Abstract
Transcript elongation by polymerase II paused at the Egr1 promoter is activated by mitogen-activated protein kinase phosphorylation of the ternary complex factor (TCF) ELK1 bound at multiple upstream sites and subsequent phospho-ELK1 interaction with mediator through the MED23 subunit. Consequently, Med23 knockout (KO) nearly eliminates Egr1 (early growth response factor 1) transcription in embryonic stem (ES) cells, leaving a paused polymerase at the promoter. Med23 KO did not, however, eliminate Egr1 transcription in fibroblasts. Chromatin immunoprecipitation analysis and direct visualization of fluorescently labeled TCF derivatives and mediator subunits revealed that three closely related TCFs bound to the same control regions. The relative amounts of these TCFs, which responded differently to the loss of MED23, differed in ES cells and fibroblasts. Transcriptome analysis suggests that most genes expressed in both cell types, such as Egr1, are regulated by alternative transcription factors in the two cell types that respond differently to the same signal transduction pathways.
INTRODUCTION
The eukaryotic mediator of transcription complex (mediator) functions as a molecular bridge that links regulatory signals from DNA-bound transcription factors to RNA polymerase II (Pol II) and its several attendant initiation factors [general transcription factors (GTFs)] through direct protein interactions. The assembly of ~30 individual subunits into an active mediator complex about the size of a ribosomal subunit creates a large surface area that could potentially facilitate numerous simultaneous interactions with different transcriptional regulators that each influence transcription initiation. Understanding these interactions on a molecular level may provide insight into why the mediator evolved diverse transcription factor interaction sites, how mediator integrates regulatory signals from multiple regulatory factors, and, for vertebrates in particular, why so many small gene families evolved encoding transcription factors with similar amino acid sequences.
The interaction of several transcription factor activation domains (ADs) with specific mediator subunits has been reported (table S1). Some ADs have been reported to contact more than one subunit. In Saccharomyces cerevisiae, the transcriptional activator Gcn4 binds to a mediator subcomplex, made up of MED2, MED3, and MED15 (1, 2). Activation of some, but not all, Gcn4-regulated genes also depends on the MED16 and MED22 subunits (3); moreover, subunits MED20, MED21, and MED17 have been reported to bind a glutathione S-transferase–Gcn4 fusion protein in vitro (4). The mammalian glucocorticoid receptor (GR) likewise has been reported to interact with several mediator subunits. Although MED1 provides the dominant GR-binding surface (5), MED14 is also involved in expression of some GR-regulated genes (6), and, when GR is expressed in yeast, its preferred binding partner is MED15 (7). The viral activator VP16 binds not only MED25 (8, 9), but also MED17 (10). Apparently, a single activator can contact multiple mediator-binding sites and thus could potentially activate transcription from the same promoter differently depending on which contacts are made.
The MED23 mediator subunit is vital for transcription of Egr1 (early growth response factor 1) in mouse embryonic stem (ES) cells (11, 12). The zinc finger EGR1 transcription factor is involved in growth regulation, cell differentiation, apoptosis, and cell transformation (13–15). Several regulatory serum response elements (SREs) are clustered near its core promoter region (16, 17) that are bound cooperatively by the MADS box protein serum response factor (SRF) and one member of the ternary complex factor (TCF) family that includes ELK1, SAP1 (SRF accessory protein–1), and NET [new ets (E twenty-six) factor] (18). Orthologous TCF factors are expressed in humans and mice, and similar factors have been identified in Xenopus and zebrafish (19–21). These TCFs constitute a subfamily of the larger ets superfamily of DNA binding transcription factors (22). Evidence of their conservation highlights the important function these factors have in vertebrate biology (23–25).
TCFs activate many early response genes that are rapidly induced by mitogen-activated protein kinase (MAPK)–mediated signaling pathways in the absence of de novo protein synthesis. For example, the c-fos promoter is constitutively bound by SRF and a TCF (26–28), and, within the Egr1 promoter, TCFs form biologically active complexes at SREs (29, 30). Growth-factor signals that trigger the MAPK pathway activate terminal effector kinases, such as the extracellular signal–related kinase (ERK), the c-Jun N-terminal kinase (JNK), and p38 (31), and stimulate their import into nuclei where they phosphorylate target factors, including TCFs. In serum-depleted cultured mammalian cells, serum addition triggers the MAPK cascade and stimulates Egr1 expression within minutes via phosphorylation of TCFs by ERK (16, 32–34). At the Egr1 promoter, phosphorylation of SRE-bound ELK1 stimulates gene transcription (11, 35, 36). In ES cells, ELK1 phosphorylation induces mediator binding to phospho-ELK1 through an interaction with the MED23 subunit (11, 12). Thirty to 40 min after exposure to serum, dephosphorylation of promoter-bound ELK1, probably by a protein phosphatase 3 family member (PPP3CA, B, or C, formerly known as PP2Bs and calcineurins) (37), eliminates this interaction linking mediator to the promoter, and Egr1 transcription diminishes greatly. In mouse ES cells with a knockout (KO) of both Med23 alleles, transcriptional activation by ELK1 is greatly reduced and Egr1 expression is inhibited. These results connect the MED23 mediator subunit with MAPK induction of Egr1 expression in mouse ES cells through ELK1.
Given our experience with Egr1 expression in ES cells, we examined the biological effect of a Med23 KO in a more differentiated cell type. Unexpectedly, we found that expression of Egr1, which was markedly reduced in Med23 KO ES cells, was far less affected in KO mouse embryonic fibroblasts (MEFs) derived from Med23−/− embryos. We wondered what cellular changes must have accompanied the differentiation process to largely circumvent the loss of MED23 and allow expression of Egr1 in its absence. Because TCFs share nearly identical DNA binding specificities (38), an exchange of these factors at the Egr1 promoter could explain our observations if there were substantial differences in the way their ADs interact with the mediator, despite the high similarity of their sequences.
We found that the protein landscape of the Egr1 promoter does change with respect to TCF factor occupancy during the development of MEF cells from ES cells and that, despite a high similarity of amino acid sequence, the TCF ADs make different interactions with the mediator complex. Our results establish a functional link between TCF factor exchange at the Egr1 promoter, the effect on mediator recruitment, and subsequent differences in the magnitude and duration of Egr1 transcription in response to MAPK signaling in different cell types. Transcriptome analysis suggests that this is a common occurrence at genes expressed in both ES cells and fibroblasts. Our results also show that transcript elongation by Pol II at the Egr1 promoter is regulated by TCF AD–mediator interactions.
RESULTS
Med23 KO affects transcription differently in mouse ES and MEF cells
Studies in ES cells knocked out for the gene encoding the mediator MED23 subunit revealed that transcription of the Egr1 immediate response gene largely depends on this subunit (12). As for many genes in mammalian cells (39, 40), before induction of Egr1 by serum growth factors, histone H3 lysine 4 is trimethylated in the promoter region; Pol II and other GTFs are bound to the Egr1 promoter region, but Pol II does not efficiently transcribe into the gene (11). MED23 is required for mediator complex recruitment to the Egr1 promoter region in response to MAPK phosphorylation of ELK1 prebound to the Egr1 control region. The binding of phospho-ELK1 to MED23 stimulates further Pol II and GTF assembly into preinitiation complexes on Egr1 promoters and triggers postrecruitment steps in transcription initiation that stimulate Pol II transcription away from the promoter region and into the gene (11).
To analyze the requirement of MED23 for serum-induced gene control in differentiated cells, we prepared mouse embryo fibroblasts (MEFs) from wild-type (WT) and Med23 KO embryo littermates from a mating of phenotypically normal Med23+/− adult mice. Although the development of Med23 KO embryos was delayed, and mutant embryos died between embryonic day 9 (E9) and E10.5, all three germ layers developed and early organogenesis was initiated before death, which was likely caused by systemic circulatory failure (table S2 and fig. S1). MEF cultures were established from WT and KO sibling embryos harvested on E9.5.
