Abstract
A secreted CC chemokine homolog, encoded by the MC148 gene of molluscum contagiosum virus, potently interfered with the chemotaxis of human monocytes, lymphocytes, and neutrophils in response to a large number of CC and CXC chemokines with diverse receptor specificities. Evidence that the viral protein binds to human chemokine receptors was obtained by competition binding and calcium mobilization experiments. The broad spectrum chemokine antagonistic activity of MC148 can explain the prolonged absence of an inflammatory response in skin tumors that harbor replicating molluscum contagiosum virus.
Molluscum contagiosum virus (MCV), a member of the poxvirus family, produces small, papular tumors in the skin of immunocompetent children and young adults and opportunistic infections in immunodeficient AIDS patients (1). The absence of inflammatory cell infiltrates, in lesions that contain large numbers of virus particles, is a remarkable feature of the disease that distinguishes it from many other virus infections. One possible explanation is that MCV inhibits the activity of chemoattractant cytokines (chemokines), which comprise a large superfamily of small, secreted proteins that bind to specific G protein-coupled membrane receptors and recruit leukocytes to areas of infection and injury (2). Generally, the α or CXC chemokines attract neutrophils or lymphocytes, whereas the β or CC chemokines attract monocytes, lymphocytes, eosinophils, or basophils. Presently, four CXC receptors and nine CC receptors have been defined. Usually, CXC chemokines exhibit high affinity for a single receptor whereas CC chemokines frequently bind to two or more different receptors. The finding of an ORF sequence (MC148) encoding a CC chemokine homolog, within the 190,000-bp MCV genome, provided a clue to the mechanism used to prevent an inflammatory response (3). In comparison to mammalian CC chemokines, the predicted NH2-terminal region of the mature (signal peptide-cleaved) protein encoded by MC148 is shortened (Fig. 1A). Because the NH2-terminal region of chemokines is involved in receptor activation, we suggested that the MC148 protein (MC148P) may lack agonist activity and function as a chemokine antagonist (3). In support of that hypothesis, Krathwohl et al. (4) reported that MC148P blocks the chemotactic response of monocytes to one CC chemokine, macrophage inflammatory protein (MIP)-1α. Here we show that purified MC148P inhibits the attraction of multiple leukocyte subsets to CXC and CC chemokines, providing a possible explanation for the absent or delayed inflammatory response in MCV lesions.
Figure 1.
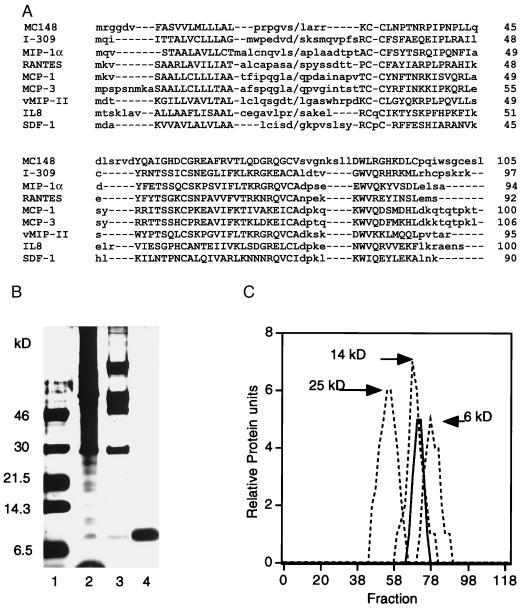
Primary structure comparisons and purification of MC148P. (A) The alignment of the deduced amino acid sequence of MC148P with those of CC (I-309, MIP1-α, RANTES, MCP-1, MCP-3) and CXC (IL8, SDF-1) chemokines and HHV 8 vMIP-II chemokine homolog was performed with the macaw multiple alignment program. Upper- and lowercase letters refer to conserved and nonconserved amino acids, respectively; dashes are alignment gaps; back slashes are signal peptide cleavage sites; and numbers on the right are amino acid numbers. The gap in the NH2-terminal region of MC148 may have special significance with regard to the absence of agonist activity. (B) Purification. SDS/PAGE and silver staining of unreduced MC148P at successive purification steps. Lanes: 1, markers; 2, clarified supernatant; 3, pooled fractions from heparin column; 4, pooled fractions from SP column. (C) Elution from a G50 Sephadex column of MC148P (solid line) and the following markers (dashed lines): chymotrypsinogen, 25 kDa; ribonuclease A, 14 kDa; insulin, 6 kDa.
