in plants called starch excess 4, SEX4 (53,54). SEX4 has a putative phosphatase domain (CX5R) followed by a domain that binds starch; while laforin has a starch-binding domain followed by a phosphatase domain (53,54). Plants also express a protein kinase known as R1 that is responsible for phosphorylating glucose residues in amylopectin (55,56). To date, our data base searches have not yielded a eukaryotic R1 equivalent. While the roles that R1 and SEX4 play in the storage and utilization of plant starch are currently not well understood, they are both clearly involved in starch metabolism (50,57). In fact, SEX4 mutant plants display a starch excess phenotype reminiscent of the accumulation of LBs in Lafora disease (53).
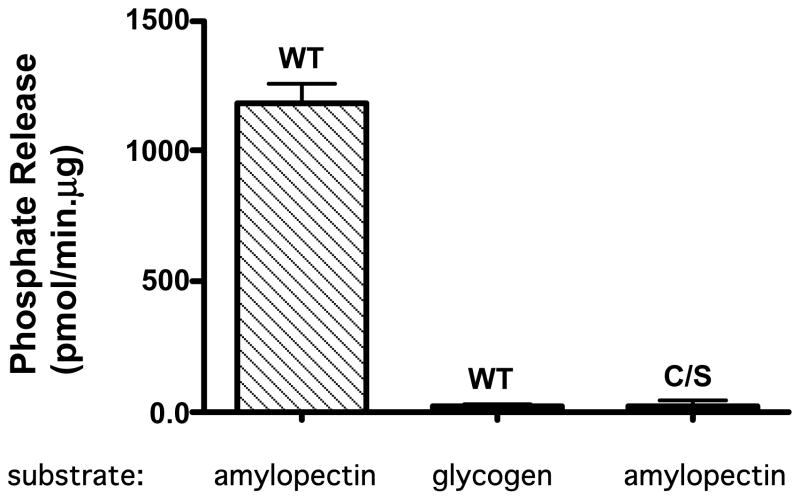
Since LBs are most similar to amylopectin and both are reportedly phosphorylated, we tested potato amylopectin as a potential substrate for laforin using the malachite green assay. This assay is highly sensitive for detecting inorganic phosphate (30). WT laforin displayed robust phosphatase activity towards potato amylopectin (Fig. 3). This activity is not the result of a co-purifying enzyme as catalytically inactive laforin (C/S) is not able to catalyze this reaction. In addition, laforin does not remove phosphate residues from glycogen in our assay. This is most likely a result of the fact that normal cellular glycogen does not contain an appreciable quantity of phosphate residues and our assay conditions may not be able to detect this low a level of phosphate release. Because of the unusual nature of this activity, we also tested SEX4, the plant protein that contains a phosphatase and starch-binding domain (53), for its ability to remove phosphate residues from amylopectin. SEX4 is also capable of dephosphorylating amylopectin (Gentry and Dixon, unpublished results). In light of these results, we hypothesize that laforin’s role is to maintain proper glycogen metabolism by removing phosphate residues during either glycogen synthesis or degradation. In the absence of laforin, we predict that LBs, unlike glycogen, would contain phosphate; indeed this has been reported on several occasions in the literature (26,58,59).
Figure 3.
Laforin dephosphorylates amylopectin. WT and C/S His-tagged laforin were expressed in and purified from bacteria. Standard malachite green assays were performed containing 100 ng of enzyme and 45 μg of amylopectin or glycogen as described in Materials and Methods. Phosphate release was calculated from the change of A620. Error bars represent the standard error of the mean (SEM).
