Abstract
How transforming growth factor-β (TGF-β) signaling elicits diverse cell responses remains elusive, despite the major molecular components of the pathway being known. We contend that understanding TGF-β biology requires mathematical models to decipher the quantitative nature of TGF-β/Smad signaling and to account for its complexity. Here, we review mathematical models of TGF-β superfamily signaling that predict how robustness is achieved in bone-morphogenetic-protein signaling in the Drosophila embryo, how changes in receptor-trafficking dynamics can be exploited by cancer cells and how the basic mechanisms of TGF-β/Smad signaling conspire to promote Smad accumulation in the nucleus. These studies demonstrate the power of mathematical modeling for understanding TGF-β biology.
Towards a systems biology understanding of TGF-β signaling
Transforming growth factor-β (TGF-β) is the prototypical molecule of a superfamily of ligands that regulate diverse aspects of cellular homeostasis, including proliferation, differentiation, migration and death. The superfamily includes several ligands such as TGF-β itself, activin and the bone morphogenetic proteins (BMPs). In almost three decades of research, the principal components and the major molecular events that comprise TGF-β signal transduction have been characterized. Owing to the complexity and quantitative nature of TGF-β signaling, a systems biology understanding of TGF-β signaling is now desired. Mathematical modeling is an important tool in this regard, and several models of TGF-β superfamily signaling have recently been published. In this review, we describe the motivation for mathematical modeling studies of TGF-β signaling and discuss how first-generation models have contributed to the understanding of TGF-β biology.
The dynamics of the TGF-β/Smad signaling module: the numbers matter
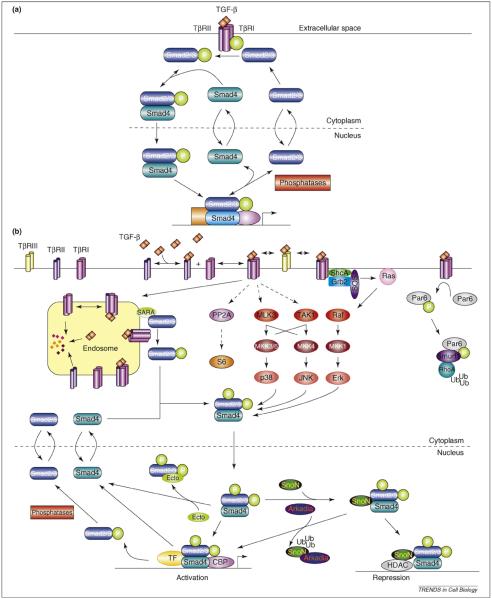
A simplified overview of canonical TGF-β signaling is depicted in Figure 1a. Briefly, TGF-β binds two receptor types, the TGF-β type I and type II receptors (TβRI and TβRII, respectively) to form the active signaling complex. The TβRII activates TβRI kinase activity by phosphorylating the TβRI, which then transmits the signal intracellularly by phosphorylating the Smad transcription factors. There are eight Smad isoforms, which are functionally classified as receptor-regulated Smads (R-Smads; isoforms 1, 2, 3, 5 and 8), common-mediator Smad (Co-Smad; isoform 4) and inhibitory Smads (I-Smads; isoforms 6 and 7). In TGF-β signaling proper, the active TβRI phosphorylates Smads 2 and 3, which facilitates complex formation with Smad4. The Smads constitutively shuttle between the cytoplasm and nucleus, but signaling causes the Smads to accumulate predominantly in the nucleus where they bind DNA and other transcriptional machinery to regulate the expression of target genes. Within the nucleus, the Smad complex can dissociate and the phosphorylated R-Smads (phospho-R-Smads) are dephosphorylated by nuclear phosphatases (such as PPM1A/PP2C [1]), such that the Smads become available for export to the cytoplasm. This cycle continues for as long as active receptors are present [2].
Figure 1.
TGF-β/Smad signaling. (a) An overview of canonical TGF-β/Smad signaling. The TGF-β signal is delivered intracellularly by the TGF-β receptors and Smad transcription factors. (b) TGF-β activates non-canonical pathways and engages in crosstalk. Depicted examples include additional proteins that are putatively involved in canonical signaling, such as the TGF-β type III receptor (TβRIII) and SARA. In addition, TGF-β signaling interacts with other major cell-signaling pathways such as MAPK signaling (i.e. p38, Jnk and ERK), whereby TGF-β is able to activate MAPK signaling but also be modulated by it through phosphorylation of the Smad linker domain. TGF-β can also regulate non-Smad signaling such as RhoA GTPase degradation via activation of Par6. In the nucleus, multiple interactions occur, including repression of Smad signaling by SnoN, which is alleviated by the E3 ubiquitin ligase Arkadia, and additional signaling mediated by interactions between the phospho-R-Smads with other transcriptional regulators, such as ectodermin (Ecto), which is also known as transcriptional intermediary factor 1-γ (TIF1γ).
Canonical signaling through the TβRI and the Smads is necessary [3-6], but not sufficient, for most cellular responses to TGF-β. TGF-β signaling is embedded in the cellular signaling network, such that it regulates non-canonical signaling pathways and engages in crosstalk [7-9] (Figure 1b). In particular, interactions with other major signaling pathways [e.g. mitogen-activated protein kinases (MAPKs)] and extensive interactions with proteins in the nucleus modulate the activities of the canonical pathway (Figure 1b). This feature of TGF-β signaling probably underlies its exceptionally pleiotropic and multifunctional nature [10,11], in which responses to TGF-β depend on context [10] (e.g. ligand concentration as discussed later, cell type, differentiation status [12] and presence of other hormones). Not surprisingly, much research is currently devoted to identifying additional molecules that are essential for TGF-β signaling.
Although such research is needed, we emphasize the need for integrative and quantitative studies to understand TGF-β biology. Quantitative analysis of TGF-β signaling is necessary because the signal itself, TGF-β concentration, is quantitative. Cells can read TGF-β concentration with high precision, as demonstrated by their ability to sense their position in concentration gradients. For example, during wound healing immune cells [13] and fibroblasts [14] chemotax according to TGF-β gradients. In development, TGF-β superfamily members form morphogen gradients to specify the fates of cells according to their position [15] (Box 1). Cells can respond in a graded manner to ligand concentration because the degrees to which proliferation [16], angiogenesis [17], extracellular matrix production and fibrosis [18-20] are regulated in cultured cells depend on ligand concentration. Cells also exhibit discrete responses to ligand concentration. For example, activin concentration can induce five distinct differentiation fates in Xenopus animal cap cells [21]. Similarly, kidney tubule cells proliferate in response to low dose BMP-7 but undergo apoptosis in response to high doses of BMP-7 [22]. Therefore, cells can read and distinguish subtle differences in TGF-β concentration and orchestrate distinct responses.
Box 1. Morphogen gradient signaling.