MATERIALS AND METHODS
Expression of the MC148 ORF.
PCR was used to amplify the MC148 ORF, preceded by an optimized translation initiation sequence (5), from DNA of MCV isolates. The products of two reactions were cloned into the transfer vector pRB21 (6) and recombined into the vaccinia virus genome to generate recombinant viruses vIDA-1 and vIDA-2. Optimization of the translation initiation sequence increased expression by 3-fold over that of a previous recombinant virus (7).
Purification of MC148P.
MC148P was purified from the culture supernatant of BS-C-1 or HeLa cells infected with 10 infectious units per cell of vIDA-2. Typically, 70–90 150-cm2 flasks containing monolayers of BS-C-1 or HeLa cells in Optimem (GIBCO/BRL) medium were used. After 30–35 h, the medium was harvested and suspended cells were removed by centrifugation at 1,000 × g for 10 min. The supernatant was adjusted to contain 0.1% Triton X-100 and clarified by centrifugation at 10,000 × g for 30 min. The high-speed supernatant then was applied to a 5-ml HiTrap heparin column (Pharmacia) that had been equilibrated with 0.05 M Tris⋅HCl, pH 7.4/0.1 M NaCl. A linear gradient of 0.1–0.6 M NaCl in 0.05 M Tris⋅HCl (pH 7.4) was used for elution. MC148P was detected by SDS/PAGE and Western blotting. Fractions containing 0.28–0.37 M NaCl were pooled and applied to a 200-ml G50 Sephadex (Pharmacia) gel filtration column that had been equilibrated with 0.02 M sodium phosphate, pH 7.4/0.2 M NaCl. The fractions eluting between 135 and 160 ml were pooled, diluted 3-fold with 0.02 M sodium phosphate (pH 7.4), and applied to a final 1-ml HiTrap SP column (Pharmacia) equilibrated with 0.02 M sodium phosphate, pH 7.4/0.05 M NaCl. The cation exchange column was developed with a linear 0.05–0.5 M NaCl gradient and MC148P eluted between 0.27 and 0.37 M NaCl. Approximately 150 μg of purified protein usually was obtained.
Chemotaxis Inhibition.
Chemotaxis of freshly elutriated human monocytes was analyzed in a 48-well microwell Boyden chamber (Neuroprobe, Cabin John, MD) fitted with a polyvinylpyrrolidone (PVP)-coated polycarbonate filter (pore size 5 μm) using standard procedures (8). Dilutions of MC148P were made in RPMI 1640 medium (Quality Biologicals, Gaithersburg, MD) containing 0.5% BSA and 25 mM Hepes, pH 7.4. After a 40-min incubation at 37°C, migrated cells were counted in five 40× fields. Chemotaxis of monocytes to I-309 also was carried out for 40 min at 37°C following the protocol of Miller and Krangel (9). Chemotaxis of neutrophils, obtained by dextran sedimentation and hypotonic lysis of random donor buffy coats, was analyzed in a chamber fitted with a 3-μm pore PVP-free polycarbonate membrane. Cells and chemokines were in Hanks’ balanced salt solution (GIBCO/BRL) containing 0.1% BSA. Migrated cells were counted after a 30-min incubation at 37°C. Chemotaxis of elutriated lymphocytes, isolated by Ficoll Hypaque gradient centrifugation, was analyzed in a chamber with mouse type IV collagen-coated, PVP-free polycarbonate filters (pore size 5 μm) and a 2-h incubation at 37°C (8).
Calcium Flux Measurements.
Cells were loaded with Fura-2 (Molecular Probes) by using standard methods (10), and fluorescence was monitored in a spectrofluorimeter (Photon Technology International, Monmouth Junction, NJ). Results were expressed as the ratio of fluorescent emission at 510 nm from repetitive, sequential excitations at 340 nm and 380 nm every 200 msec.