Laforin is unique in its ability to dephosphorylate amylopectin
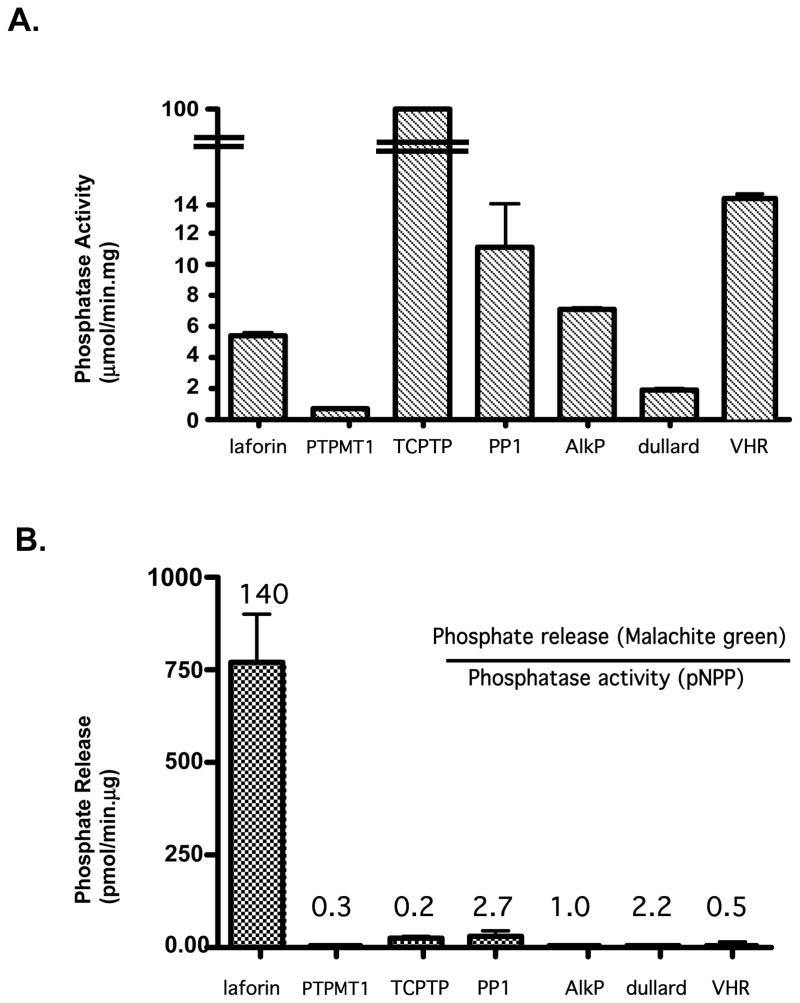
Due to the unusual nature of this activity, we sought to ascertain if other active phosphatases could indiscriminately dephosphorylate amylopectin. In order to test this hypothesis, we selected several different types of phosphatases for our analysis: PTPMT1, a dual specificity phosphatase that prefers phosphatidylinositol 5-phosphate as its substrate (60); TCPTP, a phosphotyrosine specific phosphatase (61); PP1 a very active serine/threonine phosphatase (62); alkaline phosphatase (AlkP), a more nonspecific phosphatase that can dephosphorylate DNA as well as protein substrates (63); VHR, a dual specificity phosphatase (64); and dullard, a phosphoSer/Pro directed phosphatase (65). In each case, the purified recombinant phosphatases were capable of utilizing pNPP as a substrate (Fig. 4A). However, only laforin was capable of removing phosphate from amylopectin (Fig. 4B). As mentioned previously, since amylopectin can be phosphorylated on both the 3′ and 6′-OH groups, the substrate is heterogenous. This precludes us from undertaking more detailed analyses to determine Km or Kcat values for this substrate. To obtain an assessment of the relative activity of laforin toward amylopectin, we generated a relative measure of an enzyme’s ability to remove phosphate from amylopectin versus its activity against pNPP (Fig. 4B, numbers above bars). Using this criteria, laforin is 50–700 times more efficient at removing phosphate from amylopectin than the other phosphatases. This suggests that removal of phosphate from amylopectin is not a property common to phosphatases in general, but requires a specific orientation of the phosphatase active site to the phosphorylated sugar. Roach and co-workers recently measured the activity of laforin against pNPP in the presence of glycogen and amylopectin (66). They noted that addition of glycogen to the reaction caused potent inhibition of pNPP hydrolysis and that the less branched glucose polymers, amylopectin and amylose, were more potent inhibitors. They hypothesized that laforin undergoes a conformational change that blocks its active site upon binding a complex carbohydrate. In light of our results, this inhibition may more likely be a result of competition for laforin’s active site.
Figure 4.
Laforin is unique in its ability to dephosphorylate amylopectin. (A) All enzymes were subjected to standard pNPP assays as described in Materials and Methods. Phosphatase activity was calculated from the change of absorbance at 410nm. (B) Malachite green assays were performed as described in Materials and Methods. The numbers positioned above the bars represent the ratio of phosphate release (malachite green assay) to phosphatase activity (pNPP assay). Phosphate release was calculated from the change of A620. Error bars represent the SEM.