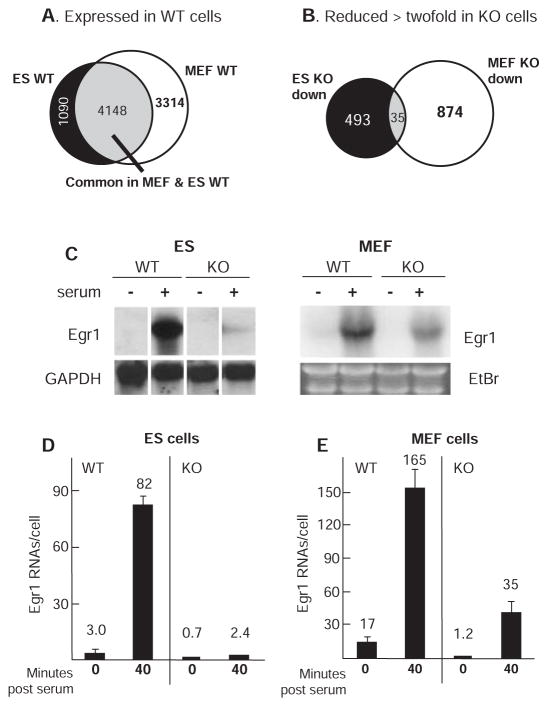
Microarray RNA expression analysis detected reliable RNA signals (>300 intensity units; excluding genes expressed at low level) from ~5200 genes in WT ES cells and ~7500 genes in MEFs, 4148 of which were expressed in common (Fig. 1A). Of those genes expressed in WT ES cells, ~10% showed reduced expression (528 genes) and another ~ 10% showed increased expression (557 genes) by a factor of 2 or more in Med23 KO ES cells (Fig. 1B and fig. S2A). Remarkably, even though ~56% of the genes expressed in WT MEFs were also expressed in WT ES cells, <1% of those genes were changed in common by twofold or more by the loss of MED23 (Fig. 1B and fig. S2). For example, Egr1 transcription in KO ES cells was reduced to ~3% of WT levels, but in KO MEFs it was ~21% of WT levels; a sevenfold difference in Egr1 expression between KO cell types (Fig. 1, C to E). This differential sensitivity to the loss of MED23 was not unique to the Egr1 gene. Based on the overlap of genes expressed in common and showing reduced expression in the KO cells, 793 other genes are sensitive to the loss of MED23 in one cell type but not the other. Changes in Egr1 messenger RNA (mRNA) abundance in response to serum and Med23 KO were due primarily to differences in the rate of Egr1 transcription rather than alterations in RNA stability, because comparable changes were observed for Egr1 nascent (unspliced) transcript assayed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) with primers specific for the single Egr1 intron (fig. S3). These results raised the question of whether mediator function makes a less prominent contribution to the overall level of Egr1 transcription in differentiated MEFs compared to undifferentiated ES cells.
Fig. 1.
Gene expression profiles and Egr1 transcript levels in ES and MEF cells. Total RNA from cells induced with 20% FBS for 30 min was harvested for (A) microarray gene expression profiling of WT cells. The intersection indicates genes expressed in both cell types. Analysis of Med23 KO cells treated in the same way showed the overlap of genes reduced twofold or more in KO ES and MEF cells (B). (C) Northern blot of Egr1 mRNA from WT and KO ES and MEF cells serum-starved for 16 hours (− serum) or stimulated for 30 min in 20% serum. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) Egr1-specific qRT-PCR with complementary DNA (cDNA) from ES cells with and without 40 min of serum stimulation. y axis, Egr1 RNA molecules per cell. Error bars indicate SD from four independent experiments. (E) Egr1 message levels in MEFs prepared as in (D).
Mediator recruitment in WT and Med23 KO MEFs and ES cells
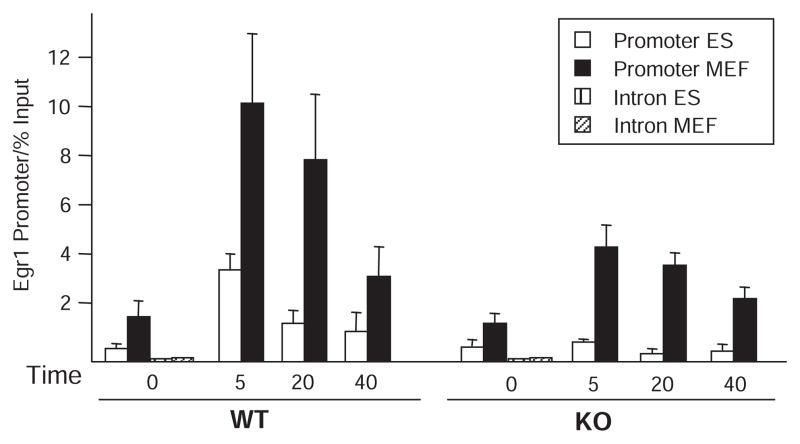
The principal defect in Egr1 transcription in Med23 KO ES cells is the failure of mediator to associate with the Egr1 promoter region in response to serum (11). To address whether transcription in MEFs is activated by mechanisms less dependent on mediator function than in ES cells, we analyzed mediator recruitment to the Egr1 promoter in WT and KO cells by chromatin immunoprecipitation (ChIP). Egr1 promoter ChIP was analyzed by real-time PCR and plotted as the percent of total input chromatin precipitated (Fig. 2). As internal controls for these ChIPs, the promoter region of the highly transcribed translation elongation factor 2 (EF2) gene, which is expressed in all cells, was assayed in the same immunoprecipitates (fig. S4). Signal for mediator association with the Egr1 promoter region in uninduced, serum-starved cells was substantially above the background observed at the Egr1 intron for both cell types. In WT ES and MEF cells, mediator association increased after serum addition. However, the degree of mediator occupancy in response to serum was several times higher and was maintained for a longer period in WT MEFs compared to WT ES cells (Fig. 2). The increased and prolonged association of mediator with the Egr1 promoter in WT MEFs compared to WT ES cells correlated with the increased abundance of Egr1 mRNA observed in MEF compared to that in ES cells (Fig. 1, D and E).
Fig. 2.
Time course of mediator binding to the Egr1 promoter in ES and MEF cells. ChIP for mediator at the Egr1 promoter in ES (white bars) and MEF (black bars) WT and KO cells in replicate. Sheared chromatin was immunoprecipitated with antibodies to both MED1 and MED17 to maximize the mediator ChIP signal. Samples were quantitated by real-time PCR and normalized to percent of signal relative to input chromatin. Egr1 intron PCRs at t = 0 for ES (left) and MEF cells (right) show the background, nonspecific signal from the ChIP. After 16 hours of serum-starvation, 20% FBS was added for 0, 5, 20, or 40 min before formaldehyde cross-linking. Error bars indicate SD from four independent experiments.
In contrast to the Med23 KO ES cells, Med23 KO MEFs showed a clear and greater increase in mediator association with the Egr1 promoter in response to serum and followed a time course of mediator binding similar to that in WT MEFs, although recruitment in KO was reduced to about one-third of that in WT MEFs (Fig. 2). This is similar to the reduction in Egr1 mRNA in KO MEFs compared to that in WT MEFs (Fig. 1E). These results also confirmed the prior observation that the failure of Egr1 induction in Med23 KO ES cells correlates with a defect in mediator recruitment to the promoter region (11). For Med23 WT and KO ES and MEF cells, Egr1 transcription was approximately proportional to the extent and duration of mediator recruitment. Thus, it appears that Egr1 transcription is as dependent on mediator in MEFs as in ES cells. However, these results raise the question: Why is mediator recruitment to the Egr1 promoter in response to serum so much more dependent on MED23 in ES cells compared to MEFs?
TCF association with the Egr1 control region in ES and MEF cells
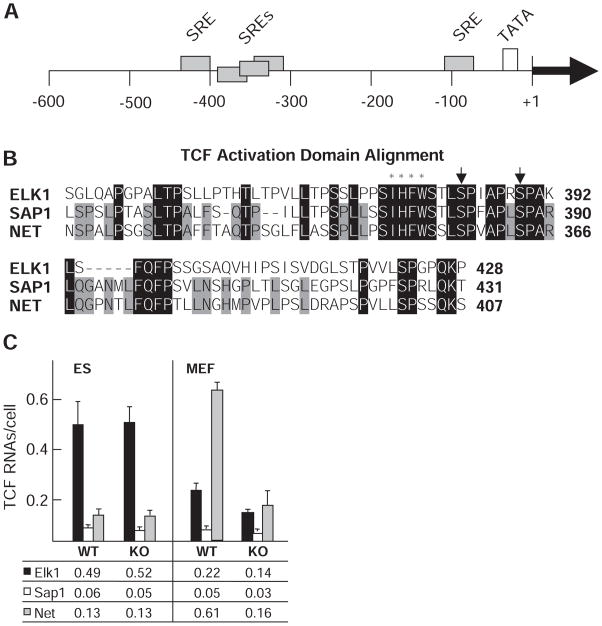
We hypothesized that differences in mediator recruitment between Med23 KO ES cells and MEFs might be due to differences in the transcription factors that are bound to the Egr1 control region in these two cell types. Egr1 induction in fibroblasts by serum growth factors is controlled by TCFs (ELK1, SAP1, and NET) that bind cooperatively with SRF to SREs in the Egr1 transcription control region (Fig. 3A). The DNA binding domains of these TCFs are 73% identical, and all three have indistinguishable in vitro c-fos SRE binding affinities (38). The ADs of the TCFs are also highly conserved, although less than their DNA binding domains (Fig. 3B). The TCFs share clusters of sequence identity, which include two serine residues whose phosphorylation is required for activation (35, 41, 42). SAP1 and NET share 56% identity; ELK1 is the most divergent. Consequently, we considered the possibility that differences in Egr1 induction between Med23 KO ES cells and Med23 KO MEFs might be due to differences in which TCFs are associated with the Egr1 control region in the two cell types.