Chemokine-Binding Studies.
One million HEK 293 cells bearing the CCR2B receptor (10, 11) in RPMI buffer containing 0.5% BSA and 25 mM Hepes, pH 7.4, were dispensed to triplicate tubes containing 0.1 nM 125I-labeled MCP-1 (2,200 Ci/mmol; DuPont/NEN) and either unlabeled MCP-1, MC148P, or interleukin 8 (IL-8) at indicated concentrations. Binding was allowed to occur at 4°C with gentle agitation for 2 h. To separate cell-bound from unbound radioactivity, the cells were centrifuged for 5 min at low speed in a microfuge. The pellet was resuspended in 0.2 ml of binding buffer and layered over a 1-ml cushion of 10% sucrose in PBS. After centrifugation at 10,000 rpm in a microfuge, the supernatant was aspirated and the cell associated radioactivity was quantitated in a gamma counter.
RESULTS
Expression and Purification of MC148P.
The lack of a cell culture system to grow MCV led us to express the chemokine homolog by introducing the MC148 coding sequence into the genome of vaccinia virus, a prototype poxvirus that is distantly related to MCV. Two recombinant viruses, vIDA-1 and vIDA-2, containing identical MC148 genes except for a natural Arg/Lys polymorphism at the third codon after the predicted signal peptide cleavage site were isolated. The MC148 sequence in Fig. 1A shows the Arg polymorphism. Cells infected with either recombinant virus secreted a protein that reacted with antiserum raised against an MC148 peptide (not shown). The purification of MC148P was monitored by PAGE. After three steps, a single 10-kDa silver-stained band was detected after denaturation of the purified protein in the absence (Fig. 1B) or presence (not shown) of reducing agent. The NH2-terminal protein sequence was obtained and it confirmed the predicted signal peptide cleavage site. A potential N-glycosylation site is apparently not utilized because the mobility of the protein was unchanged after digestion with N-glycosidase (data not shown). MC148P eluted from a G-50 Sephadex column between markers of 13.5 and 6.5 kDa, consistent with it being largely monomeric (Fig. 1C).
Inhibition of Chemotaxis.
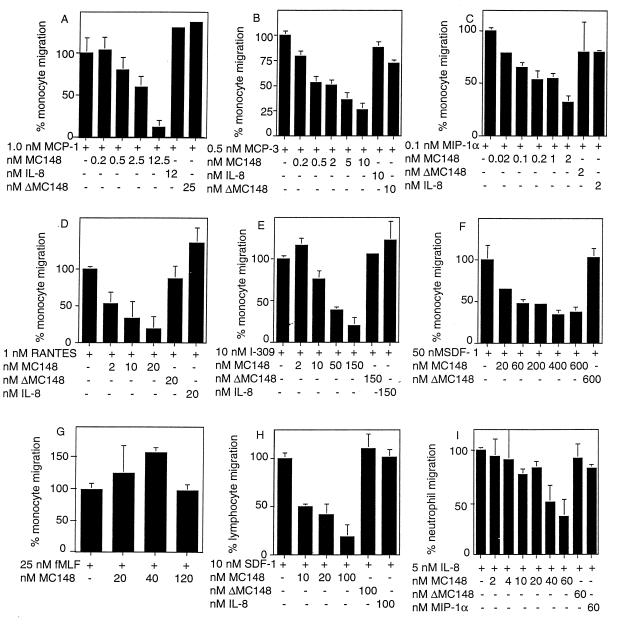
Chemotaxis was measured by using a modified Boyden cell migration system in which the cells are placed in the upper chambers and the chemoattractant is placed in the lower ones. Initial experiments indicated that MC148P itself had no effect on the migration of monocytes, lymphocytes, or neutrophils. The concentration of human chemokine that provided half-maximal cell migration then was mixed with several concentrations of MC148P in the lower wells of the chamber. In Fig. 2A–E, the effect of MC148P on the chemotaxis of freshly prepared human monocytes to the CC chemokines MCP-1, MCP-3, MIP-1α, RANTES, or I-309 is shown. MC148P exhibited dose-dependent antagonistic activity to a maximum of 70–80% over a 0.2–20× molar concentration relative to each active chemokine. Half-maximal antagonistic activity usually occurred at a 1–2× concentration of MC148P. The negative controls, MC148P that was heat treated at 96°C for 15 min (ΔMC148P) and the CXC chemokine IL-8, had no chemotactic inhibitory activities.
