Summary
YOD1 is a highly conserved deubiquitinating enzyme of the ovarian tumor (otubain) family, whose function has yet to be assigned in mammalian cells. YOD1 is a constituent of a multiprotein complex with p97 as its nucleus, suggesting a functional link to a pathway responsible for the dislocation of misfolded proteins from the endoplasmic reticulum. Expression of a YOD1 variant deprived of its deubiquitinating activity imposes a halt on the dislocation reaction, as judged by the stabilization of various dislocation substrates. Accordingly, we observe an increase in polyubiquitinated dislocation intermediates in association with p97 in the cytosol. This dominant negative effect is dependent on the UBX and Zinc finger domains, appended to the N- and C-terminus of the catalytic otubain core domain, respectively. The assignment of a p97-associated ubiquitin processing function to YOD1 adds to our understanding of p97’s role in the dislocation process.
Introduction
In eukaryotes, the Ubiquitin (Ub)/proteasome system (UPS) is the major pathway responsible for the destruction of misfolded proteins. Even though the UPS machinery is confined to the cytosol, it can also degrade secretory, membrane, or luminal proteins that reside in the endoplasmic reticulum (ER). This type of destruction requires the translocation of substrates into the cytosol, a process referred to as dislocation or retrotranslocation. It can be divided into several steps (Raasi and Wolf, 2007; Vembar and Brodsky, 2008): substrates need to be recognized as misfolded, recruited into a protein-conducting channel, and dislocated into the cytosol. Derlin-1 and Sec61 may contribute to the construction of the relevant protein conducting channels (Lilley and Ploegh, 2004; Scott and Schekman, 2008; Wiertz et al., 1996b; Ye et al., 2004), but alternative strategies for substrate passage to the cytosol have been suggested (Ploegh, 2007). In mammalian cells, there are in all likelihood multiple exit strategies from the ER, which may then converge on the UPS. The emergence of a glycoprotein substrate in the cytosol coincides with the removal of N-linked glycans by the action of N-glycanase, and the ubiquitination via an E1-E2-E3 cascade, which tags the substrate for proteasomal destruction. Ub is utilized not only as degradation tag, it also serves as handle for cytosolic ATPases to exert a pulling force on the substrate, thus facilitating the movement of dislocation substrates into the cytosol (Flierman et al., 2003).
Two distinct multiprotein complexes can contribute to the mechanical force that drives dislocation: the p97/Valosin-containing protein (VCP, or Cdc48 in Saccharomyces cerevisiae) complex and the 19S cap of the 26S proteasome. Although different in their composition, both complexes contain functionally similar elements, namely adaptor proteins required for Ub recognition and ring-shaped, hexameric ATPase modules (Elsasser and Finley, 2005). The ATPases that are part of the VCP and 19S complexes are members of the same family, designated AAA (ATPases associated with a variety of cellular activities) ATPases (Neuwald et al., 1999) and unfold the substrate in ATP-dependent fashion (Navon and Goldberg, 2001). Removal of the Ub chain from the substrate by proteasome-associated deubiquitinating enzymes (DUBs) is key to allow the passage of the unfolded polypeptide through a narrow constriction into the proteolytic chamber of the proteasome core particle, where proteolysis ensues (Pickart and Cohen, 2004). Ub removal also allows recycling of this essential modifier. Mutations in the proteasome-associated DUB Rpn11 that disrupt its catalytic activity stall the processive substrate degradation by the proteasome and eventually lead to cell death (Verma et al., 2002). The 19S cap associates with the Sec61 channel, and in purified form supports ER dislocation in vitro, suggesting that it could indeed contribute force, and so couple dislocation and degradation (Kalies et al., 2005; Lee et al., 2004; Ng et al., 2007).