A morphogen is a signaling molecule that diffuses away from a localized source such that its concentration decreases with increasing distance from the source (hence the term morphogen ‘gradient’) [58]. Cells are thus exposed to different concentrations of the morphogen depending on their position relative to the source, which determines their developmental fate [58]. Morphogens are considered the principal mechanism for specifying positional information to cells during embryonic development [58].
TGF-β-superfamily members are morphogens, the best studied of which are the BMP-2/4 homolog, Dpp in Drosophila, and activin and BMP in Xenopus. Dpp specifies Drosophila embryo dorsal patterning [15] and larval wing imaginal-disc development [59] by signaling through the Smad1 homolog, Mad. Activin signals through Smad2 to help mediate Xenopus mesoderm formation [15] and BMP-4 contributes to dorsal-ventral patterning [60].
Not only is the extracellular signal quantitative but cells are also quantitatively tuned to respond to TGF-β signals, such that perturbing the concentrations of the intracellular signaling components can affect responses to TGF-β. Heterozygous mutant mice that express TGF-β-superfamily-member ligands, receptors, or the Smads below normal levels often display haploinsufficient phenotypes [23,24]. In tumor cells, reduced TβRI expression correlates with attenuated Smad phosphorylation levels, which leads to abrogated expression of genes required for growth inhibition [25]. Even the ratio between the abundances of Smad2 and Smad3 can affect the TGF-β-mediated cytostatic response [26]. Therefore, for basic TGF-β signal transduction, the numbers matter, so much so that a benchmark goal in achieving a systems biology understanding of TGF-β signaling should be a quantitative map of TGF-β signaling in the cell. We provide a sample quantitative map of canonical TGF-β signaling and supply estimates of selected parameters in Box 2.
Box 2. A quantitative perspective of TGF-β/Smad signaling.
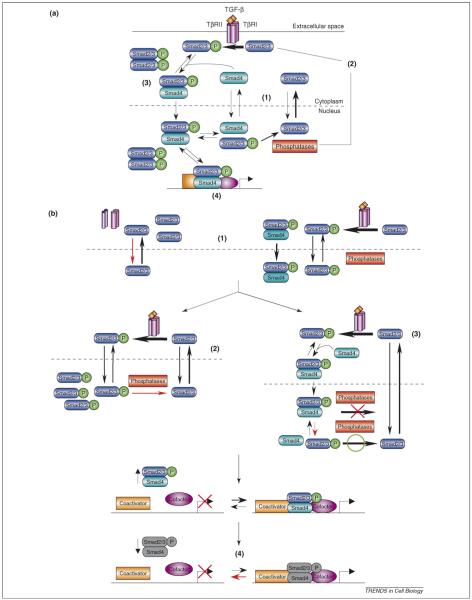
In Figure I, we present a more comprehensive view of TGF-β/Smad signaling, emphasizing the kinetics of the underlying mechanisms, with estimates for selected parameters displayed in Table I. Under basal conditions, the Smads shuttle between the nucleus and cytoplasm, with the rate of nuclear export being higher than the rate of import, such that the Smads are predominantly localized to the cytoplasm [39]. The receptors are also in constant motion, trafficking between the plasma membrane and endosomal compartments [36], with the abundance at the cell surface determined by the balance of receptor production and recycling rates with internalization and degradation rates. The receptors are internalized via two different routes, clathrincoated pits and caveolae [35], with receptors internalized via clathrin-coated pits trafficked to the endosomal compartment for recycling or constitutive degradation through the lysosomal pathway [36] or via caveolae degraded by way of the ubiquitin-proteosomal pathway [35]. Signaling begins with the sequential binding of a TGF-β dimer to homodimers of the type II and type I receptors (TβRII and TβRI) to form the active signaling complex [61]. Within the complex, the constitutively active TβRII kinase phosphorylates the TβRI, which activates the TβRI kinase [62]. The TβRI phosphorylates Smads 2 and 3 at their distal C-terminal serines [63,64], which promotes the reversible interaction between the R-Smads and Smad4 [65,66]. Receptor activity is negatively controlled by dephosphorylation and degradation. Dephosphorylation of the TβRI is mediated by protein phosphatase 1c (PP1c) and growth arrest and DNA damage protein 34 (GADD34) which are recruited to the receptor by Smad7 and enhanced by the protein Smad anchor for receptor activation (SARA) [67,68]. Ligand-induced receptor degradation is thought to occur via the caveolar pathway [35]. Constitutive Smad nucleocytoplasmic shuttling continues during signaling but phosphorylation of the R-Smads and complex formation inhibits their ability to interact with the export machinery [57]. The phospho-R-Smads therefore accumulate in the nucleus because of rate-limiting dephosphorylation and sequestration due to binding to retention factors [39,41,43]. Meanwhile, Smad4 molecules bound to phospho-R-Smad cannot bind to CRM1 [55], a protein required for Smad4 nuclear export [69], such that Smad4 also accumulates in the nucleus. However, a constant rate of phospho-R-Smad dephosphorylation and dissociation of Smad4 from Smad complexes [39,41] ensures transient nuclear residence for each Smad molecule, such that the Smads remain available to continually monitor the state of receptor activation in the cytoplasm [2]. Smad signaling is therefore a dynamic cyclical process; the Smads continuously cycle between the cytoplasm and nucleus and signaling shifts the predominant localization of the Smads to the nucleus.
Figure I.
A dynamic view of canonical TGF-β/Smad signaling. The abbreviations associated with the rate constants for the depicted reactions identify the process and in most cases, the species involved, separated by an underscore. Process abbreviations: a, association; deg, degradation; d, dissociation; exp, nuclear export; imp, nuclear import; int, internalization; kcat, enzyme turnover number; KM, Michaelis-Menten constant; prod, production; rec, recycling. Species abbreviations: TGF-β, transforming growth factor-β ligand; TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor; L-TβRII, ligand-TβRII complex; LRC, ligand-TβRII-TβRI heterotetrameric complex; M2, Smad2 or Smad3; M4, Smad4; Phosphatases, nuclear phosphatases (e.g. PPM1A/PP2C [1] and possibly others).
Based on this discussion, it is reasonable to hypothesize that the diversity of cellular responses to TGF-β signaling stems from the dynamics of many molecules acting together (for a list of molecules that interact with canonical TGF-β signaling, see Ref. [27]). Exploring this hypothesis will require mathematical and computational modeling because such models represent a natural framework for studying quantitative system properties and because they can efficiently manage complexity. Moreover, the predictive power of modeling can help overcome experimental obstacles by inferring the dynamics of molecules that are experimentally inaccessible and by simulating network dynamics for conditions that would otherwise require a prohibitive number of experiments. In this way, mathematical and computational models serve as powerful and efficient tools for investigating TGF-β biology. Accordingly, several studies of TGF-β-superfamily signaling that rely on mathematical models have recently been published. The remainder of this review is devoted to discussing studies of BMP-signaling robustness in Drosophila development and studies of TGF-β/Smad signaling dynamics, which comprise the bulk of modeling studies of TGF-β signaling. Several modeling studies focusing on morphogen gradient formation [with specific applications to Decapentaplegic (Dpp) signaling] have been reviewed elsewhere [28]. For those seeking a better understanding of the modeling process, we provide a basic description in Box 3.