Figure 2.
Inhibition by MC148P of CC and CXC chemokine-induced migration of human monocytes, neutrophils, and lymphocytes. Cell migration was determined by using a modified Boyden chamber. Additions to the lower well are indicated below each bar. Each condition was tested in triplicate, and the data are presented as the mean ± SE. The percentage of cell migration was determined from the ratio of the number of cells on the underside of the filter (in five 40× fields) in the presence of chemokine and indicated concentrations of MC148P, and the number of cells on the underside of the filter in the presence of chemokine alone (in five 40× fields). Each experiment was performed at least three times, and results from representative ones are shown. The average number of cells and the calculated SEM counted in the absence of MC148P were: MCP-1, 533 ± 98; MCP-3, 502 ± 19; fMLF, 165 ± 15; IL-8, 305 ± 15; RANTES, 169 ± 11; MIP-1α, 217 ± 25; SDF-1 (lymphocytes), 877 ± 37; SDF-1 (monocytes), 276 ± 45; I-309, 571 ± 37.
To extend these observations, we determined the effect of MC148P on the chemotaxis of monocytes (Fig. 2F) and lymphocytes (Fig. 2H) to the CXC chemokine SDF-1. A 70–80% inhibition occurred at 10- to 12-fold molar ratios of MC148P to SDF-1 with a half-maximal effect at a 1- to 2-fold molar ratio. Neither of the negative controls, ΔMC148P nor IL-8, inhibited chemotaxis of monocytes or lymphocytes in response to SDF-1. We also tested the effect of MC148P on the ability of the CXC chemokine IL-8 to attract neutrophils. MC148 inhibited chemotaxis in a dose-dependent manner (Fig. 2I). A 12-fold molar ratio of MC148P to IL-8 inhibited chemotaxis by 70%; half-maximal inhibition occurred at a molar ratio of 2–4:1.
The antagonistic effects of MC148P appear to be specific for the chemokine family of chemoattractant molecules because there was no effect on the chemotaxis of monocytes to the classical chemoattractant fMet-Leu-Phe (fMLF) (Fig. 2G). This result also ruled out nonspecific or other effects of MC148P that might inhibit the ability of cells to respond to chemoattractants.
Inhibition of Calcium Flux.
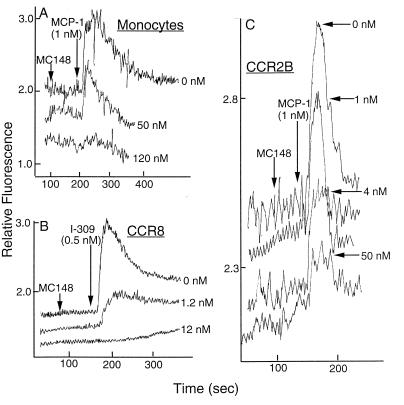
In addition to chemotaxis, the binding of chemokines to their receptors induces a rapid release of intracellular calcium. Consistent with the antagonistic chemoattractive activity, MC148P did not induce a calcium flux in primary monocytes or CCR2B-transfected HEK 293 cells but greatly diminished the responses of these cells upon subsequent incubation with MCP-1, a specific CCR2B agonist (Fig. 3 A and C). Moreover, the extent of inhibition was directly related to the amount of MC148P used. Similarly, incubation of MC148 with CCR8-transfected murine pre-B cells (12) prevented the intracellular calcium release generated by I-309, a CCR8 agonist (Fig. 3B). In other experiments (not shown), MC148P prevented submaximal amounts of SDF-1α from mobilizing calcium in murine pre-B cells, which have an endogenous SDF-1 receptor, and in HOS cells transfected with the human SDF-1 receptor CXCR4 (13).
Figure 3.