P97 nucleates a number of distinct protein complexes, variable in composition and function. Of relevance for dislocation is a complex that includes NPL4 and UFD1. Both associate to form a heterodimeric adaptor that binds to Ub and to p97’s N-terminal domain (Meyer et al., 2000), thus contributing to p97’s ability to associate with dislocation substrates and enabling p97 pulling substrate (Ye et al., 2001, 2003). It has been proposed that p97/Cdc48 can be recruited to the site(s) of ER dislocation by UBXD2 and/or UBXD8, two UBX domain containing proteins embedded in the ER membrane (Liang et al., 2006; Mueller et al., 2008). In yeast, Ubx2 not only binds to Cdc48, but also to the ER-resident Ub ligases Doa10 and Hrd1. Substrate ubiquitination and Cdc48 recruitment are thus spatially and temporally coordinated to facilitate substrate transfer (Neuber et al., 2005; Schuberth and Buchberger, 2005). A collective of substrate-processing cofactors associate with p97/Cdc48 to limit, promote, or reverse the ubiquitination of p97/Cdc48-associated substrates. Ubiquitination is therefore highly dynamic and carefully controlled (Jentsch and Rumpf, 2007; Raasi and Wolf, 2007). Shuttling factors, e.g. Rad23 and Dsk2 in budding yeast, finally transfer the substrate from p97 to the proteasome, where it is ultimately degraded (Elsasser et al., 2004; Medicherla et al., 2004; Richly et al., 2005).
Here we address the function of YOD1 (also known as OtuD2 or DUBA8; gene ID: 55432) in mammalian cells, a ubiquitin-specific protease equipped with a UBX domain, considered a hallmark of p97-associated proteins (Schuberth and Buchberger, 2008). YOD1 is the closest homolog of S. cerevisiae Otu1, which associates with Cdc48, to regulate the processing of the ER-membrane embedded transcription factor Spt23, a crucial component of the OLE pathway (Rumpf and Jentsch, 2006). Although highly conserved, the function of YOD1 is not known in higher eukaryotes. The human genome lacks a bona fide homolog of Spt23, suggesting that YOD1 participates in other, presumably conserved, cellular processes. Given the established involvement of p97 in ER dislocation, we reasoned that YOD1 might serve as p97-associated Ub processing factor in the context of protein dislocation from the ER. We now show that YOD1 is indeed a constituent of a p97 complex that drives ER-dislocation. A dominant negative YOD1 variant stalls the dislocation of various misfolded, ER-resident proteins. These substrates accumulate as ubiquitinated intermediates, establishing an important function for a deubiquitinating activity in the context of ER-dislocation.
Results
Identification of YOD1 interaction partners links YOD1 to the p97 complex
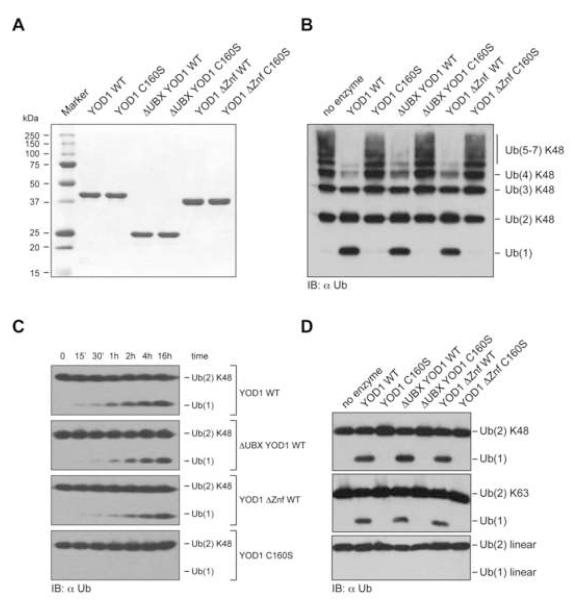
To determine its possible functions, we first identified interaction partners of human YOD1 by immunopurification. We identified not only YOD1 itself, as expected, but also p97, NPL4 and UFD1 as unique hits with good sequence coverage when compared to the corresponding control data set (Fig. S1). We cloned suitably tagged versions of p97 and YOD1 to allow their expression in 293T cells. In addition, we engineered an active site mutant of YOD1 (C160S) to address whether and how its catalytic activity is essential for biological function. According to Pfam predictions (Finn et al., 2008), YOD1 comprises three domains: An N-terminal UBX domain, a central otubain domain, and a C-terminal C2H2-type Zinc finger (Znf) domain. To study the role of these domains, we created a variant lacking the C-terminal Znf domain (YOD1 Znf), a version in which the N-terminal UBX domain was deleted (ΔUBX YOD1) or replaced by green fluorescent protein (ΔUBX GFP YOD1), and their combinations with the active site mutation (Fig. 1 A).
Figure 1. YOD1 associates with p97 via the N-terminal UBX domain.
(A) Domain organization of YOD1 and its mutant derivatives.
