SUMMARY
DNA polymerase δ (Pol δ) is one of the main replicative DNA polymerases in human cells and therefore is a critical determinant of the overall accuracy of DNA synthesis. Here we document the fidelity of a human Pol δ holoenzyme and systematically score the types of mutations that the enzyme generates in a forward mutation assay. We find that human Pol δ is highly accurate, catalyzing less than one nucleotide mis-insertion per 220,000 nucleotides polymerized. Inactivation of proofreading or mutation of a conserved active site residue significantly elevates the frequency of incorporation errors, demonstrating the contribution of both the base selection and proofreading domains to the overall accuracy of synthesis by Pol δ. The highly selective nature of the polymerase active site is also indicated by the stalling of Pol δ upon encountering multiple types of DNA lesions. However, DNA damage is not an absolute block to Pol δ progression. We propose that partial lesion bypass by Pol δ represents a balance between stalling to allow for repair of mutagenic lesions by specialized repair proteins and bypass of damage to allow for successful completion of DNA synthesis by Pol δ in the presence of weakly blocking DNA adducts.
Keywords: DNA polymerase delta, DNA replication fidelity, proofreading, lesion bypass synthesis
INTRODUCTION
While human cells express at least 14 distinct DNA polymerases, almost all nuclear DNA synthesis is carried out by the two replicative nuclear polymerases, DNA polymerase δ (Pol δ) and DNA polymerase ε (Pol ε) [1]. Pol δ and Pol ε possess both DNA polymerase activity and 3'–5' exonuclease proofreading activity, which is encoded in a separate protein domain. Current evidence suggests that Pol δ carries out the majority of synthesis on the lagging strand of the replication fork [2] and is utilized for resynthesis during mismatch repair [3], base excision repair [4], and homologous recombination [5], whereas Pol ε carries out leading strand synthesis. While the catalytic activity of Pol ε is dispensable in yeast [6], Pol δ activity is absolutely required for viability [7], suggesting that Pol δ can substitute for Pol ε in leading strand synthesis. Consistent with the key role of Pol δ in DNA replication, inactivation of Pol δ proofreading or mutation of critical Pol δ residues involved in base selection results in a dramatic increase in replication errors in both yeast [8] and mice [9, 10]. Therefore the accuracy of Pol δ is a critical determinant of the overall fidelity of DNA replication.
The human Pol δ holoenzyme is a heterotetramer of subunits with molecular weights of 125, 66, 50, and 12 kDa. The 125 kDa subunit contains both the polymerase and exonuclease domains, while the smaller subunits increase the activity and DNA binding properties of the enzyme. The small subunits also modulate interactions between Pol δ and accessory replication factors, most notably the circular DNA binding clamp, PCNA (reviewed in ref. [11]).
Initial studies of eukaryotic Pol δ base selection were performed with a Pol δ complex that was purified from fetal calf thymus [12]. Results of this work indicated that the enzyme possessed high fidelity, although somewhat lower than that of Pol ε. However, partial degradation of the studied Pol δ complex during the purification process left only two of the four subunits intact and the potential effect of loss of the remaining subunits on the enzyme’s fidelity could not be determined. The most detailed fidelity analysis was conducted with purified Pol δ complexes from S. cerevisiae [13] [14] and S. pombe [15]. These studies indicated that Pol δ is accurate for base selection but prone to slippage events, catalyzing an elevated frequency of both single nucleotide and multi-base deletions. These studies yielded important information regarding the yeast enzyme, but their relevance to the human enzyme is unknown. The properties of human Pol δ may differ from those of yeast Pol δ for several reasons. The amino acid sequence of the Pol δ catalytic subunit is only 50% identical between yeast and humans. The subunit structure differs as well: S. cerevisiae Pol δ consists of 3 subunits of 125, 58, and 55 kDa, and S. pombe Pol δ has 4 subunits of 125, 55, 54, and 22 kDa [16]. Moreover the overall accuracy of DNA replication is approximately 10-fold higher in humans relative to yeast [17]; this difference in the overall accuracy of synthesis is presumably due to differences in the properties of the proteins responsible for replication of the genome. Thus, the principal aim of this study is to determine the accuracy of synthesis by human Pol δ and to dissect the relative contributions of the enzyme’s base selection and proofreading activities to its overall accuracy.
An additional area of interest concerns the effect of active site mutations on the fidelity of replication by human Pol δ. In S. cerevisiae, the L612M active site variant of Pol δ has been used to delineate the role of Pol δ during chromosomal replication in vivo [18]. This study exploited the fact that S. cerevisiae Pol δ L612M possesses a biased error spectrum, catalyzing a specific mutational signature which can be detected to identify stretches of DNA that have been copied by Pol δ. This work revealed that, near a single well-defined origin of replication, S. cerevisiae Pol δ primarily copies the lagging strand of chromosomal DNA. It would be of great interest to utilize a similar strategy to determine the specific roles of Pol δ in human cells during DNA replication, recombination, and repair. However, in order to pursue such studies, it is first necessary to determine whether the analogous L606M variant of human Pol δ yields a biased error spectrum.
To determine the accuracy of human Pol δ, we utilized in the present study a human 4-subunit Pol δ holoenzyme that was isolated at high yield and purity by a recently developed bacterial purification system [19]. By purifying the enzyme in both its native form and with its exonuclease domain inactivated by a point mutation, we determined its overall fidelity in both the presence and absence of proofreading. We find that human Pol δ is highly accurate, with both faithful base selection and efficient proofreading of mis-insertions contributing to the overall fidelity of synthesis. Mutation of the highly conserved L606 residue induces a mutator phenotype, with a biased error spectrum that would be suitable for strand-specific identification of Pol δ mediated DNA transactions in vivo. Moreover, we report that Pol δ stalls upon encountering DNA damage, but retains a partial capacity for bypass synthesis. These results are discussed in comparison to work with Pol δ from other species, and in the context of the overall role of Pol δ in DNA replication.