Inhibition by MC148P of chemokine-induced calcium mobilization. MC148P (amounts indicated to the right of each curve) and MCP-1 (1 nM) or I-309 (0.5 nM) were added successively to cells that had been loaded with Fura-2. Fluorescence was measured at 510 nm after repetitive, sequential excitations at 340 nm and 380 nm and the ratios were determined. (A) Primary monocytes. (B) CCR8-transfected murine pre B cells (12). (C) CCR2B-transfected HEK 293 cells (10).
Inhibition of Chemokine Binding.
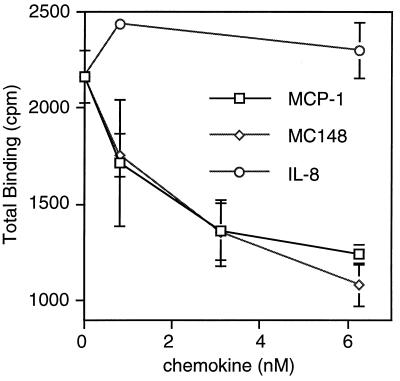
Next, we determined whether MC148P could specifically prevent or displace the binding of a chemokine to its receptor. The binding of 125I-MCP-1 to CCR2B, expressed in transfected HEK 293 cells, was prevented by equivalent concentrations of unlabeled MCP-1 or MC148P (Fig. 4). An IC50 for MC148P of approximately 1.5 nM was calculated. As expected, the negative control IL-8 had no effect on MCP-1 binding (Fig. 4). These results strongly suggest that MC148P acts by binding directly to chemokine receptors. An alternative possibility, that MC148P binds to chemokines, was investigated. Attempts to cross-link biologically active concentrations of MC148P with iodinated MCP-1 or MIP1-α by using 2.5 mM ethylene glycol bis(sulfosuccinimidyl-succinate), however, were unsuccessful.
Figure 4.
Competition between MC148P and MCP-1 for binding to CCR2B. HEK 293 cells expressing CCR2B were incubated with 0.1 nM 125I-labeled MCP-1 and indicated amounts of unlabeled MCP-1, MC148P, or IL-8. The cells were separated from unbound radioactive material, and gamma emissions were counted. Each point is the mean ± SE derived from triplicate assays of a representative experiment.
DISCUSSION
Viruses have developed a variety of methods for exploiting or combating chemokines and their receptors (14). Some herpesviruses encode chemokine receptor homologs that may exhibit constitutive or chemokine-induced signaling (15–17) and HIV and related retroviruses have envelope proteins that bind to chemokine receptors for cell entry (18–23). In contrast, some poxviruses (not including MCV) encode a secreted protein that binds avidly to CC chemokines and may interfere with their actions (24–26). Chemokine homologs are encoded by human herpesvirus (HHV) 6 (27), HHV 8 (28, 29), and murine cytomegalovirus (30). Of these, HHV 8 vMIP-II has been characterized in the most detail; it has broad-spectrum CC chemokine receptor-binding properties and inhibits chemotaxis of monocytes to several CC chemokines (31). In addition, vMIP-II binds the CXC receptor CXCR4 (31). At present, however, MC148P is the only viral chemokine homolog that has been shown to inhibit chemotaxis of multiple leukocyte subsets induced by CC and CXC chemokines. Based on the chemotaxis results, we can infer that MC148P functionally interacts with at least the following CC and CXC receptors: CCR1 and/or CCR5, CCR2, CCR8, CXCR1 and/or CXCR2, and CXCR4. Further experiments are likely to expand this list. Aside from one CC chemokine (I-309), MC148P is the only molecule shown to interact with CCR8.