(B) 293T cells were transiently transfected with the indicated constructs or empty vector and homogenized by NP40 detergent lysis 24h post transfection. Lysates were subjected to immunoblotting with anti-FLAG and anti-p97 antibodies to control for expression of YOD1 derivatives and p97, respectively (upper and middle panels). Retrieved p97 in anti-FLAG immunoprecipitates was detected by immunoblotting with anti-p97 antibodies (lower panel).
To confirm that p97 and YOD1 form a complex in a cellular context, we transfected FLAG-tagged YOD1 variants, followed by preparation of detergent extracts. All YOD1 variants were expressed to a similar level, as judged by immunoblotting (Fig. 1 B, upper panel). YOD1 and its mutant derivatives were retrieved by immunoprecipitation, and p97 association was monitored by immunoblotting using anti-p97 antibodies (Fig. 1 B, lower panel). Endogenous p97 was retrieved in a complex with YOD1 WT and C160S. This interaction was strictly dependent on the UBX domain, since the ΔUBX GFP YOD1 variant failed to interact with p97, whereas the Zn finger (Znf) domain was dispensable for interaction with p97, both in cell and in vitro, further demonstrating that YOD1 binds to the N-terminal domain of p97 by virtue of its UBX domain. (Fig. S2A).
The otubain core domain is necessary and sufficient for catalytic activity in vitro
We next tested whether the UBX domain or the Znf domain are required for enzymatic activity. Purified YOD1 WT and its truncation derivatives all hydrolyzed K48-linked poly- and di-Ub chains (Fig. 2 B, C). The otubain core domain is thus necessary and sufficient to confer basal catalytic activity (Fig. S2). All truncation mutants were covalently modified by HA-tagged Ub vinylmethyl ester (HA-UbVME), a Ub-based suicide inhibitor that forms a covalent adduct with active Ub-specific cysteine proteases (Borodovsky et al., 2002; Schlieker et al., 2007), unless such truncations were combined with the C160S active site mutation (Fig. S2 C).
Figure 2. The otubain domain confers catalytic activity independent of accessory domains.
(A) Heterologously expressed, purified YOD1 variants (10 μg each) were separated by SDS-PAGE (12%) and stained with Coomassie Blue.
(B) K48-linked poly-Ub chains (2 μg) were incubated for 16 h in a total volume of 10 μl with different YOD1 variants (7.7 μM). Poly-Ub and free Ub were detected by immunoblotting using anti-Ub antibodies.
(C) K48-linked di-Ub (0.5 μg) were incubated for indicated times at 25°C in a total volume of 10 μl with different YOD1 variants (2.58 μM). Ub was detected by immunoblotting using anti-Ub antibodies.
(D) K48-, K63-linked and genetically fused linear di-Ub (2 μg) were incubated for 16 h in a total volume of 10 μl with various YOD1 variants (5.2 μM), and detected as in (B).
Yeast Otu1 and human OtuB1, two related members of the otubain family, display a strong preference for K48 linkages (Edelmann et al., 2009; Messick et al., 2008). Since additional domains may influence the ability of YOD1 to attack isopeptide-linked Ub chains, as exemplified by IsoT (Reyes-Turcu et al., 2008), we tested if the truncation variants differed in their ability to deconjugate K48- or K63-linked Ub chains. YOD1 WT and the truncation variants released similar quantities of free Ub in the same period of time (Fig. 2 B, S2 D). Do any of the additional domains influence the activity towards di-Ub chains of different linkage? YOD1 and its mutant derivatives all produced similar amounts of free Ub in the same period of time when assayed on K48- and K63-linked chains (Fig. 2 D, Fig. S2 D). YOD1 is isopeptide-linkage specific as neither variant cleaved linear Ub chains (Fig. 2 D, lower panel). Thus, the core domain is necessary and sufficient to confer specificity, although we cannot exclude the formal possibility that either domain is important to discriminate between other linkage types, e.g. K11 versus K48/K63 linkages.