Box 3. The basics of modeling analysis.
Models are first formulated as a diagram similar to that shown in Box 2, Figure I. Each molecule features arrows directed towards and away from it. These arrows represent chemical reactions that either increase or decrease the abundance of the given molecule. The arrows are also associated with a rate constant (indicated by a subscripted k), which is necessary for quantifying the rate at which the reaction occurs. In general, three types of chemical reactions are modeled: association and dissociation reactions (such as protein-protein interactions), enzyme-catalyzed reactions and transport reactions (e.g. nuclear import or export), each of which is described by a rate law. Typically, association and dissociation reactions are described by mass-action kinetics [i.e. the rate is directly proportional to the concentrations of the species involved], whereas enzyme-catalyzed reactions are usually described by Michaelis-Menten kinetics. Handling of transport reactions depends on whether ordinary or partial differential equations are used. Most of the models reviewed here employ ordinary differential equations in which time is the sole independent variable. In these cases, the rates of transport reactions are usually described using mass-action kinetics. Some of the BMP-signaling models (specifically, those proposed in Refs [31] and [34]) use partial differential equations (characterized by two independent variables: time and space) to model transport effects more realistically. Once the rate laws are specified, a differential equation is written for each molecule that describes the overall rate of change of its concentration, which equals the sum of the rates of the individual reactions in which the molecule participates. The resulting system of differential equations is solved using numerical methods implemented using a computer.
Obtaining insight typically involves analyzing the model output (i.e. the kinetics of the molecular concentrations) as a function of the parameter values. Parameters include the model initial conditions, that is, the concentration of each molecule at time zero, and rate constants of each reaction (selected parameter value estimates are listed in Box 2, Table I). Model analysis can be achieved analytically, whereby the equations are manipulated to express the modeled variables as a function of the parameters, or numerically, whereby many simulations are performed with parameter values randomly sampled from reasonable ranges, followed by statistical analyses to determine the relationships between model variables and the parameters. The latter approach is especially useful when parameter estimates are either unavailable or uncertain, which is often the case in models of TGF-β signaling.
For those seeking more in-depth understanding of modeling in cell-signaling research, several excellent reviews and tutorials are available elsewhere [70-74].
BMP morphogen signaling in Drosophila dorsal patterning
BMP ligands have major roles in Drosophila embryo development. For dorsal patterning, the principal ligands are Dpp and Screw (Scw), which signal through a common type II receptor, Punt, and through separate type I receptors, Thickveins (Tkv) and Saxophone (Sax) (Figure 2). The active type I receptors phosphorylate Mad, the Drosophila Smad1 homolog, which carries the signal to the nucleus along with Medea (the Smad4 homolog) to regulate the expression of target genes that control cell differentiation.
Figure 2.
BMP signaling in Drosophila dorsal-ventral patterning. (a) Molecular-level events in BMP signaling at the dorsal surface of the Drosophila embryo. BMP ligands consist of homodimers of either Dpp or Scw or a heterodimer of Dpp and Scw. These ligands (generically represented with gray coloring) form ternary complexes with the proteins Sog and Tsg, which prevents binding of these ligands to their receptors. Tolloid (Tld), a metalloprotease, degrades Sog, the loss of which promotes dissociation of the complex and liberation of the BMP ligand and Tsg. Free Dpp can bind the type II receptor, Punt, and its type I receptor, Tkv, whereas Scw binds to Punt and another type I receptor, Sax. The active type I receptors within the receptor complex phosphorylate the Drosophila Smad1 homolog, Mad. Phospho-Mad forms a complex with the Smad4 homolog, Medea. The phospho-Mad-Medea complex shuttles into the nucleus and regulates the transcription of target genes. (b) Sites of production of the BMP ligands and putative extracellular modulators of BMP signaling. Represented is a transverse cross-section of the Drosophila embryo. Dpp, Scw, Tld, and Tsg are broadly expressed in the dorsal region whereas Sog is expressed ventro-laterally [53]. In the dorsal region, BMP receptors and Mad are expressed uniformly [53]. (c). A simplified view of the dynamics of the extracellular events in BMP signaling (for simplicity, only Dpp is only shown). Solid arrows indicate biochemical reactions, broken arrows represent transport by diffusion. The basic mechanism operates as follows: Dpp diffuses down its concentration gradient ventro-laterally (i); Sog counteracts the movement of Dpp by binding to it and diffusing down its own concentration gradient (ii and iii) transporting Dpp dorsally (iv). Tsg facilitates complex formation between Sog and Dpp. Tld cleaves Sog, which liberates Dpp and Tsg from the complex (v) so that Dpp is free to bind its receptors to initiate signaling (vi). Degradation of Sog by Tld also provides a ‘sink’ for Sog that maintains its concentration gradient. The net result is a sharp phospho-Mad (P-Mad) gradient and downstream signaling in cells located near the dorsal midline.
Embryonic development is robust, implying that the body plan is precisely specified even in the face of biological variability and environmental noise [29]. In the case of BMP signaling in Drosophila, mechanisms operating at the cell exterior ensure that BMP levels are robust to perturbations in order for the gradient of phosphorylated Mad (phospho-Mad) to reliably take shape. Specifically, at least three extracellular proteins are involved: short gastrulation (Sog), which binds Dpp and Scw ligands and inhibits their activities; twisted gastrulation (Tsg), which functions as a binding cofactor in the Sog-Dpp-Scw complex; and tolloid (Tld), a metalloprotease that cleaves Sog (Figure 2). Each of these components is secreted in different locations in the embryo (Figure 2b) such that concentration gradients of each molecule are established. Dpp and Scw diffuse along their gradients towards the dorsal midline where the bulk of BMP signaling occurs (Figure 2c), as visualized by immunostaining of phospho-Mad. Genetic studies established the necessity of these molecules for proper dorsal patterning and biochemical studies characterized the individual reactions between these molecules. Since then, mathematical modeling has been used to integrate these data to understand how these molecules work together to ensure robust BMP signaling in vivo.
Mechanisms of BMP signaling robustness
Restricting diffusion of BMP ligands by receptor-mediated ligand internalization
Experiments using heterozygous fly mutants show that the shape of the phospho-Mad gradient at the dorsal midline is robust to perturbations in gene dosage of sog, tld and scw, but not dpp [30]. To understand the basis for this observed robustness, Eldar et al. [30] devised a simple mathematical model that included only three species: a generic BMP ligand, Sog, and Tld. In their model, BMP and Sog could form a complex; BMP, Sog, and BMP-Sog could also diffuse freely, and Tld could cleave both the free and bound formsof Sog. To identify robust networks, the authors ran simulations in which the model equations were solved using a parameter set consisting of values randomly selected from a range of reasonable values (no experimental estimates were available at the time). Four simulations were carried out per parameter set; the first simulation employed wild-type concentrations for sog, tld and the BMP ligand, and one simulation was run for each molecule in which its concentration was reduced by half (to simulate a heterozygous mutant). The properties of the predicted BMP gradients from the four simulations were compared and, if they were sufficiently similar, the network was considered robust. This set of simulations was repeated for thousands of parameter sets. The subset of parameter values that led to robust networks was then statistically analyzed, prompting the authors to infer two network properties that confer robustness: (i) preferential cleavage by Tld of bound Sog over free Sog and (ii) restricted diffusion of free BMP. Eldar et al. [30] then confirmed that these network properties conferred robustness in a more comprehensive model of BMP signaling, indicating that these properties underlie the mechanism of the experimentally observed robustness.