The broad, potent chemokine antagonist activity of MC148P and vMIP-II was unexpected in view of the binding specificities of chemokines and the absence of any example of a CC and a CXC chemokine using the same leukocyte receptor (2). Despite the distinct receptor-binding specificities of CC and CXC chemokines, these chemokines have similar three-dimensional monomeric structures comprising an NH2-terminal unstructured region, three antiparallel β-strands connected by loops, and a COOH-terminal α-helix (32). The receptor activation domain resides within the unstructured NH2 terminus, and chemokine antagonists have been engineered by truncation or extension of the NH2-terminal sequence of several chemokines (33–38). Inspection of the viral chemokine homolog sequences suggested that the antagonistic activity of MC148P resulted at least in part from a shortened NH2-terminal activation domain (Fig. 1A). The antagonistic activity of HHV 8 vMIP-II, however, must have a different basis because the length of the NH2 terminus is similar to that of CC chemokines (Fig. 1A). The ability of MC148 to inhibit neutrophil chemotaxis correlates with the conservation of the leucine, 16 aa after the second cysteine (Fig. 1A), which has been shown to be important for IL-8 binding to CXCR1 and CXCR2 (39, 40). vMIP-II lacks this conserved leucine and does not bind known IL-8 receptors.
The specificity of chemokines for one or a few receptors is clearly useful for the host because it allows selective activation of different cell types. For viruses, however, broad antagonistic activity may be advantageous. We can speculate that both poxviruses and herpesviruses pirated a chemokine gene from their hosts and that mutations conferring binding to multiple receptors provided a selective advantage to the viruses. Structure–function studies of the viral proteins may lead to a greater understanding of chemokine–receptor interactions and provide useful lead compounds for therapeutics that broadly target chemokine signaling systems. vMIP-II has been shown to inhibit interactions of HIV-1 with chemokine receptors (28, 29, 31, 41); because MC148P antagonizes a variety of chemokines including those that bind to HIV-1 coreceptors on macrophages and T lymphocytes, broad antiviral activity is expected.
At present, the role of MC148P in MCV evasion of the immune system cannot be determined directly because the virus has never been grown in cell culture and there are no useful animal models (42, 43). Nevertheless, the absence of homologs of other genes whose products interact with chemokines or chemokine receptors and the ability of MC148P to inhibit chemokine-induced migration of multiple leukocytes subsets imply that MC148P has a major role in preventing an inflammatory response to MCV infection.
Acknowledgments
We thank T. Senkevich for MCV DNA, E. Koonin for help with computer alignments, J. Sisler for DNA sequencing, and J. Farber for advice and cell migration chambers. J. Farber and E. Berger provided helpful comments on the manuscript.
ABBREVIATIONS
- MCV
molluscum contagiosum virus
- MIP
macrophage inflammatory protein
- IL
interleukin
- HHV
human herpesvirus
References
- 1.Gottlieb S L, Myskowski P L. Int J Dermatol. 1994;33:453–461. doi: 10.1111/j.1365-4362.1994.tb02853.x. [DOI] [PubMed] [Google Scholar]
- 2.Baggiolini M, Dewald B, Moser B. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 3.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 4.Krathwohl M D, Hromas R, Brown D R, Broxmeyer H E, Fife K H. Proc Natl Acad Sci USA. 1997;94:9875–9880. doi: 10.1073/pnas.94.18.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 6.Blasco R, Moss B. Gene. 1995;158:157–162. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- 7.Bugert J J, Lohmuller C, Damon I, Moss B, Darai G. Virology. 1998;242:51–59. doi: 10.1006/viro.1997.9001. [DOI] [PubMed] [Google Scholar]
- 8.Leonard E J, Sylvester I, Yoshimura T, Taub D, Oppenheim J, Wang J M, Lloyd A. In: Measurement of α and β Chemokines. Coligan J E, Kruisbeek A M, editors. Vol. 1. New York: Wiley; 1995. pp. 6.12.1–6.12.28. [Google Scholar]
- 9.Miller M D, Krangel M S. Proc Natl Acad Sci USA. 1992;89:2950–2954. doi: 10.1073/pnas.89.7.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Combadiere C, Ahuja S K, Van Damme J, Tiffany H L, Gao J L, Murphy P M. J Biol Chem. 1995;270:29671–29675. doi: 10.1074/jbc.270.50.29671. [DOI] [PubMed] [Google Scholar]