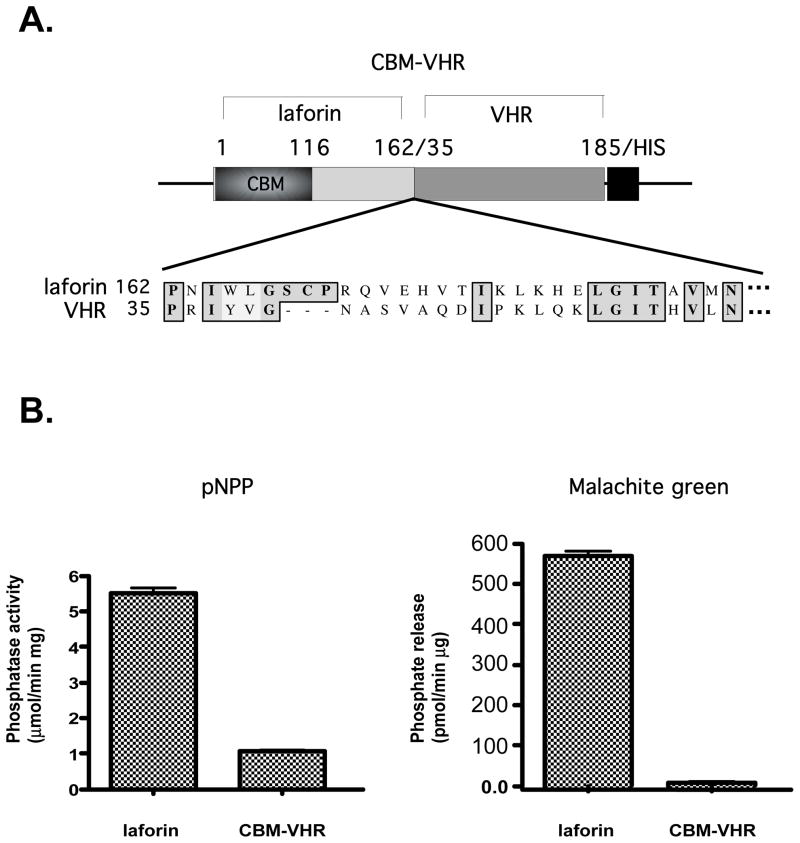
VHR containing a CBM is not able to dephosphorylate amylopectin
Our experiments utilizing amylopectin were performed in vitro and the possibility existed that since laforin was the only phosphatase tested that contained a CBM, it was the only one capable of binding the potential substrate. It occurred to us that attaching the CBM of laforin to another phosphatase would allow the fusion protein to bind to amylopectin, possibly conferring activity onto its phosphatase domain. In order to test this hypothesis, we aligned laforin’s phosphatase domain with VHR and fused the aligned portion in frame to laforin’s CBM (Fig. 5A). We then expressed and purified the fusion protein (CBM-VHR) from bacteria and tested it for phosphatase activity against pNPP and amylopectin. CBM-VHR retains approximately 10% of the wild type VHR activity when pNPP is used as a substrate and is capable of binding glycogen (data not shown). However, the CBM-VHR fusion protein was not capable of dephosphorylating amylopectin (Fig. 5B). Thus, we conclude that the active site of laforin is unique in its ability to utilize a phosphorylated complex carbohydrate as a substrate.
Figure 5.
VHR containing a CBM is not able to dephosphorylate amylopectin. (A) Amino acids 1–162 of laforin containing laforin’s CBM were fused in frame to amino acids 35–185 of VHR followed by a 6His tag. The alignment of laforin with that of VHR is shown at the fusion point. (B) Phosphatase assays utilizing laforin and CBM-VHR were performed using pNPP as a substrate (left graph) or amylopectin as a substrate (right graph). Error bars represent the SEM.
Although we cannot preclude the possibility that laforin also has a proteinaceous substrate, we have demonstrated that laforin displays robust activity against the phosphorylated complex carbohydrate amylopectin. Moreover, we demonstrate that activity against amylopectin is not a common property of phosphatases in general. While it was previously reported that cellular glycogen contains phosphate mono- and diester substitutions at the C6 position of some glucose units, there is no compelling explanation for the function of phosphate on glycogen (67). However, it is possible that glycogen can serve as a substrate for a glucose-phosphate-transferring enzyme as suggested by Lomako and colleagues. (68). Additionally, these researchers have postulated that the phosphate content could be linked to branching and glycogen synthesis. Indeed, there is precedence for this idea in plants where a tight relationship between starch phosphorylation and the degree of starch branching exists (51).