Since then, the conclusions of Eldar et al. [30] have been contested, especially the rather stringent condition of restricted BMP diffusion. Mizutani et al. [31] proposed an alternative model for robust BMP activity gradient formation. The goals of the model were to discover the minimal conditions necessary to capture the shape and dynamics of the phospho-Mad gradient. An important difference that distinguishes the models of Mizutani et al. [31] and Eldar et al. [30] is the inclusion of receptor-mediated ligand degradation in the former - that is, the loss of ligand that occurs when receptors bound to ligand are internalized and degraded (Box 2, Figure I). The models are otherwise similar, with the basic species and mechanisms of BMP gradient formation incorporated, including BMP ligand, BMP receptor, Sog, Tsg, the BMP-receptor complex, the Sog-Tsg complex and the BMP-Sog-Tsg complex. Key reactions included production (i.e. de novo biosynthesis) of each species, receptor-mediated BMP degradation, reversible interactions for the complexes listed, and Tld-mediated Sog degradation (restricted to Sog in the BMP-Sog-Tsg complex). The model output was defined as the abundance of BMP-receptor complex, which was used as a surrogate measure of phospho-Mad levels for comparison to the experimental data of Mizutani et al. [31].
The model of Mizutani et al. [31] successfully captures the basic qualitative features and dynamics of the phospho-Mad gradient, although quantifying success in such cases is difficult because the experimental data consist solely of stained embryos that one must subjectively compare with the model output. We do note, however, that this model was only partially successful in predicting the phospho-Mad gradient when sog gene dosage was reduced by half (to simulate a heterozygous mutant). The model predicted a robust response in the phospho-Mad gradient, whereas the experimental data showed wider phospho-Mad distribution in sog-/+ mutant embryos [31]. Nevertheless, the model can account for two perplexing experimental observations. First, the model explains how reducing sog dosage in a dpp-/+ embryo can partially restore the phospho-Mad gradient, despite both mutants reducing the phospho-Mad signal intensity when introduced individually. In dpp-/+ embryos, the phospho-Mad signal takes longer to achieve maximal amplitude, whereas the phospho-Mad signal peaks earlier in sog-/+ embryos compared with wild type. Combining the two mutations restores the timing of maximum phospho-Mad amplitude. Second, the model accounts for the extended phospho-Mad activity gradient in the presence of Sog. This counter-intuitive observation, given that Sog is a putative inhibitor of Dpp activity, is explained by Sog sequestering Dpp from receptor-mediated ligand degradation. This observation explains why Eldar et al. [30] concluded that the BMP-Sog complex diffuses faster than BMP alone; it was a consequence of their model omitting receptor-mediated ligand degradation. Therefore, the simple model of Mizutani et al. [31] reveals the important consequence of receptor-mediated ligand degradation on phospho-Mad activity dynamics.
Ligand heterodimerization and positive feedback
The model of Mizutani et al. [31] illustrates how a simple model can lead to insight. However, to better reflect reality, models are modified to account for new data as they emerge. Such was the case for Shimmi et al. [32], who observed that Scw and Dpp formed a heterodimer and that Scw helps localize Dpp at the dorsal midline [32]. Moreover, the Dpp-Scw heterodimer causes stronger phospho-Mad activity and promotes Sog cleavage by Tld more potently than either the Scw or Dpp homodimers. These observations indicated that ligand heterodimerization is important for phospho-Mad gradient formation. To address whether ligand heterodimerization could enhance the robustness of BMP signaling, Shimmi et al. [32] turned to mathematical modeling. Specifically, a simple model was devised that included the production of Dpp and Scw, all possible dimerization reactions (both homo- and heterodimers) and degradation of all species to examine how heterodimer abundance changes as a function of reduced Dpp and Scw production rates (to simulate the effect of reduced gene dosage). This model revealed that heterodimerization effectively buffers reductions in Scw production rate. Whereas the driving force for homodimerization is substantially impacted by monomer production, the driving force for heterodimerization is partially preserved because the interacting partner levels are unchanged. This effect depends on the normal rate of Scw production being in excess of that of Dpp, which the authors claim is the case [32]. Therefore, robustness via heterodimerization is asymmetric, favoring the interacting partner produced at the faster rate. This explains why the phospho-Mad signal is robust in scw+/- mutants but not in dpp+/- mutants.
Around the same time that the models of Mizutani et al. [31] and Shimmi et al. [32] were published, experimental evidence emerged indicative of a positive-feedback circuit that could enhance BMP ligand binding to the cell surface in response to signaling [33]. Positive feedback is a common way to induce bistability, whereby a dynamic system adopts one of two stable steady-states based on the input. In this case, phospho-Mad activity is induced either strongly or weakly in response to BMP concentration. Umulis et al. [34] performed a modeling study to evaluate the potential impact of positive feedback on shaping the phospho-Mad gradient in response to BMP. Their model featured a mechanism in which cells expressed a cell-surface BMP-binding protein that potentiated the binding of BMP to its receptor. Receptor-mediated endocytosis, which leads to ligand degradation, was also included. The authors then simulated the spatio-temporal dynamics of phospho-Mad gradient formation in response to BMP signaling (Dpp-Scw heterodimers constituted the principal BMP ligand). They found that positive feedback could replicate the observed sharpening of phospho-Mad activity at the dorsal midline that occurs during the latter stages of BMP signaling. The basis for this behavior is as follows: (i) initially, a broad BMP gradient leads to a similarly broad phospho-Mad gradient of low amplitude; (ii) as signaling progresses, cells express the cell-surface BMP-binding protein in a manner proportional to the amount of BMP to which they were exposed, leading to the opposing effects of a higher number of active receptor complexes and also to the removal of higher amounts of BMP from the immediate area via receptor-mediated ligand internalization and degradation; and (iii) competition for binding limited amounts of BMP ensues, whereby BMP diffuses to areas with higher numbers of cell-surface BMP-binding receptors (i.e. towards the dorsal midline), which enhances BMP signaling at the midline but reduces it away from the midline, thus sharpening the phospho-Mad gradient. The model of Umulis et al. [34] also captures the degree of experimentally observed robustness to gene dosages of sog, tsg, tld and scw. Therefore, BMP-mediated positive feedback serves as a feasible mechanism for robustly shaping the phospho-Mad gradient.