MATERIALS AND METHODS
Materials
Plasmids and strains for purification of human Pol δ are described elsewhere [19]. S. cerevisiae 3-subunit Pol δ was a kind gift of P.M.J. Burgers [20]. Materials for the M13 gapped DNA forward mutation assay were prepared as described [21]. HPLC-purified oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA), and lesion-containing oligonucleotides were from Midland Certified Reagent Company (Midland, TX).
Expression and purification of human Pol δ
4-subunit human Pol δ was purified as described [19], except that the SP HP cation exchange column was eluted with a linear gradient from 0.2 to 1.0 M NaCl rather than a stepwise gradient. The Pol δ mutants were prepared by the same method as the wild-type Pol δ after creating the D402A and L606M mutations in the pET303-hpolD1 plasmid with the Stratagene QuickChange kit.
Polymerase activity assay
Reactions (10 µL) contained 40 mM Tris-HCl (pH 7.5), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 2 µg BSA, 1 mM DTT, 50 µM of each dNTP, 1 µCi [α-32P]dTTP (3 Ci/µmol), 1 µg of activated calf thymus DNA, and 200 fmol of polymerase. Reactions proceeded for 15 minutes at 37 °C. Products were bound to a filter, washed to remove unincorporated [α-32P]dTTP, and quantitated by PhosphorImager analysis.
Exonuclease activity assay
The single-stranded exonuclease substrate was prepared by extending poly-dA-oligo-dT (Amersham Pharmacia) with 5 units Klenow exo- (New England Biolabs) in the presence of 50 µCi [α-32P]dTTP (3 Ci/µmol) for 15 minutes at 37 °C. The extended DNA was melted, and the enzyme inactivated, by incubating 5 minutes at 95 °C. The reaction was immediately placed on ice, and unincorporated [α-32P]dTTP removed by Sephadex G-50 filtration followed by ethanol precipitation.
Exonuclease reactions (10 µL) contained 40 mM Tris-HCl (pH 7.5), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 2 µg BSA, 1 mM DTT, 75 pmol labeled single stranded DNA substrate and 200 fmol of polymerase. The reaction was allowed to proceed 15 minutes at 37 °C and was stopped by adding EDTA to a final concentration of 10 mM. The substrate was ethanol precipitated, and released [α-32P]dTTP was detected in the supernatant by scintillation counting.
Fidelity assay
Gap-filling reactions (25 µL) contained 40 mM Tris-HCl (pH 7.5), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 5 µg BSA, 1 mM DTT, 250 µM of each dNTP, 1.5 nM gapped M13mp2 substrate and 300 nM polymerase. The reaction was incubated for 1 hour at 37 °C. Complete gap filling was confirmed by agarose gel electrophoresis. DNA products were introduced into E. coli strain MC1061 and plated on a lawn of CSH50 α-complementation cells in the presence of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside and isopropyl β-D-1-thiogalactopyranoside, as previously described [21]. Accurate synthesis results in plaques having a dark blue color phenotype, with synthesis errors resulting in light blue or colorless plaques. Mutant plaques were sequenced with primer 5'-TTC GGA ACC ACC ATC AAA C-3'. Error rates were calculated as previously described [21], with the equation ER=[(Ni/N)*MF]/[D*0.6], where Ni=the number of mutations of a specific category, N=the total number of mutations, MF=background corrected LacZ mutant frequency (MF = MF(observed) – MF(background) where MF(background)=0.7 × 10−3 [21]), D=the number of detectable sites for the specific mutation, and 0.6=the probability of expressing a LacZ polymerase error in E. coli.
Lesion bypass assay
Reactions used 32P 5' end-labeled primer 5'-CGC GCC GAA TTC CCG CTA GCA ATA TTC T-3' annealed to the template 5'-TTG GCN GCA GAA TAT TGC TAG CGG GAA TTC GGC GCG -3' where N indicates the nucleotide analog: either dT, 8-oxoguanine, 1,N6-ethenoadenine, O6-methylguanine, O4-methylthymine, or a synthetic abasic site (tetrahydrofuran). Reactions (10 µL) proceeded for 5 minutes at 37 °C and contained 40 mM Tris-HCl (pH 7.5), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 2 µg BSA, 1 mM DTT, 250 µM of each dNTP, 5 nM primer-template, and 60 nM polymerase. Reactions were quenched with an equal volume of 95% formamide/20 mM EDTA, heated to 95 °C for 5 minutes, and separated on a 14% polyacrylamide-urea gel. The gel was scanned by PhosphorImager, and the lanes aligned and assembled in Adobe Photoshop.