- 11.Charo I F, Myers S J, Herman A, Franci C, Connoly A J, Coughlin S R. Proc Natl Acad Sci USA. 1994;91:2752–2756. doi: 10.1073/pnas.91.7.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiffany H L, Lautens L L, Gao J L, Pease J, Locati M, Combadiere C, Modi W, Bonner T I, Murphy P M. J Exp Med. 1997;186:165–170. doi: 10.1084/jem.186.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J-L, Murphy P M. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 14.Wells T, Schwartz T W. Curr Opin Biotech. 1997;8:741–748. doi: 10.1016/s0958-1669(97)80129-2. [DOI] [PubMed] [Google Scholar]
- 15.Gao J L, Wynn T A, Chang Y, Lee E J, Broxmeyer H E, Cooper S, Tiffany H L, Westphal H, Kwon-Chung J, Murphy P M. J Exp Med. 1997;185:1959–1968. doi: 10.1084/jem.185.11.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahuja S K, Murphy P M. J Biol Chem. 1993;268:20691–20694. [PubMed] [Google Scholar]
- 17.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Nature (London) 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 18.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 19.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 20.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Paarmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 22.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 24.Lalani A S, Graham K, Mossman K, Rajarathnam K, Clark-Lewis I, Kelvin D, McFadden G. J Virol. 1997;71:4356–4363. doi: 10.1128/jvi.71.6.4356-4363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham K A, Lalani A S, Macen J L, Ness T L, Barry M, Liu L-Y, Lucas A, Clark-Lewis I, Moyer R W, McFadden G. Virology. 1997;229:12–24. doi: 10.1006/viro.1996.8423. [DOI] [PubMed] [Google Scholar]
- 26.Smith C A, Smith T D, Smolak P J, Friend D, Hagen H, Gerhart M, Park L, Pickup D J, Torrance D, Mohler K, et al. Virology. 1997;236:316–327. doi: 10.1006/viro.1997.8730. [DOI] [PubMed] [Google Scholar]
- 27.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 28.Moore P S, Boshoff C, Weiss R A, Chang Y. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H-G, Reitz M S, Hayward G S. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald M R, Li X-Y, Virgin H W, IV. J Virol. 1997;71:1671–1678. doi: 10.1128/jvi.71.2.1671-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnson A H, Alouani S, Power C A, Lüttichau H R, Gerstoft J, Clapham P R, et al. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 32.Clore G M, Gronenborn A M. FASEB J. 1995;9:57–62. doi: 10.1096/fasebj.9.1.7821760. [DOI] [PubMed] [Google Scholar]
- 33.Gong J-H, Clark-Lewis I. J Exp Med. 1995;181:631–640. doi: 10.1084/jem.181.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S A, Dewald B, Clark-Lewis I, Baggiolini M. J Biol Chem. 1997;272:16166–16169. doi: 10.1074/jbc.272.26.16166. [DOI] [PubMed] [Google Scholar]
- 35.Moser B, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. J Biol Chem. 1993;268:7125–7128. [PubMed] [Google Scholar]
- 36.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 37.Wells T N, Power C A, Lusti-Narasimhan M, Hoogewerf A J, Cooke R M, Chung C W, Peitsch M C, Proudfoot A E. J Leukocyte Biol. 1996;59:53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y J, Rutledge B J, Rollins B J. J Biol Chem. 1994;269:15918–15924. [PubMed] [Google Scholar]
- 39.Lusti-Narasimhan M, Power C A, Allet B, Alouani S, Bacon K B, Mermod J J, Proudfoot A E, Wells T N. J Biol Chem. 1995;270:2716–2721. doi: 10.1074/jbc.270.6.2716. [DOI] [PubMed] [Google Scholar]
- 40.Lusti-Narasimhan M, Chollet A, Power C A, Allet B, Proudfoot A E, Wells T N. J Biol Chem. 1996;271:3148–3153. doi: 10.1074/jbc.271.6.3148. [DOI] [PubMed] [Google Scholar]
- 41.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schwickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, et al. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 42.Buller R M L, Chen J B W, Kreider J. Virology. 1995;213:655–659. doi: 10.1006/viro.1995.0037. [DOI] [PubMed] [Google Scholar]
- 43.Fife K H, Whitfeld M, Faust H, Goheen M P, Bryan J, Brown D. Virology. 1996;226:95–112. doi: 10.1006/viro.1996.0631. [DOI] [PubMed] [Google Scholar]