A catalytically inactive YOD1 mutant impairs the degradation of truncated ribophorin, a misfolded, ER-resident glycoprotein
Could the interaction with p97 allow us to place YOD1 in the context of a particular cellular function? P97 is involved in homotypic membrane fusion (Hetzer et al., 2001; Meyer et al., 2000), activation of transcription factors (Rape et al., 2001), mobilization of a kinase from chromatin (Ramadan et al., 2007), and extraction of misfolded proteins from the ER (Bays et al., 2001; Ye et al., 2001). This relies on a set of adaptors, which link the common ATPase module p97 to specific molecular targets and confer specificity (Raasi and Wolf, 2007; Schuberth and Buchberger, 2008). For example, homotypic membrane fusion relies on p97 in concert with the adaptor p47 (Hetzer et al., 2001), whereas Spt23 activation requires a distinct heterodimeric adaptor, UFD1/NPL4 (Rape et al., 2001). In yeast, some of the molecular machinery involved in Spt23 activation is also required to extract misfolded proteins from the ER: both pathways employ p97, UFD1, and NPL4. Since YOD1, p97, NPL4 and UFD1 constitute a multiprotein complex in mammalian cells (Fig. 1 B and Fig. S1), we asked whether the activity of YOD1 is required for extraction of proteins from the ER.
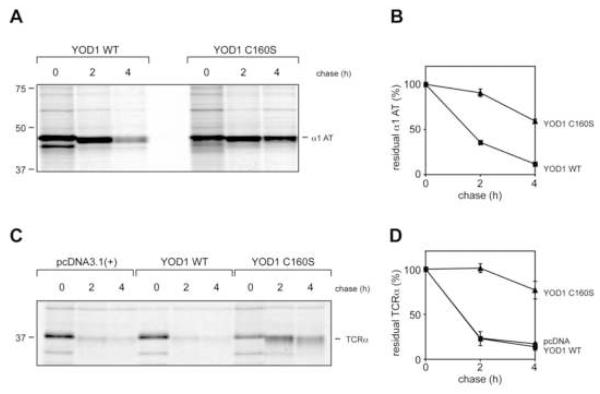
We tested whether YOD1 or its mutants affected degradation of truncated ribophorin, RI332, a misfolded ER-resident glycoprotein rapidly degraded by the ubiquitin-proteasome system (UPS) upon its arrival in the cytosol (Kitzmuller et al., 2003). If YOD1 is indeed involved, overexpression of either wildtype or catalytically inactive YOD1 should affect the degradation of RI332. 293T cells were co-transfected with RI332 and either YOD1 WT, YOD1 C160S, or empty vector, and the stability of RI332 was determined by pulse-chase analysis (Fig. 3). Introduction of YOD1 C160S markedly stabilized RI332 while overexpressed YOD1 WT did not affect the degradation of RI332 (Fig. 3 A, B). The stability of endogenous full-length ribophorin was not affected by either construct (Fig. 3 A).
Figure 3. Catalytically inactive YOD1 C160S impairs the dislocation of truncated ribophorin.
(A) 293T cells were co-transfected with RI332 and empty vector, YOD1 WT or YOD1 C160S. 24 hours after transfection, cells were pulse-labeled with 35S for 10 min, chased for the indicated time points, lysed in 1% SDS, and the lysates were subjected to immunoprecipitation with anti-ribophorin antibodies. The eluates were resolved by 12% SDS-PAGE and visualized by autoradiography (upper panel). Unbound material was immunoprecipitated with anti-FLAG antibodies to verify equal expression of the YOD1 constructs (lower panel).
(B) Densiometric quantification of RI332 levels. Plotted are the mean values from three independent experiments. Error bars depict the standard deviation.
(C) 293T cells were co-transfected with a cytosolic variant of RI332 lacking its N-terminal signal sequence (ΔSS RI332) and with either empty vector (pcDNA), YOD1 WT or catalytically inactive YOD1 C160S, and processed as in (A).
(D) ΔSS RI332 stability was quantified as in (B).
(E) Cells co-transfected with YOD1 C160S and RI332 were metabolically labeled as in (A). Cell extracts were prepared in hypotonic buffer by homogenization in absence of detergent. The homogenate was incubated on ice in presence and absence of 100 μg/ml proteinase K. 0.5% NP40 was added as indicated. Proteinase K was inactivated after 15 min by inclusion of PMSF. The resulting material was solubilized with 1% SDS, immunoprecipitated with anti-ribophorin antibodies and processed as in (A).
Is the dominant negative effect on RI332 degradation imposed by YOD1 C160S due to a specific role for YOD1 in protein dislocation from the ER, or could it be a mere consequence of non-specific stabilization of all Ub-conjugated substrates destined for proteasomal degradation? To resolve this issue, we engineered an RI332 variant that lacks the N-terminal signal sequence (ΔSS-RI332), thus creating a soluble, cytosolic version that is no longer coupled to ER dislocation. ΔSS-RI332 is rapidly degraded in UPS-dependent fashion (data not shown), but neither YOD1 WT nor its catalytically inactive counterpart stabilized ΔSS-RI332 (Fig. 3 C, D).