RESULTS
Purification of wild-type D402A and L606M human Pol δ
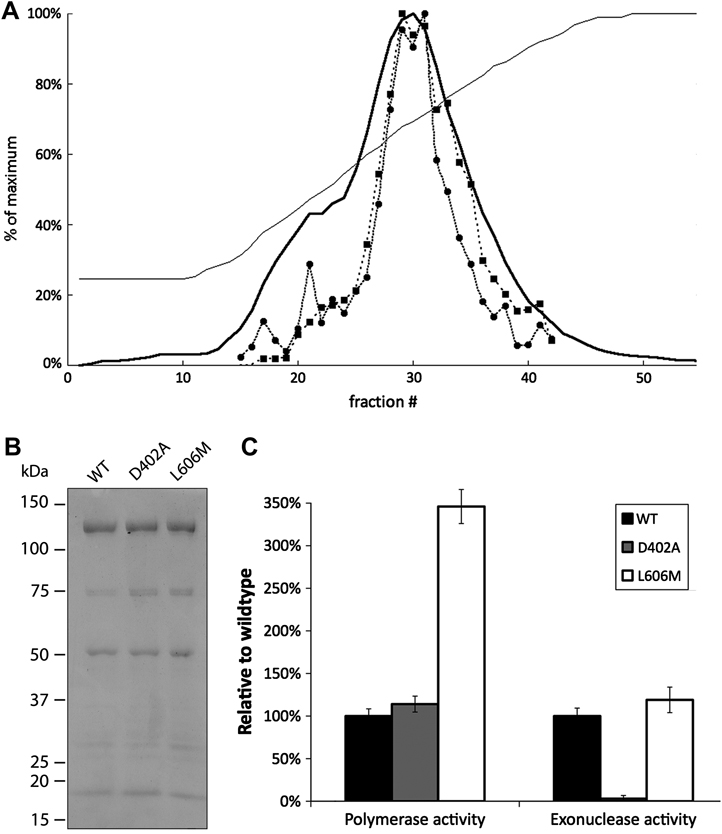
We obtained highly purified 4-subunit wild-type, exonuclease-deficient and L606M human Pol δ through the use of a recently developed E. coli expression system. This purification scheme achieves co-expression of all four Pol δ subunits by expressing the subunits on two different plasmids, followed by purification via sequential chromatography on nickel affinity and cation exchange columns. The purity of the final preparation was verified by eluting the protein from the cation exchange column with a linear salt gradient and assaying each fraction for both polymerase and exonuclease activity. The relative activity of the peak fractions was constant with respect to protein concentration (Fig. 1A). SDS-PAGE analysis with Coomassie blue staining demonstrated that the peak fraction contained all 4 Pol δ subunits (Fig. 1B). Wild-type and exonuclease-deficient Pol δ had approximately equal polymerase activity (Fig. 1C); in assays with activated DNA as a template, wild-type Pol δ incorporated 1.4 +/− 0.12 pmol dNMPs per minute per pmol enzyme, and D402A Pol δ incorporated 1.6 +/− 0.13 pmol dNMPs per minute per pmol enzyme. The L606M mutant possessed significantly elevated DNA polymerase activity, incorporating 4.8 +/− 0.28 pmol dNMPs per minute per pmol enzyme. The increased specific activity resulting from the L606M mutation in human Pol δ is consistent with the ~2-fold elevation in specific activity observed in the analagous S. cerevisiae L612M Pol δ mutant [22].
Figure 1. Purification of human Pol δ.
(A): Elution profile of wild-type human Pol δ on cation exchange media. The UV absorbance (▬) and conductivity (─) were continuously monitored as Pol δ was eluted from HP SP media (GE Life Sciences) with a linear salt gradient from 0.2 to 1.0 M NaCl. Each fraction was then assayed for polymerase (●) and exonuclease (■) activity. The peak fraction (fraction # 30) was dialyzed into storage buffer and used for further study. (B): 5 pmol of purified wild-type, exonuclease-deficient or L606M human Pol δ was separated on a 4–15% SDS-PAGE gel and stained with Coomassie Blue R-250. Molecular weight standards are indicated. The bands correspond to the p125, p66, p50, and p12 subunits. The predicted molecular weight of the tagged p12 subunit is 18 kDa. The faint band near 30 kDa is a degradation product of the p125 subunit [19]. (C): Enzymatic activities of wild-type and exonuclease-deficient human Pol δ. Assays were performed in triplicate.
Wild-type Pol δ possesses readily detected exonuclease activity on single-stranded DNA (Fig. 1C), with 1 pmol of enzyme releasing 0.84 +/− 0.079 pmol dNMPs per minute per pmol enzyme, whereas the D402A mutant does not exhibit significant exonuclease activity, confirming that mutation of the catalytically essential aspartic acid residue fully inactivated the exonuclease domain. In contrast to the elevated polymerase activity seen in L606M Pol δ, the exonuclease activity of the L606M mutant was only slightly increased relative to wild-type, releasing 1.0 +/− 0.013 pmol dNMPs per minute per pmol enzyme.
Overall fidelity of human Pol δ
The accuracy of human Pol δ was analyzed using the M13 LacZ gapped DNA forward mutation assay [21]. This assay utilizes double-stranded M13mp2 DNA which contains a single-stranded gap of 407 nucleotides in the sequence encoding the LacZα fragment. The gapped region is filled with purified polymerase in vitro and complete extension is verified by agarose gel electrophoresis. The products are transformed into E. coli, and plated on a lawn of α-complementation cells containing X-gal and IPTG. Accurate synthesis by the polymerase results in a dark blue plaque, while polymerase errors yield a light blue or colorless plaque. The ratio of mutant to wild-type plaques reveals the overall fidelity of the polymerase, while sequencing of mutant plaques allows for determination of the error spectrum.