Fig. 3.
Egr1 control regions, TCF ADs, and TCF transcript abundance. (A) Serum response elements (SREs) within the Egr1 promoter are marked by gray rectangles. The +1 indicates the transcription start site relative to these elements. The TATA box is shown in white. (B) Alignment of the three TCF ADs. Identical residues are marked by black rectangles. Amino acid residues exclusive to SAP and NET are marked by gray rectangles. Arrows indicate conserved serine residues needed for activation, and asterisks mark hydrophobic resides that disrupt function when mutated. (C) TCF mRNA abundance determined by qRT-PCR shown as copies per cell for Elk1 (black), Sap1 (white), and Net (gray) RNAs in WT and KO cells. Mean values from three independent determinations are listed at the bottom. Error bars indicate SD. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Analysis by qRT-PCR of the mRNAs encoding the three TCFs (Fig. 3C) showed that Elk1 was the dominantly expressed TCF in ES cells, regardless of Med23 status. In contrast, MEFs showed an increase in Net transcript compared to ES cells, and in the WT MEFs Net was the dominant TCF message. Less Net mRNA was expressed in the Med23 KO MEFs, suggesting a partial or indirect requirement of MED23 for Net expression. Net was expressed at a level similar to that of Elk1 in KO MEF cells; Elk1 was no longer the dominantly transcribed TCF. Although TCF message abundance may not reflect that of the encoded proteins, these data do indicate that ES cells and MEFs regulate expression of the three TCFs differently.
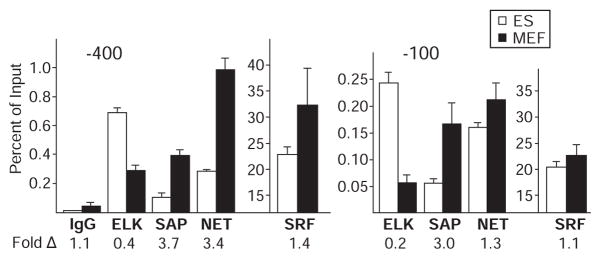
To determine if different relative amounts of the three TCFs are associated with the Egr1 control region in MEFs and ES cells as suggested by TCF mRNA expression levels, we used TCF-specific antibodies in ChIP assays. ChIPs for TCF used primer sets designed to detect the −100 and −400 SRE clusters (Fig. 3A). In this assay, it is not possible to directly compare the extent of promoter occupancy between different TCFs because of probable differences in antibody affinities and the efficiency of formaldehyde cross-linking. This is shown by comparing SRF to TCF ChIP signals (Fig. 4). Twenty to 30% of input DNA was precipitated by antibody to SRF. But for TCF ChIP, at most 1% of input DNA from the same cross-linked chromatin preparation was precipitated by antibody to TCF, even though the two proteins bind to the SRE sequence cooperatively, and consequently, both were likely bound to the Egr1 SREs in vivo, as they are at the Fos SRE (26). The higher extent of SRF cross-linking probably occurs in part because SRF has a substantially larger interface with DNA than the TCF in the ternary complex of SRF, SAP1, and an SRE (43). Despite this limitation for direct comparison of control region binding between different TCFs, we can compare the binding of a given TCF factor between cell types when using the same antibody to ChIP cross-linked chromatin from ES cells and MEFs.
Fig. 4.
TCF ChIPs at the Egr1 promoter region in Med23 KO ES and MEF cells. ChIP for TCF factors at the SRE clusters in the Egr1 promoter was done in steady-state serum conditions in replicate (n = 4) with KO ES and MEF cells. ES cell (white bars) and MEF (black bars) ChIP was done with anti-TCF, anti-SRF, or preimmune IgG for background control. Precipitated DNA was analyzed by qRT-PCR with primers for each SRE cluster and normalized as the percent of signal relative to input chromatin. Error bars indicate SD from four determinations. The fold difference between ES and MEF cells appear below each ChIP condition. Note that TCFs and SRF ChIP are plotted on different scales. This TCF ChIP was also done with WT cells and showed the same trends in factor binding (fig. S5).
The anti-TCF ChIPs (Fig. 4) showed enrichment of ELK1 at both the −400 and −100 Egr1 SRE clusters in Med23 KO ES cells compared to Med23 KO MEFs. Conversely, there was more binding of SAP1 and NET to these regions in KO MEFs relative to KO ES cells. Similar results were observed for the WT cells (fig. S5). We were not able to measure the concentrations of the TCF proteins in these cells because of their low expression (encoded by less than one transcript per cell; Fig. 3C). However, the relative abundance of TCF mRNAs in these two cell types (Fig. 3C) and the TCF ChIP data (Fig. 4) indicate that in the Med23 KOs, the extent and distribution of TCF binding to the Egr1 SRE regions were approximately proportional to the relative concentration of each TCF within the nucleus. Because the relative amounts of the three TCFs bound to the Egr1 SREs differs between ES and MEF cells, we asked if activation by the three TCF ADs respond similarly to the loss of MED23.
TCFs with similar sequences have different requirements for MED23
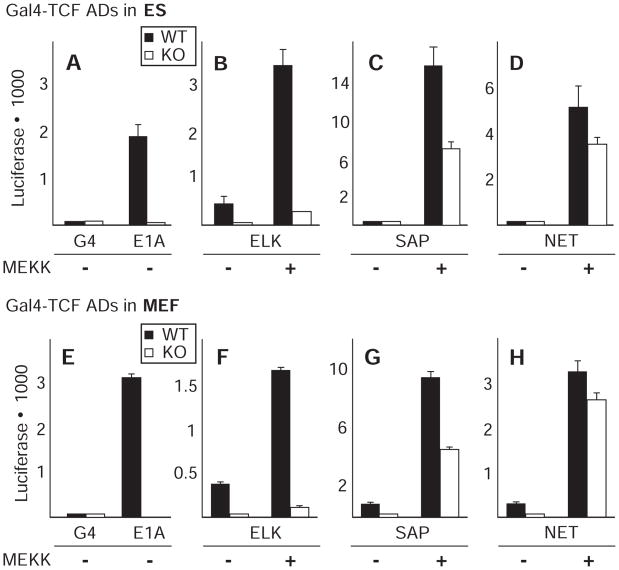
To determine the extent to which the ELK1, SAP1, and NET ADs depend on MED23 for activation, each TCF AD was fused to the S. cerevisiae Gal4-DNA binding domain and assayed for activation of a Gal4-luciferase reporter in transient transfections of WT and Med23 KO ES and MEF cells. Gal4-ELK1 activation was severely affected by Med23 KO in ES cells, as was activation by the control Gal4-E1A AD (CR3) fusion, which is completely dependent on MED23 for activation (12) (Fig. 5, A and B). In marked contrast, activity of the NET AD, although similar in sequence to the ELK1 AD (Fig. 3B), was only mildly affected by Med23 KO in ES cells (Fig. 5D). SAP1 showed an intermediate effect from the loss of MED23 (Fig. 5C), with an approximately twofold reduction in activation potential. Notably, the sensitivity of activation by each of the three TCF ADs to the loss of MED23 was identical in ES and MEF cells (Fig. 5, E to H).
Fig. 5.
TCF AD fusion to Gal4-DBD for luciferase assay in WT and KO cells. Gal4-DNA binding domain fusions to various ADs were tested for ability to activate transcription in ES and MEF WT (black) and KO (white) cells after transfection under serum-starved (0.5% FBS) conditions in the presence or absence of a cotransfected expression vector for dominant active MEKK. Error bars equal standard deviations from two independent experiments done in triplicate.