The studies discussed here serve as excellent examples of how modeling can help us understand complex biological mechanisms. Preferential cleavage of bound Sog, receptor-mediated ligand degradation, ligand heterodimerization and positive feedback all seem to be viable mechanisms underlying the robustness of BMP signaling in Drosophila dorsal patterning. The next step will be to clarify the respective roles of each of these mechanisms in BMP-signaling robustness. Experiments that would be particularly helpful in this regard include quantifying the absolute numbers of phospho-Mad molecules in the cells of the Drosophila embryo, which could help to discriminate between competing models by determining the models that predict these numbers most accurately, and identifying the putative positive feedback factor.
The duplicity of TGF-β in cancer: a role for receptor dynamics?
Recent experimental evidence indicates that TGF-β receptors are subject to continual trafficking events, such as internalization from the plasma membrane to the endosome, recycling back to the plasma membrane, and degradation through the endolysosomal pathway (via clathrin-coated pits) and the caveolar pathway [35,36] (Box 2, Figure I). However, the data are not without controversy because the importance of the putative degradation routes has not been definitively resolved. Quick resolution of these issues might be warranted, given the recent results from mathematical models of TGF-β signaling that predict important signal processing consequences of differential receptor trafficking.
Vilar et al. [37] explored a model of TGF-β receptor trafficking that includes production, internalization, recycling and constitutive and ligand-induced degradation. Formal mathematical analysis and computer simulations revealed that the ratio of the rate constants for constitutive and ligand-induced receptor degradation is crucial for regulating signal kinetics and fidelity (i.e. the degree to which the signal reflects the input) [37]. When constitutive receptor degradation dominates, the signal will faithfully reflect the input (defined as ligand concentration over time) [37], such that a sustained input will result in a sustained signal. Conversely, dominant ligand-induced receptor degradation leads to transient receptor activity, even if the TGF-β signal is sustained [37]. Importantly, a later comprehensive model of TGF-β signaling shows that these signal processing effects at the receptor level are also transmitted to the Smad signal [38]. Furthermore, the interplay between these rates can cause interesting dynamic properties. For example, if two ligands signal through a common receptor [e.g. TGF-β and BMP-7 signaling through activin-like kinase 2 (Alk2)], then the ratio between the constitutive and ligand-induced receptor degradation rates determines the degree to which the signals are coupled (i.e. the degree to which the signals interact), with higher coupling associated with dominant ligand-induced receptor degradation [37] (Figure 3). Such coupling, in which the common receptor is degraded by the action of one ligand, could underlie the reversal of TGF-β from tumor suppressor to tumor promoter [37] (Figure 3), which is an important but poorly understood feature of TGF-β biology. Although interesting, this prediction remains speculative because it depends on several untested assumptions, including whether tumor cells have dominant ligand-induced receptor degradation and whether its rate is sufficient to markedly deplete the common receptor.
Figure 3.
A potential mechanism by which TGF-β changes from a tumor suppressor to tumor promoter. TGF-β superfamily members signal through common receptors in a combinatorial manner [54]. In this example, TGF-β and BMP-7 signal through different type II receptors but through the same type I receptor, Alk2 (a). The degree of signal coupling (i.e. the degree to which the signals interact) depends on the ratio of the rates of constitutive degradation and ligand-induced degradation of the receptors. If constitutive degradation dominates, then the signals are independent of each other - compare the number of active receptor complexes for BMP-7 signaling in (b) and (c) - presumably, because the number of receptors is kept constant through balanced production and degradation independently of signaling. Conversely, if ligand-induced degradation is the dominant mechanism for negatively regulating receptor activity, then one signal can limit the other and vice versa because the common receptor becomes depleted during signaling (d). The potential for TGF-β signaling to deplete a shared receptor could underlie its switch from tumor suppressor to tumor promoter, because, in the presence of TGF-β, Alk2 is degraded, which limits the ability of BMP-7 to exert its effects on the cell. The tumor cell would therefore develop resistance to any ligand that shares a common receptor with TGF-β.
Smad nuclear accumulation: a shared responsibility
The key intracellular signal in TGF-β signaling is the concentration of Smad complexes in the nucleus, yet, definitive identification of the mechanism causing Smad nuclear accumulation remains elusive. Mathematical models of Smad signaling have provided essential insight into the mechanisms of Smad nuclear accumulation.
Simple mathematical models have been used to interpret fluorescence imaging data to estimate rate constants for Smad nuclear import and export [39,40] (Box 2, Table I). In the absence of TGF-β, a slower Smad nuclear import rate versus that of export has been shown to cause the Smads to localize mainly in the cytoplasm in cultured mammalian cells [39], whereas slower nuclear export was shown to lead to predominant nuclear localization of Mad in Drosophila muscle cells [40]. During signaling in both these types of cells, the rate of Smad nuclear import did not change, whereas the observed rate of Smad nuclear export decreased [39,40]. The decrease in the observed export rate correlated with a decrease in the mobility of Smads in the nucleus [39,40], leading to the conclusion that the Smads are sequestered in the nucleus by binding to (as yet unidentified) retention factors, thus causing Smad nuclear accumulation [39,41]. Although these models have provided a necessary clue as to what might be occurring at the molecular level, we question the interpretation of this result. The reduced Smad mobility in the nucleus merely coincides with TGF-β signaling and might not be causal for Smad nuclear accumulation. It is plausible that an increase in Smad nuclear concentration caused by an alternative mechanism could drive binding of the Smads to factors in the nucleus that would reduce the mobility of the Smads. Furthermore, the ‘retention-factor hypothesis’ implies strong prolonged binding of the Smads to the retention factors. However, the strength and time of binding cannot be inferred directly from the fluorescence imaging data. Indeed, reduced nuclear mobility could reflect transient binding [42], which implies that a fraction of rapidly exchanging unbound Smads would always be available for export. Given that export continually depletes the pool of unbound Smads, prolonged sequestration of the Smads is unlikely, such that retention factors cannot be the sole causal mechanism for Smad nuclear accumulation. To uncover alternative possible mechanisms of Smad nuclear accumulation, we turned to mathematical modeling.