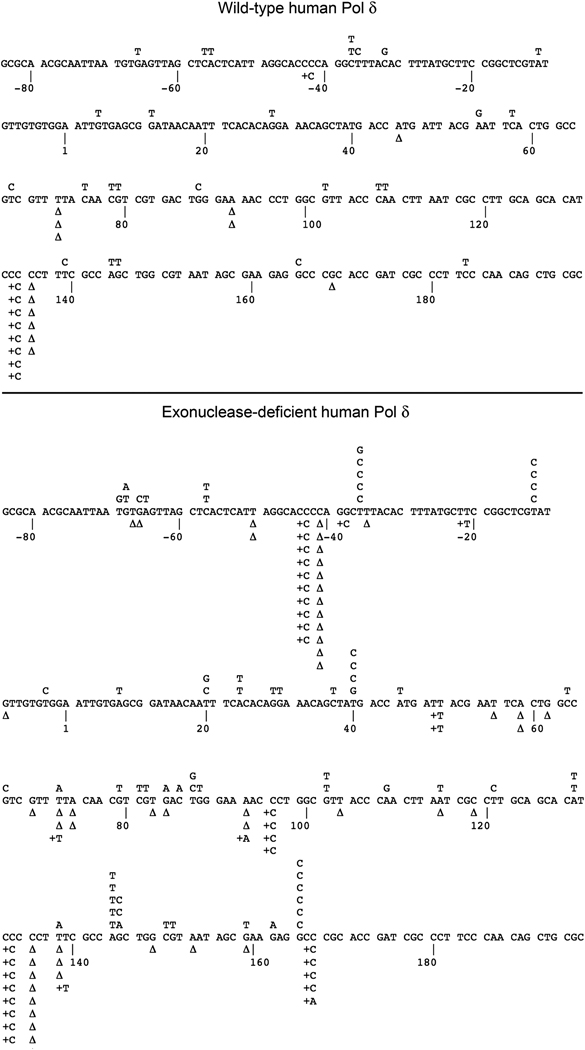
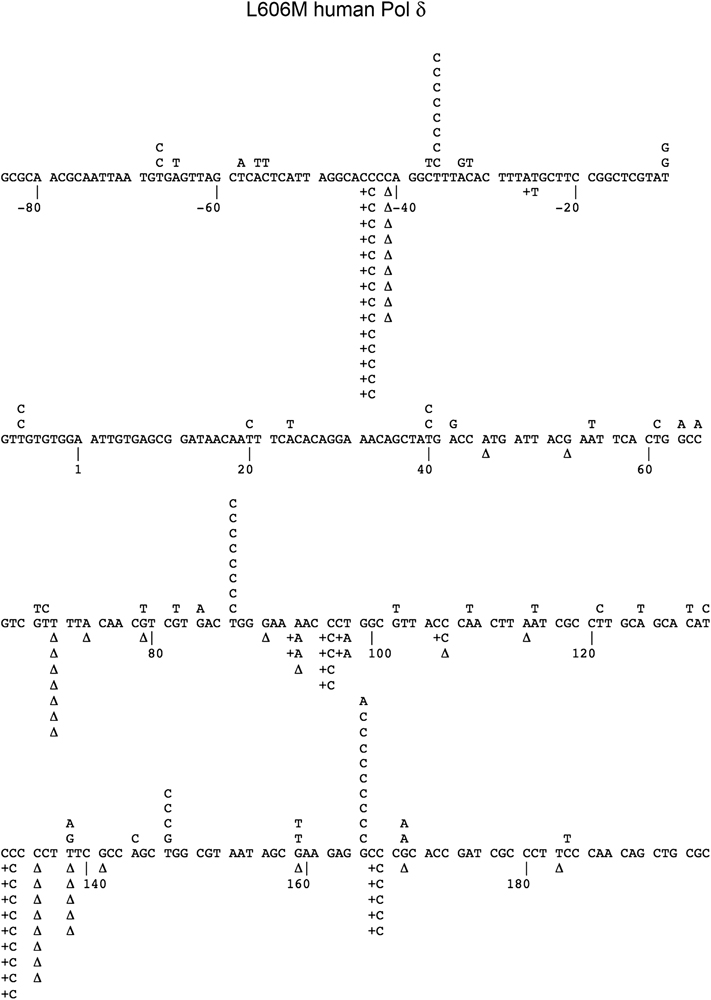
All three Pol δ preparations completely filled the gapped DNA substrate, as verified by agarose gel electrophoresis (data not shown). Sequencing of mutant plaques revealed base substitution errors, insertions, and deletions distributed throughout the LacZ target sequence for the wild-type, exonuclease-deficient, and L606M enzymes (Fig. 2 and Fig. 3); the presence of mutations throughout the entire target sequence substantiates that the substrate was filled to completion.
Figure 2. Mutation spectrum of wild-type and exonuclease-deficient human Pol δ.
Wild-type and exonuclease-deficient Pol δ were analyzed in the M13 LacZ gapped DNA forward mutation assay. The target sequence of the LacZ fragment is shown, with detected single base substitutions indicated above the target sequence and single nucleotide deletions (Δ) and insertions below.
Figure 3. Mutation spectrum of L606M human Pol δ.
The L606M human Pol δ mutant was analyzed in the M13 LacZ gapped DNA forward mutation assay. The target sequence of the LacZ fragment is shown, with detected single base substitutions indicated above the target sequence and single nucleotide deletions (Δ) and insertions below.
Wild-type human Pol δ gave an overall LacZ mutant plaque frequency of 1.2×10−3 (data pooled from 2 independent experiments, 95% confidence interval of 0.29×10−3 – 1.8×10−3) (Table I and Fig. 4A). This value is close to the background LacZ mutant frequency of 0.7×10−3, indicating that Pol δ copies the DNA template with high accuracy. The exonuclease-deficient D402A mutant had a markedly elevated LacZ mutant frequency of 8.6×10−3 (2 independent experiments, 95% confidence interval 7.4×10−3 – 9.8×10−3), indicating that proofreading contributes substantially to the fidelity of Pol δ. The L606M catalytic site mutant yielded a further elevated LacZ mutant frequency of 13×10−3 (2 independent experiments, 95% confidence interval 12×10−3 – 15×10−3). The fact that the LacZ mutant frequency of L606M Pol δ exceeds that of the exonuclease-deficient Pol δ variant indicates that the L606M mutation in part diminishes the base selection capability of the enzyme, rather than simply impairing the ability of the enzyme to proofread errors. As a control experiment, we also performed gapped DNA fidelity measurements with wild-type S. cerevisiae Pol δ under reaction conditions identical to those used for the human Pol δ. This experiment yielded 20,979 total plaques with 79 mutants. This mutant plaque frequency of 3.8×10−3 is in good agreement with published results [13].