These results can explain the ability of MEFs to maintain a greater degree of Egr1 expression in the absence of MED23 than ES cells. The percentage of Egr1 SREs bound by NET and SAP1 in MEFs is much higher than in ES cells (Fig. 4), and transcriptional activation by these TCFs is less sensitive than ELK1-dependent activation to the loss of MED23 (Fig. 5). Collectively, the data indicate that the difference in Egr1 expression in Med23 KO MEFs compared to KO ES cells is not due to intrinsic differences in the way the general transcription machinery responds to the same AD, but rather to functional differences in the ADs of the TCFs associated with the Egr1 control region, despite their similar sequences. Consistent with this explanation, overexpression of full-length NET and SAP1, but not ELK1, in Med23 KO ES cells increased transcriptional activation from the Egr1 promoter, making them more like Med23 KO MEFs (fig. S6A). Similarly, overexpression of ELK1 in Med23 KO MEFs reduced expression from the Egr1 promoter in MEF KO cells, making them more like ES KO cells (fig. S6B). This inhibition did not result from squelching due to increased expression of a TCF AD, but rather from increased expression of ELK1 specifically, because over-expression of NET had a much smaller effect (fig. S6C), even though it was expressed at similar concentration (fig. S7). Overexpression of ELK1 probably inhibits transcription from the Egr1 promoter because phospho-ELK1 displaces NET and SAP1 at the Egr1 SREs and requires the missing MED23 to activate transcription.
Analysis of TCF AD interactions with mediator in vivo
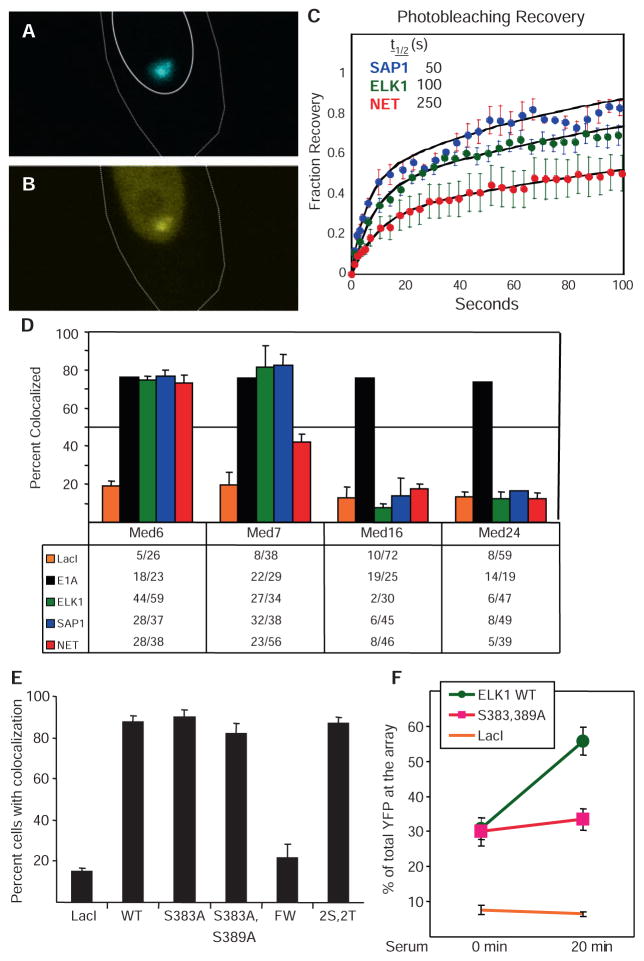
To confirm that there are differences in the way the three TCF ADs interact with mediator complexes, and to analyze the AD-mediator interactions in vivo in greater depth by direct physical methods, we used fluorescence recovery after photobleaching (FRAP) analysis in living cells to determine the in vivo stabilities of macromolecular interactions (44, 45). Chinese hamster ovary (CHO)–derived A03.1 cells contain an integrated array of ~10,000 lac operator (lacO) sites at a single chromosome locus that was generated by coamplification with a dihydrofolate reductase (DHFR) expression cassette (46). When a cyan fluorescent protein (CFP)–tagged lac repressor (LacI) was fused to a TCFAD, the fusion protein (e.g., CFP-LacI-ELK1) appeared as a single distinct focus in the nuclei of living A03.1 cells visualized by confocal microscopy (Fig. 6A). No foci were observed in parental CHO cells that lack the binding array. Coexpression with a yellow fluorescent protein (YFP)–tagged MED6 mediator subunit, located in the mediator head domain (47), and DA-MEKK [a dominant-active form of MAPK kinase kinase (MEKK)], to maintain MAPKs in the activated state, resulted in colocalization of the YFP and CFP signals in ~75% of cells expressing CFP-LacI-ELK1 (Fig. 6, B and D). Similar results were observed for CFP-LacI-E1A (Fig. 6D). These ADs probably do not bind the YFP-MED6 subunit directly, because they each fail to bind mediator complexes lacking the MED23 subunit but containing the MED6 subunit (12) and fail to activate transcription in Med23−/− cells (Fig. 5). We infer that these ADs show an interaction with YFP-MED6 in vivo because the YFP-MED6 fusion protein was assembled into multiprotein mediator complexes that interacted with these ADs via their MED23 subunits. Similarly, colocalization of YFP-MED6 with CFP-LacI-SAP1 and CFP-LacI-NET also probably is a direct visualization of the interaction between these ADs and assembled mediator complexes that had incorporated the tagged MED6 subunit.
Fig. 6.
FRAP of mediator in living cells and factor colocalization imaging. Live A03.1 cells cotransfected with expression vectors for CFP-LacI-ELK1 AD and YFP-MED6 fusion proteins and DA-MEKK show localization (A) of the CFP-LacI-ELK1 AD fusion protein to the array as a single, prominent focus. YFP-MED6 also localized to this focus (B), but was also present throughout the nucleoplasm. Recovery of YFP-MED6 fluorescence after photobleaching with each TCF AD was plotted (C) as fluorescence intensity over time after bleaching (bleached signal set to 0%; prebleach signal set to 100%). (D) A03.1 cells were cotransfected with expression vectors for DA-MEKK and CFP-LacI-TCF AD fusion proteins or CFP-LacI-E1A, and YFP fusions to the indicated MED subunits. Fixed cells were scored for robust colocalization of CFP and YFP signals in two to three independent experiments. The table indicates the fraction of cells with a YFP mediator signal colocalized with a CFP-LacI AD fusion. (E) Expression vectors for CFP fused to LacI or fused to LacI plus the indicated WT or mutant ELK1 ADs were cotransfected with YFP-MED6 into A03.1 cells, and colocalization of YFP with CFP nuclear foci was scored in serum-starved cells. Mutant 2S,2T contains mutations T363A, T368A, S383A, and S389A. Mutant FW contains F378A and W379A. (F) A03.1 cells were transfected with expression vectors for CFP-LacI-fused to the WT ELK1 AD, to the S383A, S389A double mutant, or to CFP-LacI alone. The cells were serum-starved for 22 hours and fixed without further treatment, or 20 min after addition of 20% FBS. Fixed cells were analyzed by confocal microscopy. The ratio of YFP signal at the lacO array to total nuclear YFP signal was quantified with ImageJ software to determine the percent increase of YFP intensity (y axis) at the array after serum addition in 30 to 40 cells from two independent experiments. Movie 1 shows time-lapse fluorescence confocal microscopy of the binding of YFP-MED23 to the focus of CFP-LacI-ELK1 in the nucleus of a living A03.1 cell after addition of serum to serum-starved cells.
The kinetics of mediator recruitment in living A03.1 cells was evaluated by imaging in real time the serum-induced activation of ELK1 and the subsequent enhancement of YFP signal at the binding array (movie S1). In this cell and most cells viewed, the YFP signal at the lacO array marked by CFP began to increase ~90 s after addition of serum to the medium. YFP intensity at the lacO array increased over the next several minutes, reached maximum intensity at ~15 min after serum addition, remained at this intensity for ~15 min, and then decreased to a low stable intensity between 30 and 40 min after addition of serum. This kinetics of YFP-MED23 interaction with the ELK1 AD in living A03.1 cells is similar to the kinetics of phosphorylation of ELK1 at the Egr1 promoter in ES cells (11) and the kinetics of mediator association with the endogenous Egr1 promoter in ES and MEF cells as assayed by ChIP (Fig. 2). These observations indicate that the AD-mediator interaction observed directly at the lacO array in A03.1 cells is analogous to the interaction that occurs between TCFs and mediator on the control regions of the endogenous Egr1 gene.
For FRAP experiments, expression vectors for a CFP-LacI AD fusion protein, DA-MEKK, and YFP-MED6 were cotransfected into A03.1 cells. Living cells displaying colocalized foci of CFP and YFP fluorescence with diffuse YFP signal in the nucleoplasm were selected for FRAP to assure that free, labeled mediator was available to replace bound mediator after photobleaching. After laser bleaching at the lacO array, recovery of YFP fluorescence was monitored. For each recovery curve (Fig. 6C), the kinetics of recovery were fit by two first-order equations, an initial fast rate corresponding to free YFP-MED6 in the bleached volume, and a second slow phase corresponding to the kinetics of replacement of bound YFP-labeled molecules [(48); see Supplemental Methods]. The mean half-time (t1/2) of recovery for each TCF AD–mediator interaction was determined from the slow-phase t1/2 calculated by curve fitting the data from several independent replicate experiments (Fig. 6C).