Table I. Estimates for selected parameters involved in TGF-β signaling.
| Parameter | Units | Estimated value | Refs |
|---|---|---|---|
| [Smad2] | M | 6 × 10-10 | [75] |
| Molecules cell-1 | 3.6 × 105 | [38] (calculated from [75]) | |
| Molecules cell-1 | 8.5-12 × 104 | [43] | |
| [Smad3] | Molecules cell-1 | 1.1-20 × 104 | [43] |
| [Smad4] | M | 1.4 × 10-9 | [75] |
| Molecules cell-1 | 8.4 × 105 | [38] (calculated from [75]) | |
| Molecules cell-1 | 9.3-12 × 104 | [43] | |
| Number of TβRI at cell surface | Molecules cell-1 | <4000 | [76] |
| Number of TβRII at cell surface | Molecules cell-1 | <4000 | [76] |
| Number of TβRIII at cell surface | Molecules cell-1 | <100000 | [76] |
| TGF-β1-TβRII association rate constanta | M-1s-1 | (2.3 ± 0.2) × 107 | [77] |
| TGF-β1-TβRII dissociation rate constanta | s-1 | (1.5 ± 0.2) × 10-4 | [77] |
| Smad2-Smad4 dissociation constant | M | 270 ± 66 × 10-9 | [78] |
| Phospho-Smad2-Smad4 dissociation constantb | M | 79 ± 18 × 10-9 | [78] |
| Smad2 nuclear import rate constant | s-1 | 2.7 ± 0.4 × 10-3 | [39] |
| Smad2 nuclear export rate constant | s-1 | 5.8 ± 0.7 × 10-3 | [39] |
| Receptor internalization rate constant (clathrin-coated pit pathway) | min-1 | 0.33 | [35,37,38] |
| Receptor internalization rate constant (caveolar pathway) | min-1 | 0.25-0.33 | [35,37,38] |
| Receptor recycling rate constant | min-1 | 0.033 | [36-38] |
| TβRI degradation rate constant (constitutive, via endosomal-lysosomal pathway) | min-1 | 0.005 | [38,79] |
| TβRII degradation rate constant (constitutive, via endosomal-lysosomal pathway) | min-1 | 0.025 | [38,79] |
We have developed a kinetic model of canonical Smad signaling that includes R-Smad (i.e. Smad2 and Smad3) phosphorylation, heterodimerization with Smad4 and nucleocytoplasmic shuttling steps [43]. To shed light on the principal mechanisms controlling Smad nuclear accumulation, we statistically analyzed sets of parameter values to determine the parameters to which Smad nuclear accumulation is most sensitive. Perturbing the rate constants for R-Smad phosphorylation, R-Smad dephosphorylation, and R-Smad/Smad4 complex dissociation in the nucleus caused the largest changes in Smad nuclear accumulation [43]. Further analyses revealed flaws with the hypothesis that retention factors are the cause of Smad nuclear accumulation; specifically, that physically unrealistic parameter values would be necessary for retention factors to compete with rapid dephosphorylation [43]. Our modeling analysis prompted two hypotheses for Smad nuclear accumulation: (i) rate-limiting phospho-R-Smad dephosphorylation in the nucleus relative to the rate of R-Smad phosphorylation in the cytoplasm determines the degree of Smad nuclear accumulation and (ii) participation of the Smads in complexes with cytoplasmic binding factors that shuttle as a complex into the nucleus could protect the phospho-R-Smads from rapid dephosphorylation to promote Smad nuclear accumulation [43] (Figure 4). Both mechanisms ensure that the degree of Smad nuclear accumulation is directly proportional to receptor activity, which has been experimentally demonstrated in activin signaling [44,45]. The first hypothesis is reinforced by analyses of more comprehensive models of TGF-β/Smad signaling, which also show that the rate constants associated with R-Smad phosphorylation and dephosphorylation are important for determining model behavior [38,46,47]. In addition, experimental data exist that support a primary role for oligomerization, because oligomerization-incompetent Smad2 does not accumulate in the nucleus upon TGF-β signaling [39]. Therefore, both mechanisms might cooperate to cause Smad nuclear accumulation.
Figure 4.
Mechanisms of Smad nuclear accumulation. (a). Summary of the four postulated mechanisms for Smad nuclear accumulation in the context of canonical TGF-β/Smad signaling. (b). Smad nuclear accumulation results from several mechanisms. Red arrows indicate the rate-limiting reactions that could promote Smad nuclear accumulation. The thickness of the arrows indicates the relative reaction rates. Species that would exist at very low abundance under the assumed conditions are colored gray. In the absence of TGF-β, the Smads localize predominantly in the cytoplasm because the rate of nuclear export exceeds the rate of nuclear import [39] (1). In the presence of TGF-β signaling, enhanced nuclear import of Smad complexes compared to monomeric Smads might occur [47] (1), but most evidence to date indicates that it does not [39,40,55,56]. Furthermore, the nuclear-export machinery neither recognizes phosphorylated R-Smads strongly [57] nor Smad4 contained within Smad complexes [47,55], thereby contributing to Smad complex nuclear accumulation (1). Therefore, dissociation of Smad complexes and dephosphorylation of phospho-R-Smad are probably prerequisites for nuclear export, such that these two mechanisms represent potential causes of Smad nuclear accumulation during signaling. If dephosphorylation by the nuclear phosphatase(s) is rate-limiting, then Smad nuclear accumulation occurs if the rate of R-Smad phosphorylation is higher than that of dephosphorylation [43] (2). If dephosphorylation is not rate-limiting, then Smad oligomerization in the cytoplasm, for example with Smad4, could protect the phospho-R-Smads from the phosphatase upon nuclear translocation [43] (3). In this case, the rate at which the Smad complex dissociates in the nucleus would primarily determine the degree of Smad nuclear accumulation. A third possibility is that both mechanisms contribute substantially to Smad nuclear accumulation, which seems to be the case in vivo [47]. Finally, Smad nuclear accumulation caused by either (2) or (3) promotes the reversible binding of nuclear Smads to transcriptional cofactors, coactivators and corepressors, and DNA. Binding to such nuclear-retention factors could further sequester the phospho-R-Smads from dephosphorylation and Smad4 from the nuclear-export machinery [39,41] (4). As signaling ends, the rate of R-Smad phosphorylation decreases, which decreases the driving force for Smad complex formation. Specifically, continual phospho-R-Smad dephosphorylation depletes the concentration of monomeric phospho-R-Smads such that reversible binding reactions are driven towards dissociation, thus reducing R-Smad oligomer abundance and the abundance of Smads bound to nuclear-retention factors. The rates at which the Smad complexes dissociate probably contribute to determining the observed rate of Smad nuclear export.
Recently, a model of Smad signaling dynamics used in conjunction with fluorescence imaging data has provided further insight into the mechanism of Smad nuclear accumulation. Enabling a faster nuclear-import rate constant for Smad complexes (phospho-R-Smad homodimers and phospho-R-Smad-Smad4 heterodimers) compared with that of monomeric Smads improved the data fits [47], indicating that faster nuclear import of Smad complexes might be necessary for Smad signaling dynamics. Although published data indicate a possible mechanism for differential nuclear import [48], this result contradicts those described here that showed no differences in the rate of Smad nuclear import during signaling [39,40]. Alternatively, the result might reflect the presence of an additional parameter in the model. Models with more adjustable parameters are, by default, more capable of fitting a given dataset because each parameter confers a degree of flexibility to the model. Resolving this issue could perhaps be accomplished by performing fluorescence recovery after photobleaching experiments using Smad constructs fused to complementary fragments of a fluorescent protein (cf. [49]) that depend on oligomerization of the target proteins for fluorescence to appear, such that nuclear import rates of Smad oligomers could be specifically measured. Furthermore, the model of Schmierer et al. [47] predicts that Smad heterodimers are the most abundant Smad species in the nucleus during signaling, the absolute abundance of which is most sensitive to rate constants describing R-Smad phosphorylation, phospho-R-Smad dephosphorylation, and Smad complex affinity. Further analysis has revealed that a combination of rate-limiting Smad complex dissociation and phospho-R-Smad dephosphorylation conferred the best data fit [47], which indicates that Smad nuclear accumulation is a function of many molecular mechanisms acting together rather than of a single dominant mechanism (Figure 4b).