Table 1.
Mutation rates in the LacZ gapped DNA forward mutation assay.
| WT | D402A | L606M | Exo−/Exo+ ratio | ||||
|---|---|---|---|---|---|---|---|
| Number detecteda | Error rateb (×10−5) | Number detected | Error rate (×10−5) | Number detected | Error rate (×10−5) | ||
| Total Plaques | 43,615 | 18,004 | 12,216 | ||||
| Total Mutants | 54 | 155 | 159 | ||||
| Mutant Frequency | 1.2 × 10−3 | 8.6 × 10−3 | 13 × 10−3 | ||||
| Base Substitutions | 26 | ≤0.45 | 69 | 4.4 | 73 | 6.7 | 7.2 |
| +1 Insertions | 9 | 0.077 | 30 | 1.2 | 42 | 2.4 | ≥9.8 |
| −1 Deletions | 13 | 0.11 | 51 | 2.0 | 45 | 2.6 | 16 |
| Large Deletions | 3 | 6.9 (MF)c | 13 | 72 (MF) | 7 | 57 (MF) | 18 |
| Other Mutationsd | 2 | 2 | 1 | ||||
The total mutants reported may be greater or less than the sum of the different types of mutations because of the infrequent failure of a sequencing reaction, or the occasional presence of more than one distinct mutation in an individual plaque. Mutations were considered to be distinct as long as they were separated from each other by 3 or more nucleotides.
Background mutation frequencies are 3.4 × 10−4 for base substitutions and 0.55 × 10−4 for single base insertions and deletions, corresponding to background error rated of 0.45 × 10−5 for base substitutions and 0.046 × 10−5 for single base insertions and deletions [21].
The number of scorable sites for large deletion errors is not defined; therefore this value is given as a mutant frequency (MF) rather than an error rate.
Other mutations Pol δ were: WT, 2 occurrences of TTT to CTCT at +137 to +139; D402A, deletion of TA at −34 to −33, insertion of AC at −56 to −55; L606M, deletion of AC at −56 to −55.
Figure 4. Comparison of human and yeast Pol δ fidelity.
(A): LacZ mutant frequency in the M13 gapped DNA forward mutation assay. (B): Base-pair error rates for exonuclease-deficient and L606M human Pol δ and S. cerevisiae Pol δ in the forward mutation assay. An asterisk indicates less than or equal to values, due to failure to detect any mutations of the indicated type. Data concerning S. cerevisiae exonuclease-deficient and L612M Pol δ variants are from refs. [13] and [22]. The S. cerevisiae L612M Pol δ was assayed in the presence of the accessory factors RPA, RFC and PCNA, which have been shown to slightly reduce the frequency of errors by Pol δ [24].
Base substitutions
The base substitution error rate of wild-type Pol δ (single nucleotides mis-incorporated per nucleotide synthesized) was approximately equal to the background of the assay (4.5×10−6), indicating that most or all base substitutions detected could be background events and not the result of synthesis by Pol δ. This observation indicates that wild-type Pol δ is highly accurate, catalyzing on average less than one base substitution error per 220,000 nucleotides polymerized. Exonuclease-deficient Pol δ, however, mis-incorporated nucleotides at a 10-fold higher rate, catalyzing approximately one mis-incorporation per 22,000 nucleotides synthesized. The difference in base substitution error rate between the wild-type and the exonuclease-deficient polymerase indicates that Pol δ proofreads ≥ 90% of nucleotide mis-insertion events.
The elevated base substitution frequency of exonuclease-deficient human Pol δ allowed for the spectrum of specific mis-insertion events to be analyzed in detail (Fig. 4B). Of the 12 possible single-base mutations, only two events were not scored—namely A→C and C→G mutations. This is not due to a lack of scorable sites in the assay as these events have been reported at 9 and 17 distinct locations, respectively, in the gapped DNA substrate [21]. The complete absence of these events is in contrast to the large number of other mutation types —for example 17 detected T→C mis-pair events, which was the most common mutation detected in the assay. This observation indicates that the active site of human Pol δ specifies the types of nucleotides mis-inserted.
The frequent T→C event, resulting from a T-dGMP mis-pair, was observed at 6 independent template locations within different sequence contexts, indicating that the elevated frequency of T-dGMP mis-pairing is likely an inherent property of the polymerase base-selection active site rather than being due to the specific sequence context involved. These events were not seen above background levels for the exonuclease-proficient human Pol δ, indicating that this mis-pair is proofread with high efficiency.
The L606M mutant had a mutation spectrum closely paralleling that seen for the exonuclease-deficient mutant, with the exception of the T-dGMP mis-pair event, which occurred nearly 3-fold more frequently in L606M relative to D402A (Fig. 4B). Relative to wild-type Pol δ, The L606M Pol δ mutant was approximately 70-fold more likely to mis-insert dGMP opposite template T, resulting in a T→C mutation. L606M Pol δ catalyzes T→C mutations with a rate elevated more than 10-fold relative to the reciprocal A→G mutation, which would be caused by an A-dCMP mis-pair.