For each TCF AD, the bleached array recovered its original fluorescence by 10 min after-bleaching, indicating that most YFP-tagged mediator at the lacO array was mobile. Fast recovery indicates a less stable AD-mediator interaction, such as SAP1, which had the fastest t1/2 of ~ 50 s. SAP1 also had the highest activity in the Gal4-luciferase reporter activation assay (Fig. 5). [Differences in luciferase activity induced by the three Gal4-TCF AD fusion proteins were not due to differences in their expression (fig. S7).] Mediator binding to ELK1 was slower to recover than binding to SAP1, with a t1/2 of ~100 s, and thus made a more stable interaction with mediator. The kinetics of recovery of mediator binding to NET were noticeably slower, t1/2 of ~250 s. These differences in stabilities of the in vivo interactions between mediator and the three TCF ADs (Fig. 6C) confirm that the three TCFs interact with mediator differently, despite the high similarity between their sequences (Fig. 3B).
Mediator subunit fusions influence their colocalization with TCFs
During the course of these FRAP experiments we noted another difference in how TCF ADs interacted with mediator. YFP-fusion to the N terminus of certain mediator subunits inhibited their colocalization with some but not all LacI AD fusion proteins (Fig. 6D). In cells cotransfected with expression vectors for a YFP-mediator fusion protein and CFP-tagged LacI alone without a fused AD, colocalization of YFP with CFP at the lacO array occurred in ~10 to 20% of cells (Fig. 6D). This may have been due to association of mediator with the active coamplified DHFR expression cassette used for derivation of the A03.1 cells. However, when CFP-LacI fused to the E1A AD was coexpressed with YFP fusions to the N terminus of MED6, MED7, MED16, or MED24, colocalization of YFP with CFP was observed in ~75% of cells (Fig. 6D). As discussed above, the E1A AD interacts with mediator exclusively via its MED23 subunit. Consequently, we interpret the increased colocalization of each of the YFP-mediator subunit fusion proteins with the E1A AD (Fig. 6D) to indicate that each can assemble into intact mediator complexes. The failure of all three CFP-LacI-TCFAD fusion proteins to stimulate colocalization of YFP-MED16 or YFP-MED24 with the lacO array (Fig. 6D) thus strongly suggests that YFP fusion to these mediator subunits interferes with TCF AD–mediator interactions, even though they do not block the E1A-mediator interaction. MED16 is located in the tail domain of S. cerevisiae mediator (47), Med24 KO eliminates the association of both MED16 and MED23 with mouse mediator (49), and Med23 KO decreases the association of MED16 and MED24 with mouse mediator (12), suggesting that the MED16, MED23, and MED24 subunits probably form a subcomplex in the mediator tail. Even though the E1A and phospho-ELK1 ADs interact with the mediator primarily by binding the MED23 subunit, they probably interact with this large subunit (~1400 amino acids) differently because YFP fusions to MED16 and MED24 block binding to phospho-ELK1, but not to E1A (Fig. 6D).
YFP fusion to MED7 did not inhibit mediator interaction with the E1A, ELK1, or SAP1 ADs, but it partially reduced the mediator interaction with the NET AD (Fig. 6D). Consequently, these results indicate that the NET AD interacts with mediator differently from the ELK1 and SAP1 ADs, further supporting the conclusion that the three TCF ADs interact with mediator differently, despite their high sequence similarity. As discussed below, we interpret the inhibition of mediator association by fusion of YFP to the N terminus of a mediator subunit to be due to steric inhibition of the AD-mediator interaction.
To test the functional significance of the colocalization of mediator with TCF ADs in A03.1 cells, we analyzed the colocalization of YFP-MED6–tagged mediator with foci of CFP-LacI fused to TCF ADs with mutations known to inhibit their ability to stimulate transcription in response to MAPK activation. Mutation of the two serines in each of these domains that are required for activation by MAPKs had no affect on the percentage of cells showing colocalization of mediator with the AD under conditions of high (fig. S8) or low (Fig. 6E) MAPK activity. The WT fusion proteins strongly activated expression of a lacO-luciferase reporter in these cells, whereas the serine mutants did not, as expected (fig. S8B). Additional mutation of ELK1 threonine residues 363 and 368, which are also phosphorylated by MAPKs (50), (mutant 2S,2T) had no additional affect on the high percentage of cells showing colocalization (Fig. 6E). However, the intensity of the YFP-MED23 signal at the lacO array increased in response to serum treatment of serum-starved cells when the WT ELK1 AD was bound to the array, but not when the critical phosphorylated serines were mutated (Fig. 6F). These results are in agreement with the ChIP assay data for mediator binding to the endogenous Egr1 control region in mouse fibroblasts (Fig. 2). By ChIP assay, mediator binding to the Egr1 promoter region was above background in serum-starved cells, and consequently would be visible as colocalization of CFP and YFP at the lacO array in A03.1 cells. The increase in mediator binding in response to serum observed by ChIP (Fig. 2) is visualized as increased intensity of YFP fluorescence at the lacO array in A03.1 cells (Fig. 6F).
A hydrophobic region of the TCF ADs just N-terminal to the critical phosphorylated serines (IHFW) also is required for transcriptional activation by the TCFs (38). In contrast to mutation of the critical serines in the ELK1 AD, mutation of the FWof this hydrophobic region to AA reduced mediator colocalization to the background level observed with LacI alone (Fig. 6E). As discussed below, these results suggest that hydrophobic interactions between the IHFW ELK1 sequence and MED23 provide much of the driving force for mediator binding, whereas interactions between MED23 and ELK1 phospho-Ser383 and phospho-Ser389 activate transcription by a mechanism that involves more than a simple stimulation of mediator binding to the promoter.
Pol II association with the Egr1 transcription unit
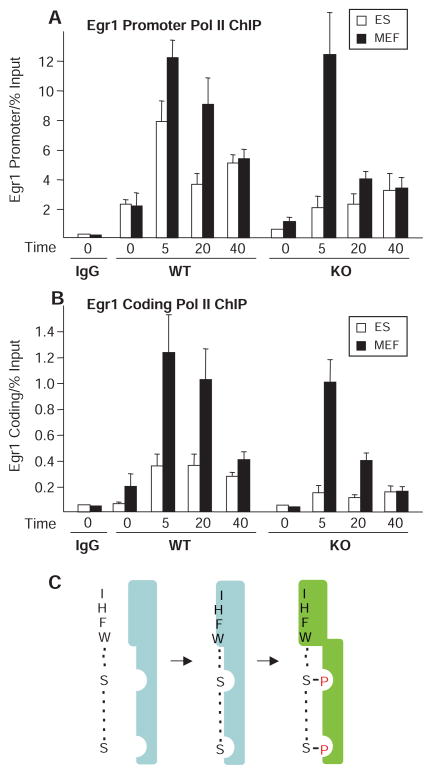
To analyze Pol II occupancy in the promoter and coding regions of the Egr1 gene, we performed ChIP for Pol II in Med23 WT and KO ES and MEF cells at different times after serum addition to serum-starved cells (Fig. 7). The amount of Pol II binding to the Egr1 coding region in WT and KO ES and MEF cells (Fig. 7B) integrated over time was proportional [after subtraction of the background signal observed with nonimmune serum (immunoglobulin G, IgG)] to the abundance of Egr1 RNA at 40 min after serum addition in these WT and mutant cell types (Fig. 1). This indicates that Pol II binding in the coding region as analyzed by ChIP was a good measure of the relative rates of Egr1 transcription in these different cell types plus and minus serum.
Fig. 7.
RNA Pol II ChIP time course in ES and MEF cells and model for ELK1 AD interaction with MED23. ChIP for Pol II at the Egr1 promoter (A) and ~4 kb downstream (B) in ES (white) and MEF (black) WT and KO cells. Background signal with preimmune IgG for WT cells at time 0 is shown. Samples were quantitated by real-time PCR and normalized to percent of input chromatin. Cells were starved and 20% FBS was added for 0, 5, 20, or 40 min before cross-linking. Error bars indicate SDs from three independent experiments. (C) Model for ELK1 AD interaction with MED23. The ELK1 IHFW sequence makes a hydrophobic interaction with MED23 (light blue) that provides much of the energy for mediator binding. MED23 interactions with phosphates added to the ELK1 regulatory serines by a MAPK enhance binding directly and also induce a conformational change in MED23 (light green) that stabilizes this interaction. This conformational change also results in increased binding of Pol II to the Egr1 promoter and an increase in the percentage of Pol II molecules that transcribe away from the promoter region.