Concluding remarks
An ultimate goal in TGF-β-signaling research is to fully account for cellular responses to TGF-β under a variety of conditions based on molecular mechanisms. Achieving this goal will require accounting for the complexity of TGF-β signaling and studying its quantitative properties, which are tasks well suited to mathematical modeling. Indeed, mathematical models of TGF-β superfamily signaling have provided insights into key questions of TGF-β biology and we expect that modeling will increase in prominence as questions that are more integrative in nature are posed. Intriguing questions that we foresee being addressed include how discrete cellular responses, for example the decision to differentiate into a particular cell type or the decision to apoptose, can arise from ligand concentration, which is a continuous variable [50]. Integrating models of TGF-β signaling with those of other signaling pathways could address questions about signaling crosstalk. Beyond TGF-β actions at the cellular level, models focusing on tissue-level effects (e.g. the model of Dallon et al. [51]) or those spanning multiple levels of hierarchy (e.g. the model of Ribba et al. [52]) will help improve understanding of the physiological roles of TGF-β. Aside from these projections, the trajectory of modeling in TGF-β biology research is difficult to predict. Because models help us understand and visualize phenomena that are beyond our intuition, they will probably inspire questions that are currently beyond the limits of our imagination.
Acknowledgements
We thank Kristen Barthel, Richard Erickson, and Scott Dixon for critical readings of the manuscript. D.C.C. was supported by a Doctoral Research Award from the Canadian Institutes for Health Research. Research in our laboratory is supported by a National Institutes of Health grant (GM083172) to X.L.
References
- 1.Lin X, et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inman GJ, et al. Nucleocytoplasmic shuttling of Smads 2, 3 and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 3.Karlsson G, et al. Gene expression profiling demonstrates that TGF-β1 signals exclusively through receptor complexes involving Alk5 and identifies targets of TGF-β signaling. Physiol. Genomics. 2005;21:396–403. doi: 10.1152/physiolgenomics.00303.2004. [DOI] [PubMed] [Google Scholar]
- 4.Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang YC, et al. Hierarchical model of gene regulation by transforming growth factor β. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10269–10274. doi: 10.1073/pnas.1834070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carcamo J, et al. Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor β and activin. Mol. Cell. Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moustakas A, Heldin CH. Non-Smad TGF-β signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 8.Rahimi RA, Leof EB. TGF-β signaling: a tale of two responses. J. Cell. Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 9.Herpin A, Cunningham C. Cross-talk between the bone morphogenetic protein pathway and other major signaling pathways results in tightly regulated cell-specific outcomes. FEBS J. 2007;274:2977–2985. doi: 10.1111/j.1742-4658.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 10.Flanders KC, Roberts A. TGFβ. In: Oppenheim JJ, et al., editors. Cytokine Reference: a Compendium of Cytokines and Other Mediators of Host Defense. Vol. 1. Academic Press; 2001. pp. 719–746. [Google Scholar]
- 11.Sporn MB, Roberts AB. Peptide growth factors are multifunctional. Nature. 1988;332:217–219. doi: 10.1038/332217a0. [DOI] [PubMed] [Google Scholar]
- 12.Mabie PC, et al. Multiple roles of bone morphogenetic protein signaling in the regulation of cortical cell number and phenotype. J. Neurosci. 1999;19:7077–7088. doi: 10.1523/JNEUROSCI.19-16-07077.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahl SM, et al. Transforming growth factor type β induces monocyte chemotaxis and growth factor production. Proc. Natl. Acad. Sci. U. S. A. 1987;84:5788–5792. doi: 10.1073/pnas.84.16.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postlethwaite AE, et al. Stimulation of the chemotactic migration of human fibroblasts by transforming growth factor β. J. Exp. Med. 1987;165:251–256. doi: 10.1084/jem.165.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurdon JB, Bourillot PY. Morphogen gradient interpretation. Nature. 2001;413:797–803. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- 16.Roberts AB, et al. Type β transforming growth factor: a bifunctional regulator of cellular growth. Proc. Natl. Acad. Sci. U. S. A. 1985;82:119–123. doi: 10.1073/pnas.82.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepper MS, et al. Biphasic effect of transforming growth factor-β 1on in vitro angiogenesis. Exp. Cell Res. 1993;204:356–363. doi: 10.1006/excr.1993.1043. [DOI] [PubMed] [Google Scholar]
- 18.Roberts AB, et al. Transforming growth factor type β: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc. Natl. Acad. Sci. U. S. A. 1986;83:4167–4171. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ignotz RA, Massague J. Transforming growth factor-β stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J. Biol. Chem. 1986;261:4337–4345. [PubMed] [Google Scholar]
- 20.Laiho M, et al. Enhanced production and extracellular deposition of the endothelial-type plasminogen activator inhibitor in cultured human lung fibroblasts by transforming growth factor-β. J. Cell Biol. 1986;103:2403–2410. doi: 10.1083/jcb.103.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green JB, et al. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–739. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 22.Piscione TD, et al. BMP7 controls collecting tubule cell proliferation and apoptosis via Smad1-dependent and -independent pathways. Am. J. Physiol. Renal Physiol. 2001;280:F19–F33. doi: 10.1152/ajprenal.2001.280.1.F19. [DOI] [PubMed] [Google Scholar]
- 23.Weinstein M, Deng C-X. Genetic disruptions within the murine genome reveal numerous roles of the Smad gene family in development, disease, and cancer. In: ten Dijke P, Heldin CH, editors. Smad Signal Transduction: Smads in Proliferation, Differentiation and Disease. Vol. 5. Springer; 2006. pp. 151–176. [Google Scholar]
- 24.Goumans MJ, Mummery C. Functional analysis of the TGF-β receptor/Smad pathway through gene ablation in mice. Int. J. Dev. Biol. 2000;44:253–265. [PubMed] [Google Scholar]
- 25.Nicolas FJ, Hill CS. Attenuation of the TGF-β-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-β-induced growth arrest. Oncogene. 2003;22:3698–3711. doi: 10.1038/sj.onc.1206420. [DOI] [PubMed] [Google Scholar]
- 26.Kim SG, et al. The endogenous ratio of Smad2 and Smad3 influences the cytostatic function of Smad3. Mol. Biol. Cell. 2005;16:4672–4683. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor IW, Wrana JL. SnapShot: the TGF β pathway interactome. Cell. 2008;133:378.e1. doi: 10.1016/j.cell.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Ibanes M, Belmonte JC. Theoretical and experimental approaches to understand morphogen gradients. Mol. Syst. Biol. 2008;4:176. doi: 10.1038/msb.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barkai N, Shilo BZ. Variability and robustness in biomolecular systems. Mol. Cell. 2007;28:755–760. doi: 10.1016/j.molcel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Eldar A, et al. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304–308. doi: 10.1038/nature01061. [DOI] [PubMed] [Google Scholar]