Frameshifts
Wild-type Pol δ catalyzed single nucleotide additions and deletions slightly above background levels, and with approximately equal efficiency. These events were most frequent within a single sequence context, namely a run of 5 cytosines at LacZ positions +132 to +136. Overall, 20 of the 22 frameshift events occurred at a run of 4 or more repeated nucleotides. Exonuclease-deficient Pol δ catalyzed single-base frameshift events at a > 15-fold elevated rate, indicating that 90–95% of insertions/deletions are corrected by proofreading. As with wild-type Pol δ, both single-base additions and deletions with the exonuclease-deficient Pol δ occurred at similar rates, and were most frequent at iterated sequences. 38 of the 81 frameshift events occurred at two sequences: a run of 4 cytosines at −36 to −39, and a run of 5 cytosines at +132 to +136. Interestingly, the LacZ target also contains runs of 4 thymidines and 4 adenines, but these runs were not a notable hotspot for slippage by the exonuclease-deficient Pol δ.
Large deletions
Several large deletions of 93 to 325 nucleotides were also detected by sequencing mutant plaques. This type of error is likely due to the primer end fraying and reannealing at a different location on the template strand having a similar DNA sequence [23]. Wild-type Pol δ created three large deletions, only one of which occurred between direct repeats. Exonuclease-deficient Pol δ created 13 large deletions, all of which occurred between direct repeats. 8 of these deletions occurred between the repeated sequence CCCGC, resulting in loss of 317 nucleotides. It has been proposed that this specific deletion occurs in the gapped DNA assay from a looped intermediate structure that is stabilized by base-pair interactions in the stem of the loop [23]. The frequency of large deletions differed between wild-type and exonuclease-deficient Pol δ by a factor of 10, suggesting that Pol δ proofreading contributes to the avoidance of large deletion errors. In addition, it has also been demonstrated in yeast that the PCNA and RPA accessory factors reduce the frequency of large deletions without affecting the fidelity of base selection. This effect is likely due to tighter binding of the Pol δ -PCNA complex to the DNA substrate, which decreases the probability of polymerase dissociation and primer fraying/reannealing [24].
Lesion bypass
To further explore nucleotide incorporation by the human Pol δ active site, we assessed the ability of wild-type Pol δ to copy DNA containing a single lesion at a defined site. Synthesis by wild-type Pol δ was efficient on an undamaged template, but was significantly stalled by the oxidative lesion 8-oxoguanine, the methyl adducts O6-methylguanine and O4-methylthymine, the cyclic 1,N6-ethenoadenine lesion, and an abasic site (Fig. 5). Stalling of Pol δ upon encountering modified template nucleotides reflects the high selectivity of the active site. The 1,N6-ethenoadenine adduct presented a complete block to Pol δ synthesis, while partial extension past the site specific adduct was observed for all other lesions investigated. This observation indicates that the Pol δ catalytic site is able to incorporate and extend opposite a modified template nucleotide. Previous reports, utilizing baculovirus-purified 4-subunit human Pol δ in complex with PCNA, have demonstrated similarly efficient bypass of O6-methylguanine [25], 8-oxoguanine, and abasic sites [26]. Our results, performed in the absence of PCNA, demonstrate that the capability for lesion bypass is an inherent property of the human Pol δ catalytic site. Partial bypass activity by Pol δ may be important to allow for a balance between accurate synthesis by allowing for repair or recruitment of specialized lesion bypass polymerases, versus efficient completion of DNA replication by allowing Pol δ to continue synthesis in the presence of low amounts of DNA damage.
Figure 5. Time course of lesion bypass synthesis by wild-type human Pol δ.
Reactions used a 5' 32P-labeled 28 nucleotide primer annealed to a 36 nucleotide template. The sequence of the template is indicated to the left of the figure, with N denoting the position of the modified nucleotide: deoxythymidine (no lesion), 8-oxoguanine (8-oxo-dG), 1,N6-ethenoadenine (eA), O6-methylguanine (O6-mG), O4-methylthymine (O4mT), or an abasic site analog (AB). The incubation time of each reaction is indicated below the figure.
DISCUSSION
Using a nearly homogeneous four subunit human Pol δ holoenzyme, we determined that human Pol δ replicates DNA with high fidelity and that its accuracy is due to both faithful base selection by the polymerase active site and efficient proofreading by hydrolysis of approximately 90% of nucleotides that are mis-inserted. The finding that human Pol δ catalyzes DNA synthesis with high accuracy in vitro may underpin the high accuracy of DNA replication in human cells where current evidence suggests Pol δ is responsible for synthesizing the lagging strand of DNA and for resynthesis during multiple DNA repair processes. We first compare the fidelity of the human enzyme to that of preparations of Pol δ from other eukaryotes. We then consider the role of human Pol δ in the context of the overall accuracy of replication in human cells. Last, we propose that the ability of Pol δ to copy past lesions in DNA that are potentially generated by endogenous processes suggests that Pol δ may have a significant role in error-prone DNA synthesis.
Accuracy of human and calf thymus Pol δ
The fidelity of mammalian Pol δ has previously been studied in the LacZ gap-filling assay with enzyme purified from calf thymus; one study [12] reported an overall LacZ mutant frequency for wild-type Pol δ as 3.6×10−3 to 4.0×10−3, representing less accurate synthesis relative to the value of 1.2×10−3 that was obtained in the present report. This difference may be explained in part by the fact that purification of the native enzyme from calf thymus tissue yielded only two of the four subunits (p125 and p48); the other two subunits were presumably degraded or lost during the labor-intensive purification process as verified by their complete absence upon Coomassie staining of the SDS-PAGE resolved enzyme (Fig. 2 in ref. [27]). The functions of the missing subunits have only been partially defined; differential effects of the missing subunits on polymerase activity, exonuclease activity, and base selection could result in alteration in the fidelity of DNA synthesis. An additional difference is that the calf thymus Pol δ complex was assayed in the presence of the processivity factor PCNA, which has been reported to decrease the fidelity of synthesis by this enzyme [28]. A second study on calf thymus DNA Pol δ, using an alternate purification method and assayed in the absence of PCNA, reported an overall LacZ mutant frequency of 1.1×10−3 [29], which is in excellent agreement with our value of 1.2×10−3. SDS-PAGE separation and Coomassie staining of this calf thymus Pol δ preparation (Fig. 4 in ref. [30]) reveals, in addition to the p125 and p48 subunits known at the time of the work, fainter bands of ~68 and 12 ~kDa which may indicate partial presence of the otherwise missing subunits and thereby may explain the discrepancy between the two studies.