Our hypothesis is that laforin removes the phosphate monoesters from glycogen allowing glycogen metabolism to proceed normally. Therefore, in the absence of laforin, glycogen accumulates more phosphate residues and longer unit chains, eventually forming LBs that resemble insoluble amylopectin. Whether laforin functions during glycogen synthesis or breakdown, our results raise the provocative and unexpected finding that laforin is capable of removing phosphate monoester residues from complex carbohydrates. Although unexpected, our data points to a heretofore over looked aspect of glycogen metabolism that may be critical in understanding the molecular etiology of Lafora disease.
Acknowledgments
We thank Drs. Gregory S. Taylor, David J. Pagliarini and Michael J. Begley for helpful discussions. This work was supported by National Institutes of Health (NIH)/National Cancer Institute Grant T32CA09523 (to M.S.G.), NIH Grant 18024 and 18849 (to J.E.D.), and the Walther Cancer Institute (J.E.D.).
References
- 1.Berkovic SF, Andermann F, Carpenter S, Wolfe LS. N Engl J Med. 1986;315(5):296–305. doi: 10.1056/NEJM198607313150506. [DOI] [PubMed] [Google Scholar]
- 2.Lafora G. Z Ges Neurol Psychiatr. 1911;6:1–14. [Google Scholar]
- 3.Minassian BA. Adv Neurol. 2002;89:199–210. [PubMed] [Google Scholar]
- 4.Lehesjoki AE. Adv Neurol. 2002;89:193–197. [PubMed] [Google Scholar]
- 5.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC, 3rd, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW. Nat Genet. 1998;20(2):171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 6.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, Grid D, Dravet C, Berkovic SF, de Cordoba SR. Hum Mol Genet. 1999;8(2):345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Stuckey JA, Wishart MJ, Dixon JE. J Biol Chem. 2002;277(4):2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 8.Ganesh S, Agarwala KL, Ueda K, Akagi T, Shoda K, Usui T, Hashikawa T, Osada H, Delgado-Escueta AV, Yamakawa K. Hum Mol Genet. 2000;9(15):2251–2261. doi: 10.1093/oxfordjournals.hmg.a018916. [DOI] [PubMed] [Google Scholar]
- 9.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. Biochem Biophys Res Commun. 2004;313(4):1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 10.Ianzano L, Young EJ, Zhao XC, Chan EM, Rodriguez MT, Torrado MV, Scherer SW, Minassian BA. Hum Mutat. 2004;23(2):170–176. doi: 10.1002/humu.10306. [DOI] [PubMed] [Google Scholar]
- 11.Ianzano L, Zhang J, Chan EM, Zhao XC, Lohi H, Scherer SW, Minassian BA. Hum Mutat. 2005;26(4):397. doi: 10.1002/humu.9376. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Sanchez ME, Criado-Garcia O, Heath KE, Garcia-Fojeda B, Medrano-Fernandez I, Gomez-Garre P, Sanz P, Serratosa JM, Rodriguez de Cordoba S. Hum Mol Genet. 2003;12(23):3161–3171. doi: 10.1093/hmg/ddg340. [DOI] [PubMed] [Google Scholar]
- 13.Gentry MS, Worby CA, Dixon JE. Proc Natl Acad Sci U S A. 2005;102(24):8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW. Nat Genet. 2003;35(2):125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 15.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Genes Dev. 2003;17(20):2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 18.Slack FJ, Ruvkun G. Trends Biochem Sci. 1998;23(12):474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 19.Chan EM, Ackerley CA, Lohi H, Ianzano L, Cortez MA, Shannon P, Scherer SW, Minassian BA. Hum Mol Genet. 2004;13(11):1117–1129. doi: 10.1093/hmg/ddh130. [DOI] [PubMed] [Google Scholar]