Pol II binding at the Egr1 promoter in WT ES and MEF cells approximately paralleled the kinetics of mediator binding to the Egr1 promoter region after serum addition (compare Figs. 2 and 7A). Pol II binding to the promoter, in both unstimulated, serum-starved cells and during the 40-min period of increased Pol II binding induced by serum, was reduced in the Med23 KO compared to WT ES and MEF cells (Fig. 7A). The decrease caused by the absence of MED23 was greater in ES cells; in MEF cells, the decrease was greater at 20 and 40 min after serum addition than at 5 min. As previously observed (11), in ES cells the reduction in Pol II binding to the promoter caused by the absence of MED23 was considerably less than the reduction in Egr1 mRNA elicited by the Med23 KO (compare Figs. 1D and 7A, open bars, WT versus KO). Rather, the decrease in Med23 transcription correlated better with the low abundance of Pol II in the Egr1 coding region (Fig. 7B).
For both WT ES and MEF cells, the amount of Pol II at the Egr1 promoter was 10 to 15 times higher than the amount in the downstream transcribed region (note y-axis scales in Fig. 7, A and B). Similar results have been observed recently for many mammalian genes (39, 40). These observations suggest that there is a delay after Pol II binding before the “paused polymerase” progresses into the coding region. In ES cells, Med23 KO decreases the fraction of promoter-bound polymerases that transcribe into the coding region (11), resulting in an accumulation of Pol II at the promoter over time (Fig. 7A, KO, white bars) and a substantial failure of Pol II transcription into the downstream region (Fig. 7B, KO, white bars). As expected from the decreased effect of the Med23 KO on Egr1 expression in MEFs compared to ES cells, the reduction in Pol II binding to the Egr1 promoter and the 3′ exon in Med23 KO MEFs was much less severe than in Med23 KO ES cells (Fig. 7, A and B).
DISCUSSION
TCF factor exchange dictates dependence of Egr1 transcription on MED23
In Med23 KO ES cells, Egr1 mRNA falls to 3% of the amount in WT ES cells (12) (Fig. 1, C and D). Consequently, it was unexpected to find that Egr1 transcription was only modestly affected in Med23 KO MEFs (Fig. 1, C to E). Furthermore, we observed that of the ~4100 genes expressed in both cell types, only a small fraction (<1%) showed reduced expression by a factor of twofold or greater in both KO cell types (Fig. 1, A and B, and fig. S1). These observations raised the possibility that mediator might function differently in ES cells versus the more differentiated MEF cells to control Egr1 and other MED23-dependent genes. However, our data argue that this difference between ES and MEF cells is not due to fundamental differences in transcription control mechanisms between these cell types. Rather, the dependence of Egr1 expression on MED23 results from cell-specific expression of alternative, closely related TCF factors that each bind equally well to Egr1 control elements, but that interact with the mediator complex differently, resulting in unequal gene expression outcomes from the loss of MED23 in the two cell types.
TCF AD interactions with the mediator are varied
Mammals encode three TCFs closely related in sequence that bind SREs cooperatively with the SRF transcription factor. Egr1 transcription is controlled principally through several SREs (Fig. 3A), making it a good model for studying gene regulation via MAPK-responsive control elements. All three TCFs were expressed in ES and MEF cells, but their relative expression levels, and consequently, their occupancy at the Egr1 control region, differed between these cell types (Figs. 3C and 4). Despite the high degree of sequence identity in their ADs (Fig. 3B), their dependence on MED23 for activating transcription varied markedly. Transcriptional activation by the ELK1 AD was almost completely dependent on MED23 in both ES and MEF cells; activation by the NET AD was nearly independent of MED23, and activation by the SAP1 AD showed an intermediate dependence on MED23 (Fig. 5). These results strongly suggest that the difference between how ES and MEF cells respond to the loss of MED23 results from changes in the occupancy of Egr1 SREs by the three alternative TCFs and not to differences in how the general components of the Pol II transcriptional regulatory apparatus respond. This conclusion was supported by experiments in which overexpression of SAP1 or NET, but not ELK1, restored Egr1 serum induction in Med23 KO ES cells, whereas ELK1 overexpression inhibited Egr1 transcription in Med23 KO MEFs, causing them to behave more like KO ES cells (fig. S6).
Fluorescence microscopy studies in living cells using mediator subunits and TCF ADs tagged with GFP derivatives indicated that, despite their high sequence similarity, the TCF ADs each contact the mediator differently (Fig. 6). MED6 and MED7 are highly conserved mediator subunits proposed to be in the head and middle domains of the mediator complex, respectively (47). YFP-fusions to MED6 colocalized equally with all three TCF ADs (and the adenovirus large E1A-CR3 AD, Fig. 6D). In contrast, when MED7 was fused to YFP, this tagged mediator showed reduced association with the NET AD but not with the ADs of the other TCFs or E1A-CR3. YFP fusions to MED16 or MED24 inhibited tagged mediator from colocalizing with all three of the TCF ADs. These YFP fusion proteins were probably incorporated into functional mediator complexes because they did colocalize with the MED23-binding E1A-CR3 AD. MED23 and MED24 probably associate with MED16 in the mediator tail domain because KO of MED24 causes a concomitant loss of MED16 and MED23 from mouse mediator complexes (49), and KO of MED23 causes reduced association of MED16 and MED24 (12). We hypothesize that fusion of the large YFP-tag to MED16 or MED24 in the mediator tail creates steric hindrances that block binding of all three TCFADs to the mediator complex. In addition, we hypothesize that YFP-fusion to MED7 in the mediator middle domain sterically restricts mediator binding by the NET AD, which functions almost independently of the MED23 tail subunit, but not the ELK1, SAP1, or E1A ADs, which are partially or completely dependent for function on MED23. The YFP tags might occlude a binding site on the mediator surface or cause local deformation of the mediator structure that indirectly interferes with TCF AD binding. The distinct FRAP kinetics of association of YFP-MED6– tagged mediator with each of the three TCF ADs (Fig. 6C) also indicate that each of the TCF ADs interacts with the mediator differently, despite their high sequence similarity.
Many ADs may interact with more than one mediator subunit
Transcriptional activation by E1A-CR3 was fully dependent on the MED23 subunit in both ES and MEF cells (Fig. 5, A and E). Activation by phospho-ELK1 was largely dependent on MED23 in both ES and MEF cells (Fig. 5, B and F), but there was still detectable activation in the KO cells. SAP1 activation was reduced ~2-fold in Med23 KO cells, but the residual ~66-fold activation above baseline was considerable (Fig. 5, C and G). Because they stimulate mediator binding to the Egr1 promoter in Med23 KO MEF cells (Fig. 2B), SAP1 and NET likely retain the ability to interact with mediator in the absence of MED23. These observations indicate that ELK1 activates transcription primarily through an interaction with MED23, whereas SAP1 activates transcription partly through an interaction with MED23, and transcriptional activation by NET1 probably involves its interaction with a mediator subunit other than MED23.
Our results with SAP1 suggest that this AD makes functionally important contacts with MED23 and at least one additional mediator subunit. This may be typical of most ADs. E1A-CR3 and ELK1 may be unusual among ADs in that they make functional interactions exclusively or primarily with only one mediator subunit. There are several reports in the literature of the same AD interacting with alternative mediator subunits. For example, the VP16 AD is reported to contact two independent human mediator subunits, MED17 (10) and MED25 (8, 9), and in S. cerevisiae, the VP16 AD has been reported to interact with Med15 (Gal11) (1). A single VP16 AD of ~90 amino acid residues is unlikely to simultaneously contact two mediator subunits in different domains of the large mediator complex (e.g., MED15 and 17 in the tail and head domains, respectively). Consequently, at least two separate VP16 ADs can probably interact simultaneously with distinct mediator subunits (table S1). Perhaps strong ADs evolved to interact with several sites on the mediator complex to augment mediator recruitment and stimulation of transcription downstream from the promoter region, as at the Egr1 promoter (11). Most gene transcription in multicellular organisms is controlled by several regulatory factors; thus, the interplay between these factors ultimately guides mediator recruitment and gene activity. In some cases, the ADs of factors regulating the same gene may interact with different, non-overlapping surfaces on the mediator simultaneously. In other cases, ADs may compete for binding to overlapping surfaces on the mediator, or two or more ADs may bind to mediator surfaces cooperatively. Thus, alternative combinations of AD-mediator interactions may contribute to the combinatorial complexity of gene control in multicellular organisms.