Accuracy of human and yeast Pol δ
Both the wild-type and exonuclease-deficient human enzymes were approximately 2.5-fold more accurate than their S. cerevisiae homologues (Fig. 4A). Our own measurement of the fidelity of yeast Pol δ yielded a mutant plaque frequency of 3.8×10−3, which is consistent with the published value of 4.7×10−3 [13]. These results suggest that human Pol δ may possess greater accuracy than yeast Pol δ. Given that the magnitude of the apparent fidelity difference is approximately the same in both the presence and absence of proofreading, it is likely that any difference in accuracy between the two species is due to enhanced base selection by the active site of human Pol δ, rather than a difference in proofreading ability or polymerase/exonuclease domain partitioning. The error rate of human exonuclease-deficient Pol δ is approximately 3-fold lower than the reported value for yeast exonuclease-deficient Pol δ when considering base substitutions, -1 deletions, and large deletions (Table I and ref. [13]).
Comparison with other eukaryotic polymerases
The other primary replicative nuclear polymerase in mammals, Pol ε, has been reported to replicate DNA with extremely high accuracy: in the M13 gapped DNA forward mutation assay, calf thymus Pol ε does not introduce mutations above background levels [12]. Our data indicate similar (i.e., close to assay background) fidelity for 4-subunit mammalian Pol δ. This result is consistent with the current model of DNA replication, in which Pol ε replicates the leading strand while Pol δ, in concert with Pol α, replicates the lagging strand. One would expect both Pol δ and Pol ε to have similarly high fidelity, so that the accuracy of replication would be constant independent of which of the two DNA strands is copied and passed on to a progeny cell. Mammalian Pol α has been reported to copy DNA at a significantly lower accuracy, yielding an approximately 10-fold higher mutation frequency than Pol δ or Pol ε [12]. This lower fidelity is due in part to the absence of an associated proofreading domain. If uncorrected, Pol α-dependent errors would cause the lagging strand to have an elevated mutation frequency relative to the leading strand since discontinuous lagging strand synthesis requires multiple Pol α-dependent priming events as opposed to the continuous synthesis on the leading strand. Thus it is plausible that the exonuclease of Pol δ (or possibly an extrinsic exonuclease) proofreads for Pol α [31] thereby elevating its ultimate fidelity. Such proofreading would then, in theory, result in identical overall error rates on both the leading and lagging strands during DNA replication. This type of proofreading in trans has indeed been demonstrated in yeast [32]. However, whether the fidelity of replication is truly identical on both the leading and lagging strands has yet to be determined.
Biased error spectrum of the L606M Pol δ Mutant
Mutator polymerases which express biased error spectrums have successfully been used to identify the strand preference of Pol δ and Pol ε in yeast near a single, well defined origin of replication [18, 33]. As a first step toward determining the specific roles of Pol δ in human cells, we studied the L606M active-site mutant of human Pol δ, which is analogous to the previously studied S. cerevisiae L612M substitution [22]. We found that the L606M mutation induces a mutator phenotype in human Pol δ in vitro, increasing the overall rate of base substitution errors ≥15-fold relative to wild-type Pol δ. Moreover, the mutation spectrum of L606M Pol δ is biased, with a particularly high rate of mis-insertion of dGMP opposite template T. This mis-insertion event would cause a T→C mutation event. In vivo, a T→C mutation could be caused by mis-insertion of dGMP opposite T, or also during replication of the opposite strand by mis-insertion of dCMP opposite template A. However, L606M Pol δ catalyzes T-dGMP errors at a >10-fold higher frequency than A-dCMP errors. Thus, L606M Pol δ expresses a strongly biased mutational signature that will allow for future studies to pursue strand-specific identification of regions of the human genome processed by Pol δ in vivo during spontaneous replication, recombination, and repair.
Lesion bypass synthesis
The fact that Pol δ is accurate in DNA synthesis and is also able to copy past adducted nucleotides in DNA is of considerable relevance to understanding the role of Pol δ in mutagenesis and carcinogenesis. It has been demonstrated that large amounts of DNA damage result in selective degradation of the p12 subunit of human Pol δ, which impairs the lesion bypass capability of Pol δ [26] and thereby may allow extensively damaged DNA to be processed by specialized repair and lesion bypass enzymes. However, in normal cells, DNA constantly undergoes lower levels of damage by a variety of reactive species generated by normal cellular process [34]. For example evidence suggests that more than 10,000 purines are lost per cell per day [35] resulting in apurinic sites that, if not repaired, will be encountered by Pol δ during DNA replication. An even larger number of nucleotide alterations occur in cellular DNA as a result of attack by reactive oxygen and nitrogen species. On the one hand stalling of Pol δ at DNA lesions indicates the need for specialized bypass polymerases and/or replication-coupled repair for efficient replication at sites of DNA damage. Failure to continue DNA synthesis would result in replication fork collapse and catastrophic failure of replication and/or large, heritable and potentially deleterious genomic rearrangements. Yet, we have also observed that Pol δ was able to synthesize past all lesions that we analyzed other than 1,N6-ethenoadenine. This limited amount of bypass synthesis may indicate some capability of Pol δ to continue replication without interruption in the presence of low amounts of DNA damage or weakly blocking adducts. While bypass of damaged DNA by replicative polymerases is frequently mutagenic [19], the resultant point mutations may be better tolerated than arrest of synthesis. Thus there could be a balance between efficient completion of DNA synthesis and highly accurate replication. It is frequently assumed that the Y-family (error-prone) DNA polymerases are responsible for copying past impediments to DNA replication. However, these enzymes are predominantly distributive and add one or a few nucleotides per catalytic event. Considering that Pol δ is responsible for synthesizing at least half of the genome, it seems likely that Pol δ also copies past many endogenous lesions including alternative DNA structures [36] and that only when blockage is complete or damage overwhelming is it necessary to recruit other bypass DNA polymerases. Thus, while the loss of the p12 fragment of Pol δ and the resultant impairment in Pol δ lesion bypass synthesis may be particularly relevant to cells exposed to extensive levels of DNA damage, the inherent lesion bypass properties of Pol δ may be critical to allow for efficient completion of replication in the presence of lower levels of damage.