- 20.Busard HL, Gabreels-Festen AA, Renier WO, Gabreels FJ, Stadhouders AM. Ann Neurol. 1987;21(6):599–601. doi: 10.1002/ana.410210613. [DOI] [PubMed] [Google Scholar]
- 21.Busard HL, Renier WO, Gabreels FJ, Jaspar HH, Slooff JL, Janssen AJ, Van Haelst UJ. Clin Neuropathol. 1987;6(1):1–6. [PubMed] [Google Scholar]
- 22.Carpenter S, Karpati G. Neurology. 1981;31(12):1564–1568. doi: 10.1212/wnl.31.12.1564. [DOI] [PubMed] [Google Scholar]
- 23.Harriman DG, Millar JH, Stevenson AC. Brain. 1955;78(3):325–349. doi: 10.1093/brain/78.3.325. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura RN, Ishak KG, Reddick R, Porter R, James S, Barranger JA. Ann Neurol. 1980;8(4):409–415. doi: 10.1002/ana.410080412. [DOI] [PubMed] [Google Scholar]
- 25.Yokoi S, Austin J, Witmer F, Sakai M. Arch Neurol. 1968;19(1):15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- 26.Sakai M, Austin J, Witmer F, Trueb L. Neurology. 1970;20(2):160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 27.Blennow A, Nielsen TH, Baunsgaard L, Mikkelsen R, Engelsen SB. Trends Plant Sci. 2002;7(10):445–450. doi: 10.1016/s1360-1385(02)02332-4. [DOI] [PubMed] [Google Scholar]
- 28.Yokoi S, Austin J, Witmer F. J Neuropathol Exp Neurol. 1967;26(1):125–127. [PubMed] [Google Scholar]
- 29.Yokoi S, Nakayama H, Negishi T. Clin Chim Acta. 1975;62(3):415–423. doi: 10.1016/0009-8981(75)90093-5. [DOI] [PubMed] [Google Scholar]
- 30.Maehama T, Taylor GS, Slama JT, Dixon JE. Anal Biochem. 2000;279(2):248–250. doi: 10.1006/abio.2000.4497. [DOI] [PubMed] [Google Scholar]
- 31.Paldi T, Levy I, Shoseyov O. Biochem J. 2003;372(Pt 3):905–910. doi: 10.1042/BJ20021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Hum Mol Genet. 2005;14(18):2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 33.Cross DA, Watt PW, Shaw M, van der Kaay J, Downes CP, Holder JC, Cohen P. FEBS Lett. 1997;406(1–2):211–215. doi: 10.1016/s0014-5793(97)00240-8. [DOI] [PubMed] [Google Scholar]
- 34.Halse R, Rochford JJ, McCormack JG, Vandenheede JR, Hemmings BA, Yeaman SJ. J Biol Chem. 1999;274(2):776–780. doi: 10.1074/jbc.274.2.776. [DOI] [PubMed] [Google Scholar]
- 35.Wang QM, Fiol CJ, DePaoli-Roach AA, Roach PJ. J Biol Chem. 1994;269(20):14566–14574. [PubMed] [Google Scholar]
- 36.Pederson BA, Csitkovits AG, Simon R, Schroeder JM, Wang W, Skurat AV, Roach PJ. Biochem Biophys Res Commun. 2003;305(4):826–830. doi: 10.1016/s0006-291x(03)00862-3. [DOI] [PubMed] [Google Scholar]
- 37.Brady MJ, Printen JA, Mastick CC, Saltiel AR. J Biol Chem. 1997;272(32):20198–20204. doi: 10.1074/jbc.272.32.20198. [DOI] [PubMed] [Google Scholar]
- 38.Doherty MJ, Young PR, Cohen PT. FEBS Lett. 1996;399(3):339–343. doi: 10.1016/s0014-5793(96)01357-9. [DOI] [PubMed] [Google Scholar]
- 39.Printen JA, Brady MJ, Saltiel AR. Science. 1997;275(5305):1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- 40.Doherty MJ, Moorhead G, Morrice N, Cohen P, Cohen PT. FEBS Lett. 1995;375(3):294–298. doi: 10.1016/0014-5793(95)01184-g. [DOI] [PubMed] [Google Scholar]
- 41.Gasa R, Jensen PB, Berman HK, Brady MJ, DePaoli-Roach AA, Newgard CB. J Biol Chem. 2000;275(34):26396–26403. doi: 10.1074/jbc.M002427200. [DOI] [PubMed] [Google Scholar]
- 42.Armstrong CG, Browne GJ, Cohen P, Cohen PT. FEBS Lett. 1997;418(1–2):210–214. doi: 10.1016/s0014-5793(97)01385-9. [DOI] [PubMed] [Google Scholar]
- 43.Krisman CR, Tolmasky DS, Raffo S. Anal Biochem. 1985;147(2):491–496. doi: 10.1016/0003-2697(85)90303-3. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, Hurley TD, DePaoli-Roach AA. J Biol Chem. 2000;275(30):22635–22644. doi: 10.1074/jbc.M003082200. [DOI] [PubMed] [Google Scholar]