Vertebrates contain many gene families of closely related transcription factors
As Gal4 DNA binding domain fusions, the three TCF ADs varied over a factor of about 5 to 10 in the extent to which they activated transcription from a Gal4 luciferase reporter (Fig. 5). We asked how these differences in activation strength and MED23 dependency related to gene regulation. Each TCFAD interacted with the mediator in different ways, as indicated by their varying dependence on MED23 for activity (Fig. 5), differences in the stabilities of their interaction with mediator in vivo (Fig. 6C), and differences in the way YFP fusion to the N terminus of MED7 influence their interactions with mediator (Fig. 6D). Pol II and mediator occupancy at the Egr1 promoter region were greater in MEFs compared to that in ES cells before stimulation with serum. In response to serum stimulation, mediator bound to the promoter region to a greater extent and was maintained for a longer period in MEFs compared to ES cells (Fig. 2). Because the different TCF ADs make distinct interactions with the mediator, we postulate that the exchange of TCFs at the Egr1 control regions in different cell types (Fig. 4) changes the extent and duration of transcriptional responses to MAPK signaling. The Med23 KO amplified the intrinsic differences in the way these closely related TCFs interact with the mediator complex, as exemplified by differences in Egr1 transcription in KO ES versus KO MEF cells.
Why did three closely related TCFs evolve in mammals? Evolutionary pressure to evolve differences in their DNA binding properties or cooperative binding with SRF seems unlikely because any such differences appear to be minimal (38). The three TCFs may have evolved partly because variations in their ADs that affect their interaction with mediator and possibly other transcriptional coactivators provide a mechanism to fine-tune transcriptional output in response to MAPK signaling. By altering the relative proportions of the three TCFs, the extent and duration of transcriptional activation in response to MAPK signaling through SREs can be fine-tuned for hundreds of genes as appropriate for different cell types. For example, in proliferating myoblast progenitor cells, ELK1 and SRF occupy control regions for smooth muscle–specific genes SM22α (51) and telokin (52), repressing their expression. Removal of growth factors from the medium triggers myoblast differentiation into myotubes, which is accompanied by the exchange of myocardin for ELK1 at these promoters, and induction of SM22α and telokin transcription. Displacement occurs because ELK1 and myocardin compete for overlapping docking sites on SRF. Similarly, in fibroblasts and other cell types, the myocardin-related factors MRTF and MAL (megakaryocytic acute leukemia) compete with TCFs for binding to SRF at promoters for genes involved in the actin cytoskeleton, regulating them in response to changes in cytoplasmic G-actin concentration (53, 54). In these examples, unrelated proteins exchange at SRF-bound sites, MLTFs for TCFs. In the work described here, closely related TCFs exchange at SRF-binding sites within the Egr1 control region. Rather than on or off regulation, this exchange modifies the duration and strength of Egr1 expression in response to MAPK signaling. Presumably, the dominance of ELK1 in ES cells allows for optimized MAPK responses in ES cells, whereas NET and SAP1 dominance in MEFs presumably optimizes gene expression for fibroblasts. Thus, different cell types could modify their transcriptional responses to MAPK signaling over a broad range of strength and duration by controlling the relative nuclear concentrations of the three TCFs.
Our genome-wide RNA expression analysis showed that of the 4148 genes expressed in both ES and MEF cells, <1% were affected similarly by a Med23 KO (Fig. 1B and fig. S2A). Indirect effects of the Med23 KO undoubtedly contribute to the differences in how genes respond to the mediator mutation. For example, all genes regulated by the EGR1 transcription factor would be affected differently by the Med23 KO in MEF cells compared to ES cells because of the modest versus severe drop in EGR1 concentration in the two cell types. Nonetheless, these results suggest that differences in the way most genes expressed in both ES and MEF cells respond to a Med23 KO occur for the same reason that differences in Egr1 expression occur: Alternative transcription factors regulate the same gene in different cell types, even in the case of so-called housekeeping genes expressed in multiple cell types. Transcription factor exchange at regulatory elements probably occurs at most genes during mammalian cell differentiation, even for ubiquitously expressed genes.
Phospho-ELK1 interaction with mediator regulates transcription elongation downstream from the promoter region
Our analysis of the influence of TCF AD mutations on mediator binding in vivo further supports our conclusion that the phospho-TCF AD– mediator interaction stimulates transcription at a step subsequent to GTF and Pol II binding (11). Mutation of the hydrophobic IHFW sequence immediately N-terminal to the critical phosphorylated serines in ELK1 eliminates activation function (38) and, accordingly, abolished the mediator interaction visualized in A03.1 cells (Fig. 6E). However, TCF AD mutants with alanine substitutions for the critical serines, whose phosphorylation is required for MAPK-activated transcription, stimulated mediator binding to the lacO array to the same extent as that of the WT TCF ADs. Mediator binding by the serine mutants and by the WT ADs in the absence of serum when the critical serines are un-phosphorylated was considerably greater than the background observed with the lac repressor alone (Fig. 6E and fig. S8A). As expected, the CFP-LacI-TCF AD fusion proteins with WT ADs induced a marked increase in expression from a lacO-luciferase reporter in A03.1 cells, whereas alanine subsitution mutants in the critical serines did not (fig. S8B). However, MAPK kinase activation caused only an approximately twofold increase in mediator concentration at the lacO array in most cells (Fig. 6F). An increase in mediator binding is expected because the additional interactions that likely occur between the phosphates added to the TCF ADs and mediator should contribute additional binding energy. But the stimulation of transcription from reporters in transient transfection assays and from the endogenous Egr1 locus resulting from MAPK phosphorylation of these serines was far greater than can be accounted for by the modest increase in mediator recruitment by a factor of 2 in most cells.
Together with our ChIP studies in WT and Med23 KO ES cells (11) and MEFs (Figs. 2 and 7), these results on mediator recruitment to the lacO array in A03.1 cells indicate that in addition to stimulating mediator binding, the phospho-TCF AD–mediator interaction also activates a post-recruitment step in transcription initiation that leads to an increase in the fraction of promoter-bound Pol II molecules that escape from the promoter region and transcribe many kilobases into the transcription unit. Because mutation of the IHFW sequence in ELK1 eliminated mediator binding (Fig. 6E), a hydrophobic interaction between this sequence and a surface of MED23 likely contributes much of the energy for mediator binding. Previous studies have shown that interactions between MED23 and the MAPK-phosphorylated serines increase the affinity of ELK1 for mediator (12). These interactions of MED23 with the added phosphates may also induce a conformational change in the mediator complex that stimulates Pol II binding and transcription downstream from the promoter region (Fig. 7C). The recent discovery that “paused polymerases” are found at the promoter region of a large fraction of mammalian genes (39, 40) has led to the suggestion that transcription may frequently be regulated by control of promoter-proximal transcript elongation through interactions between transcription factor ADs and elongation factors (39, 40). The studies presented here regarding the TCFs show that control of promoter proximal transcription elongation can also be regulated through an AD-mediator interaction.
EXPERIMENTAL PROCEDURES
Cell culture
Wild-type and Med23−/− ES cells and MEFs from single Med23−/− and Med23+/+ E9.5 embryo littermates (12) were cultured in ES media [containing leukemia inhibitory factor (LIF) for ES cells, no LIF for MEFs] + 10% fetal bovine serum (FBS). A03.1 cells (46) were cultured in F12 Ham’s medium + 10% FBS. Cells were serum-deprived by culturing for 16 hours at 37¡ in media without FBS (ES cells) or 0.5% FBS for MEFs and A03.1 cells. Serum was added to 20% at t = 0.
RNA and PCR
Total RNAs from serum-deprived and +40-min cells were analyzed with Affymetrix microarrays and Focus software (55). Specific mRNAs were quantified by qRT-PCR (Supplemental Methods). For Egr1, RNA was isolated from serum-starved and +40-min cells, for TCFs, from cells maintained in media with serum.
Chromatin immunoprecipitation
ChIP was done as previously described (11) with antibodies listed in the Supplemental Methods. For TCF ChIPs, cells were maintained in media with 10% serum and cross-linked. For mediator and Pol II ChIPs, serum-starved (t = 0) and serum + 5, 20, or 40 min were cross-linked. Immunoprecipitated DNA was assayed by quantitative real-time PCR with primer sets listed in table S3.
Microscopy
For in vivo imaging, A03.1 cells were transfected with 2 μg of CFP-LacI-TCF (ELK1, SAP1, or NET), 2 μg of YFP-mediator subunit, and, for constant MAPK stimulation, 0.05 μg of DA-MEKK 24 hours before imaging with a Leica TCS SP2 AOBS single-photon confocal microscope with a 63× 1.4 numerical aperture oil-immersion objective with stage and objective in an enclosed chamber with 5% CO2 at 37¡C. Data acquisition and statistical analysis are described in the Supplemental Methods. To score for colocalization of YFP-MED6, -7, -16, and -24 with CFP-LacI-TCFs, 24 hours after transfection cells were fixed in 1.6% formaldehyde, washed in 1× PBS, mounted onto slides, and imaged.