Failure of Pol δ to copy the genome with uninterrupted high fidelity will result in error-prone synthesis or collapse of the replication fork, leading to accumulation of mutations and potentially deleterious consequences. In accord with this concept is the demonstration that allelic substitution of mutant Pol δ that exhibits a mutator phenotype causes an increase in carcinogenesis [9] [10]. However it remains to be determined whether enhanced carcinogenesis by mutator polymerases is the result of increased base substitutions in coordination with DNA damage bypass, or fork collapse with the production of double-stranded breaks resulting in deletions.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health [R01 CA102029, R01 CA115802, P01 CA077852 to L.A.L. and F30 AG030314, T32 GM007266 to M.W.S.]. We would like to thank Michael Fry, Ashwini Kamath-Loeb and Scott Kennedy for critically reviewing the manuscript.
REFERENCES
- 1.Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair and recombination in eukaryotes. Internat. Rev. Cytology. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Burgers PMJ. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longley MJ, Pierce AJ, Modrich P. DNA polymerase δ is required for human mismatch repair in vitro. J. Biol. Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 4.Blank A, Kim B, Loeb LA. DNA polymerase δ is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1994;91:9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maloisel L, Fabre F, Gangloff S. DNA polymerase δ is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 7.Francesconi S, Park H, Wang TS. Fission yeast with DNA polymerase δ temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase δ. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 9.Venkatesan RN, Treuting P, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, Ladiges W, Preston BD, Loeb LA. Mutation at the polymerase active site of mouse DNA polymerse δ increases genomic instability and accelerates carcinogenesis. Mol. Cell. Biol. 2007;27:7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc. Natl. Acad. Sci. USA. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat. Rev. Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 12.Thomas DC, Roberts JD, Sabatino RD, Myers TW, Tan CK, Downey KM, So AG, Bambara RA, Kunkel TA. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 13.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PMJ, Kunkel TA. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto K, Shimizu K, Nakashima N, Sugino A. Fidelity of DNA polymerase δ holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42:14207–14213. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Zuo S, Kelman Z, O'Donnell M, Hurwitz J, Goodman MF. Fidelity of eucaryotic DNA polymerase δ holoenzyme from Schizosaccharomyces pombe. J. Biol. Chem. 2000;275:17677–17682. doi: 10.1074/jbc.M910278199. [DOI] [PubMed] [Google Scholar]
- 16.MacNeill SA, Baldacci G, Burgers PMJ, Hubscher U. A unified nomenclature for the subunits of eukaryotic DNA polymerase δ. Trends Biochem. Sci. 2001;26:16–17. doi: 10.1016/s0968-0004(00)01709-6. [DOI] [PubMed] [Google Scholar]
- 17.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazlieva R, Spittle CS, Morrissey D, Hayashi H, Yan H, Matsumoto Y. Proofreading exonuclease activity of human DNA polymerase δ and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37:2854–2866. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers PMJ, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 21.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Meth. Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 22.Nick McElhinny SA, Stith CM, Burgers PMJ, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J. Biol. Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 24.Fortune JM, Stith CM, Kissling GE, Burgers PMJ, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase δ. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 26.Meng X, Zhou Y, Zhang S, Lee EYC, Frick DN, Lee MY. DNA damage alters DNA polymerase δ to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–657. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng L, Tan CK, Downey KM, Fisher PA. Enzymologic mechanism of calf thymus DNA polymerase δ. J. Biol. Chem. 1991;266:11699–11704. [PubMed] [Google Scholar]
- 28.Mozzherin DJ, McConnell M, Jasko MV, Krayevsky AA, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes misincorporation catalyzed by calf thymus DNA polymerase δ. J. Biol. Chem. 1996;271:31711–31717. doi: 10.1074/jbc.271.49.31711. [DOI] [PubMed] [Google Scholar]
- 29.Shevelev IV, Ramadan K, Hubscher U. The TREX2 3'-->5' exonuclease physically interacts with DNA polymerase δ and increases its accuracy. ScientificWorldJournal. 2002;2:275–281. doi: 10.1100/tsw.2002.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hubscher U. Biochemical and functional comparison of DNA polymerases α, δ, and ε from calf thymus. J. Biol. Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 31.Perrino FW, Loeb LA. Hydrolysis of 3'-terminal mispairs in vitro by the 3'->5' exonuclease of DNA polymerase δ permits subsequent extension by DNA polymerase α. Biochemistry. 1990;29:5226–5231. doi: 10.1021/bi00474a002. [DOI] [PubMed] [Google Scholar]
- 32.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errorrs made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Pursell ZF, Isoz I, Lundstrom E, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 36.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PMJ, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase δ specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]