- 45.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Embo J. 1997;16(13):3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hudson ER, Pan DA, James J, Lucocq JM, Hawley SA, Green KA, Baba O, Terashima T, Hardie DG. Curr Biol. 2003;13(10):861–866. doi: 10.1016/s0960-9822(03)00249-5. [DOI] [PubMed] [Google Scholar]
- 47.Coleman DL, Gambetti P, Mauro SD, Blume RE. Arch Neurol. 1974;31(6):396–406. doi: 10.1001/archneur.1974.00490420062007. [DOI] [PubMed] [Google Scholar]
- 48.Gambetti P, Di Mauro S, Hirt L, Blume RP. Arch Neurol. 1971;25(6):483–493. doi: 10.1001/archneur.1971.00490060017002. [DOI] [PubMed] [Google Scholar]
- 49.Janeway R, Ravens JR, Pearce LA, Odor DL, Suzuki K. Arch Neurol. 1967;16(6):565–582. doi: 10.1001/archneur.1967.00470240003001. [DOI] [PubMed] [Google Scholar]
- 50.Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. Plant J. 2005;41(4):595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 51.Vikso-Nielsen A, Hao-Jie Chen P, Larsson H, Blennow A, Moller BL. Carbohydr Res. 2002;337(4):327–333. doi: 10.1016/s0008-6215(01)00326-3. [DOI] [PubMed] [Google Scholar]
- 52.Yu TS, Kofler H, Hausler RE, Hille D, Flugge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, Steup M, Lue WL, Chen J, Weber A. Plant Cell. 2001;13(8):1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niittyla T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, Gatehouse JA, Villadsen D, Smith SM, Chen J, Zeeman SC, Smith AM. J Biol Chem. 2006;281(17):11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 54.Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB. Plant J. 2006;46(3):400–413. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- 55.Lorberth R, Ritte G, Willmitzer L, Kossmann J. Nat Biotechnol. 1998;16(5):473–477. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- 56.Ritte G, Lorberth R, Steup M. Plant J. 2000;21(4):387–391. doi: 10.1046/j.1365-313x.2000.00683.x. [DOI] [PubMed] [Google Scholar]
- 57.Mikkelsen R, Baunsgaard L, Blennow A. Biochem J. 2004;377(Pt 2):525–532. doi: 10.1042/BJ20030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakai M, Austin J, Witmer F, Trueb L. Arch Neurol. 1969;21(5):526–544. doi: 10.1001/archneur.1969.00480170098011. [DOI] [PubMed] [Google Scholar]
- 59.Schnabel R, Seitelberger F. Pathol Eur. 1968;3(2):218–226. [PubMed] [Google Scholar]
- 60.Pagliarini DJ, Worby CA, Dixon JE. J Biol Chem. 2004;279(37):38590–38596. doi: 10.1074/jbc.M404959200. [DOI] [PubMed] [Google Scholar]
- 61.Romsicki Y, Kennedy BP, Asante-Appiah E. Arch Biochem Biophys. 2003;414(1):40–50. doi: 10.1016/s0003-9861(03)00178-4. [DOI] [PubMed] [Google Scholar]
- 62.Ceulemans H, Bollen M. Physiol Rev. 2004;84(1):1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 63.Coleman JE. Annu Rev Biophys Biomol Struct. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 64.Denu JM, Zhou G, Wu L, Zhao R, Yuvaniyama J, Saper MA, Dixon JE. J Biol Chem. 1995;270(8):3796–3803. doi: 10.1074/jbc.270.8.3796. [DOI] [PubMed] [Google Scholar]
- 65.Satow R, Chan TC, Asashima M. Biochem Biophys Res Commun. 2002;295(1):85–91. doi: 10.1016/S0006-291X(02)00641-1. [DOI] [PubMed] [Google Scholar]
- 66.Wang W, Roach PJ. Biochem Biophys Res Commun. 2004;325(3):726–730. doi: 10.1016/j.bbrc.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 67.Lomako J, Lomako WM, Whelan WJ, Marchase RB. FEBS Lett. 1993;329(3):263–267. doi: 10.1016/0014-5793(93)80234-l. [DOI] [PubMed] [Google Scholar]
- 68.Lomako J, Lomako WM, Kirkman BR, Whelan WJ. Biofactors. 1994;4(3–4):167–171. [PubMed] [Google Scholar]