Luciferase assay
Gal4-DBD-TCF AD function was assayed in transient transfections with DA-MEKK using a Gal4 luciferase reporter and the dual-luciferase assay (Promega). For overexpression studies, full-length TCF constructs with an Egr1-promoter fragment driving luciferase were used for KO cell line transfection.
Supplementary Material
Movie S1. Kinetics of mediator binding to ELK1 AD in vivo after serum addition.
Methods and Materials
Fig. S1. Phenotype of E9.5 Med23−/− mouse embryos.
Fig. S2. Overlap of genes expressed in ES and MEF cells affected by the Med23 KO.
Fig. S3. Egr1 mRNAs and Egr1 nascent pre-mRNAs (intron containing) per cell.
Fig. S4. ChIP of Pol II and mediator at the EF2 promoter in WT and KO ES and MEF cells after addition of serum to serum-starved cells.
Fig. S5. TCF ChIP in WT ES and MEF cells.
Fig. S6. Effect of overexpression of full-length TCFs in ES and MEF KO cells.
Fig. S7. Gal4-TCF AD protein levels and activation function in ES KO, H1299, and HeLa cells.
Fig. S8. Mediator colocalization with serine mutants of ELK1, SAP1, and NET with MAPK activation.
Table S1. Activator-mediator subunit interactions.
Table S2. Lethal phenotype of the homozygous Med23 knockout.
Table S3. Primers for mRNA and ChIP qRT-PCR analyses.
References
REFERENCES AND NOTES
- 1.Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drysdale CM, Jackson BM, McVeigh R, Klebanow ER, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil PA, Hinnebusch AG. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, Roeder RG. The Mediator subunit MED1/TRAP220 is required for optimal glucocorticoid receptor-mediated transcription activation. Nucleic Acids Res. 2007;35:6161–6169. doi: 10.1093/nar/gkm661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol. 2006;20:560–572. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Kim GS, Yun CH, Lee YC. Functional conservation of the glutamine-rich domains of yeast Gal11 and human SRC-1 in the transactivation of glucocorticoid receptor Tau 1 in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28:913–925. doi: 10.1128/MCB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittler G, Stühler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F, DeBeaumont R, Zhou S, Näär AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998;5:3–28. [PubMed] [Google Scholar]
- 14.Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 15.Huang RP, Liu C, Fan Y, Mercola D, Adamson ED. Egr-1 negatively regulates human tumor cell growth via the DNA-binding domain. Cancer Res. 1995;55:5054–5062. [PubMed] [Google Scholar]
- 16.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto KM, Bardeleben C, Yates KE, Raines MA, Golde DW, Gasson JC. 5′ upstream sequence and genomic structure of the human primary response gene, EGR-1/TIS8. Oncogene. 1991;6:867–871. [PubMed] [Google Scholar]
- 18.Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: Nuclear effectors of the Ras–MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- 19.Nentwich O, Münchberg FE, Frommer G, Nordheim A. Tissue-specific expression of the Ets gene Xsap-1 during Xenopus laevis development. Mech Dev. 2001;109:433–436. doi: 10.1016/s0925-4773(01)00555-x. [DOI] [PubMed] [Google Scholar]
- 20.Goldman D, Sapru MK, Stewart S, Plotkin J, Libermann TA, Wasylyk B, Guan K. Cloning and characterization of GETS-1, a goldfish Ets family member that functions as a transcriptional repressor in muscle. Biochem J. 1998;335(Pt 2):267–275. doi: 10.1042/bj3350267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown LA, Yang SH, Hair A, Galanis A, Sharrocks AD. Molecular characterization of a zebrafish TCF ETS-domain transcription factor. Oncogene. 1999;18:7985–7993. doi: 10.1038/sj.onc.1203197. [DOI] [PubMed] [Google Scholar]
- 22.Hill CS, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): A novel role for the DNA binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 24.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 25.Wasylyk C, Bradford AP, Gutierrez-Hartmann A, Wasylyk B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- 26.Herrera RE, Shaw PE, Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- 27.König H. Cell-type specific multiprotein complex formation over the c-fos serum response element in vivo: Ternary complex formation is not required for the induction of c-fos. Nucleic Acids Res. 1991;19:3607–3611. doi: 10.1093/nar/19.13.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shore P, Sharrocks AD. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol Cell Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz ME, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 32.Denhardt DT. Signal-transducing protein phosphorylation cascades mediated by Ras/Rho proteins in the mammalian cell: The potential for multiplex signalling. Biochem J. 1996;318(Pt 3):729–747. doi: 10.1042/bj3180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J Biol Chem. 2001;276:24531–24539. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann K, Thiel G. Epidermal growth factor and platelet-derived growth factor induce expression of Egr-1, a zinc finger transcription factor, in human malignant glioma cells. J Neurol Sci. 2001;189:83–91. doi: 10.1016/s0022-510x(01)00562-7. [DOI] [PubMed] [Google Scholar]
- 35.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 36.Whitmarsh AJ, Shore P, Sharrocks AD, Davis RJ. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto T, Stewart S, Guan KL. The calcium/calmodulin-dependent protein phosphatase calcineurin is the major Elk-1 phosphatase. J Biol Chem. 1997;272:29415–29418. doi: 10.1074/jbc.272.47.29415. [DOI] [PubMed] [Google Scholar]
- 38.Price MA, Rogers AE, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buratowski S. Transcription. Gene expression—Where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janknecht R, Ernst WH, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassler M, Richmond TJ. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 2001;20:3018–3028. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 45.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 46.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, Conaway JW, Conaway RC, Emmons SW, Fondell JD, Freedman LP, Fukasawa T, Gustafsson CM, Han M, He X, Herman PK, Hinnebusch AG, Holmberg S, Holstege FC, Jaehning JA, Kim YJ, Kuras L, Leutz A, Lis JT, Meisterernest M, Naar AM, Nasmyth K, Parvin JD, Ptashne M, Reinberg D, Ronne H, Sadowski I, Sakurai H, Sipiczki M, Sternberg PW, Stillman DJ, Strich R, Struhl K, Svejstrup JQ, Tuck S, Winston F, Roeder RG, Kornberg RD. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Sprague BL, McNally JG. FRAP analysis of binding: Proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Ito M, Okano HJ, Darnell RB, Roeder RG. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002;21:3464–3475. doi: 10.1093/emboj/cdf348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cruzalegui FH, Cano E, Treisman R. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene. 1999;18:7948–7957. doi: 10.1038/sj.onc.1203362. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Hu G, Herring BP. Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol Cell Biol. 2005;25:9874–9885. doi: 10.1128/MCB.25.22.9874-9885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol Cell Biol. 2008;28:732–742. doi: 10.1128/MCB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posern G, Treisman R. Actin’ together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 55.Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: A PRIM approach. Bioinformatics. 2003;19:1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- 56.This study was supported by U.S. Public Health Service grant R37CA025235 and American Cancer Society postdoctoral fellowship grant PF0804801GMC to M.A.P. FRAP experiments were performed at the California NanoSystems Institute Advanced Light Microscopy/Spectroscopy Shared Facility at University of California, Los Angeles, directed by S. Weiss, Department of Chemistry and Biochemistry. We thank D. Bachiller for performing the whole-mount embryo microscopy shown in fig. S1; S. Gallaher and J. Woo for their input; and C. Eng for technical assistance.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Kinetics of mediator binding to ELK1 AD in vivo after serum addition.
Methods and Materials
Fig. S1. Phenotype of E9.5 Med23−/− mouse embryos.
Fig. S2. Overlap of genes expressed in ES and MEF cells affected by the Med23 KO.
Fig. S3. Egr1 mRNAs and Egr1 nascent pre-mRNAs (intron containing) per cell.
Fig. S4. ChIP of Pol II and mediator at the EF2 promoter in WT and KO ES and MEF cells after addition of serum to serum-starved cells.
Fig. S5. TCF ChIP in WT ES and MEF cells.
Fig. S6. Effect of overexpression of full-length TCFs in ES and MEF KO cells.
Fig. S7. Gal4-TCF AD protein levels and activation function in ES KO, H1299, and HeLa cells.
Fig. S8. Mediator colocalization with serine mutants of ELK1, SAP1, and NET with MAPK activation.
Table S1. Activator-mediator subunit interactions.
Table S2. Lethal phenotype of the homozygous Med23 knockout.
Table S3. Primers for mRNA and ChIP qRT-PCR analyses.
References