- 31.Mizutani CM, et al. Formation of the BMP activity gradient in the Drosophila embryo. Dev. Cell. 2005;8:915–924. doi: 10.1016/j.devcel.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimmi O, et al. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YC, Ferguson EL. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature. 2005;434:229–234. doi: 10.1038/nature03318. [DOI] [PubMed] [Google Scholar]
- 34.Umulis DM, et al. Robust, bistable patterning of the dorsal surface of the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11613–11618. doi: 10.1073/pnas.0510398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Guglielmo GM, et al. Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nat. Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell H, et al. Ligand-dependent and -independent transforming growth factor-β receptor recycling regulated by clathrin-mediated endocytosis and Rab11. Mol. Biol. Cell. 2004;15:4166–4178. doi: 10.1091/mbc.E04-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilar JM, et al. Signal processing in the TGF-β superfamily ligand-receptor network. PLoS Comput. Biol. 2006;2:e3. doi: 10.1371/journal.pcbi.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zi Z, Klipp E. Constraint-based modeling and kinetic analysis of the Smad dependent TGF-β signaling pathway. PLoS ONE. 2007;2:e936. doi: 10.1371/journal.pone.0000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudu V, et al. Postsynaptic mad signaling at the Drosophila neuromuscular junction. Curr. Biol. 2006;16:625–635. doi: 10.1016/j.cub.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 41.Nicolas FJ, et al. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 2004;117:4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- 42.Beaudouin J, et al. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 2006;90:1878–1894. doi: 10.1529/biophysj.105.071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke DC, et al. Systems theory of Smad signalling. Syst. Biol. (Stevenage) 2006;153:412–424. doi: 10.1049/ip-syb:20050055. [DOI] [PubMed] [Google Scholar]
- 44.Bourillot PY, et al. A changing morphogen gradient is interpreted by continuous transduction flow. Development. 2002;129:2167–2180. doi: 10.1242/dev.129.9.2167. [DOI] [PubMed] [Google Scholar]
- 45.Dyson S, Gurdon JB. The interpretation of position in a morphogen gradient as revealed by occupancy of activin receptors. Cell. 1998;93:557–568. doi: 10.1016/s0092-8674(00)81185-x. [DOI] [PubMed] [Google Scholar]
- 46.Melke P, et al. A rate equation approach to elucidate the kinetics and robustness of the TGF-β pathway. Biophys. J. 2006;91:4368–4380. doi: 10.1529/biophysj.105.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmierer B, et al. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6608–6613. doi: 10.1073/pnas.0710134105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu L, et al. Msk is required for nuclear import of TGF-β/BMP-activated Smads. J. Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saka Y, et al. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007;134:4209–4218. doi: 10.1242/dev.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green J. Morphogen gradients, positional information, and Xenopus: interplay of theory and experiment. Dev. Dyn. 2002;225:392–408. doi: 10.1002/dvdy.10170. [DOI] [PubMed] [Google Scholar]
- 51.Dallon JC, et al. Modeling the effects of transforming growth factor-β on extracellular matrix alignment in dermal wound repair. Wound Repair Regen. 2001;9:278–286. doi: 10.1046/j.1524-475x.2001.00278.x. [DOI] [PubMed] [Google Scholar]
- 52.Ribba B, et al. A multiscale mathematical model of cancer, and its use in analyzing irradiation therapies. Theor. Biol. Med. Model. 2006;3:7. doi: 10.1186/1742-4682-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Connor MB, et al. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 55.Chen HB, et al. Nuclear targeting of transforming growth factor-β-activated Smad complexes. J. Biol. Chem. 2005;280:21329–21336. doi: 10.1074/jbc.M500362200. [DOI] [PubMed] [Google Scholar]
- 56.Xu L, et al. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nat. Cell Biol. 2000;2:559–562. doi: 10.1038/35019649. [DOI] [PubMed] [Google Scholar]
- 57.Kurisaki A, et al. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell. Biol. 2006;26:1318–1332. doi: 10.1128/MCB.26.4.1318-1332.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- 59.Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 60.Hill CS. TGF-β signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev. 2001;11:533–540. doi: 10.1016/s0959-437x(00)00229-x. [DOI] [PubMed] [Google Scholar]
- 61.Wrana JL, et al. TGF β signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 62.Wrana JL, et al. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 63.Souchelnytskyi S, et al. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J. Biol. Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 64.Liu X, et al. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagna G, et al. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 66.Chacko BM, et al. Structural basis of heteromeric smad protein assembly in TGF-β signaling. Mol. Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 67.Valdimarsdottir G, et al. Smad7 and protein phosphatase 1 α are critical determinants in the duration of TGF-β/ALK1 signaling in endothelial cells. BMC Cell Biol. 2006;7:16. doi: 10.1186/1471-2121-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itoh S, Ten Dijke P. Negative regulation of TGF-β receptor/Smad signal transduction. Curr. Opin. Cell Biol. 2007;19:176–184. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Pierreux CE, et al. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 2000;20:9041–9054. doi: 10.1128/mcb.20.23.9041-9054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sible JC, Tyson JJ. Mathematical modeling as a tool for investigating cell cycle control networks. Methods. 2007;41:238–247. doi: 10.1016/j.ymeth.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klipp E, Liebermeister W. Mathematical modeling of intracellular signaling pathways. BMC Neurosci. 2006;7(Suppl 1):S10. doi: 10.1186/1471-2202-7-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mogilner A, et al. Quantitative modeling in cell biology: what is it good for? Dev. Cell. 2006;11:279–287. doi: 10.1016/j.devcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 73.Reeves GT, et al. Quantitative models of developmental pattern formation. Dev. Cell. 2006;11:289–300. doi: 10.1016/j.devcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 74.Andrews SS, Arkin AP. Simulating cell biology. Curr. Biol. 2006;16:R523–R527. doi: 10.1016/j.cub.2006.06.048. [DOI] [PubMed] [Google Scholar]
- 75.He W, et al. Hematopoiesis controlled by distinct TIF1 γ and Smad4 branches of the TGF β pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 76.Massague J. The transforming growth factor-β family. Annu. Rev. Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 77.De Crescenzo G, et al. Transforming growth factor-β (TGF-β) binding to the extracellular domain of the type II TGF-β receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high affinity binding. J. Mol. Biol. 2003;328:1173–1183. doi: 10.1016/s0022-2836(03)00360-7. [DOI] [PubMed] [Google Scholar]
- 78.Funaba M, Mathews LS. Identification and characterization of constitutively active Smad2 mutants: evaluation of formation of Smad complex and subcellular distribution. Mol. Endocrinol. 2000;14:1583–1591. doi: 10.1210/mend.14.10.0537. [DOI] [PubMed] [Google Scholar]
- 79.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 80.Groppe J, et al. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]