We routinely observed three closely spaced, electrophoretically distinct bands for RI332. By glycosidase-treatment of immunoprecipitated RI332 and by expression of a RI332 variant (N275T) devoid of its N-glycosylation site, the nature of the three bands was revealed as glycosylated (EndoH sensitive), de-glycosylated and non-glycosylated RI332, respectively (Fig. S3). Expression of YOD1 C160S stabilized both, the glycosylated (RI332+CHO) and the non-glycosylated (RI332−CHO) form of RI332 within a membrane compartment, consistent with a ER-luminal localization, as judged by protease protection experiments (Fig. 3 E). We conclude that YOD1 C160S specifically blocks a step in the course of dislocation and/or degradation of ER-resident proteins destined for proteasomal degradation.
Both UBX and Znf domains are required for YOD1 activity in vivo
While the otubain core domain appears to be necessary and sufficient for deubiquitinating activity in vitro, the UBX and Znf domains may well play important roles in vivo. Since a construct lacking the UBX domain did not yield satisfactory expression levels (data not shown), we created a variant in which this domain was replaced by GFP (ΔUBX-GFP YOD1) to yield higher expression and so be able to investigate the role of the UBX domain of YOD1. Both ΔUBX-GFP YOD1 and its catalytically inactive counterpart (ΔUBX-GFP YOD1 C160S) were expressed to levels comparable to YOD1 WT (Fig. 4 A). To examine whether the UBX domain is required for biological function, we tested the ability of ΔUBX-GFP YOD1 C160S to stabilize RI332. ΔUBX-GFP YOD1 C160S stabilized RI332 only to some degree and this dominant negative effect was less pronounced than in the case of YOD1 C160S (Fig. 4 A and B).
Figure 4. The UBX and Znf domains are required for YOD1 activity in vivo.
(A) 293T cells were transfected with RI332 and either full-length or truncated (ΔUBX) YOD1 WT or C160S. The experiment was performed as in Fig. 3 A.
(B) Quantification of (A). The error bars represent the standard deviation of three independent experiments. The asterisk indicates a proteolytic fragment of ΔUBX YOD1.
(C) 293T cells were transfected with either full-length or truncated (ΔZnf) YOD1 WT or C160S. The experiment was performed as in Fig. 3 A.
(D) Quantification of (C). The error bars represent the deviation from the mean of two independent experiments.
Next, we tested if the Znf domain was required for the dominant negative effect of YOD1 C160S. The catalyticaly inactive variant deprived of its Znf domain domain completely lost its ability to block RI332 degradation (Fig. 4 C and D), suggesting that the Znf domain is essential for the function of YOD1 in vivo. Both the UBX and Znf domains, accessories to the catalytic core, are thus important for the function of YOD1, presumably by placing the catalytic core in the appropriate context.
YOD1 C160S affects the degradative fate of ER-resident dislocation substrates with various topologies
To put our findings on a more general footing, we tested whether the dominant negative effect imposed by YOD1 C160S applies to other substrates as well. First, we chose 1-antitrypsin Null Hong Kong (NHK), a mutant allele of α1-antitrypsin subject to dislocation and degradation via the UPS (Sifers et al., 1988), as substrate. 293T cells were co-transfected with YOD1 C160S and NHK, with YOD1 WT serving as the control. NHK was markedly stabilized in presence of YOD1 C160S (Fig. 5 A, B).
Figure 5. YOD1 C160S impairs dislocation of α1-antitrypsin and TCRα chain.
(A) 293T were cells transfected with α1-antitrypsin NHK (α1 AT), and YOD1 WT or YOD1 C160S, were pulse-labeled with 35S for 10 min and chased for the indicated time points. The cells were lysed in SDS and the lysates were immunoprecipitated with anti-α1-AT antibodies. The eluates were separated by SDS PAGE (12%) and the bands were visualized by autoradiography.
(B) Quantification of NHK levels. Plotted is the mean value of three independent experiments with the error bar corresponding to the standard deviation.
(C) 293T cells were transfected with TCRα, and either empty vector (pcDNA), YOD1 WT or YOD1 C160S. The pulse-chase experiment was performed as in Fig. 3 A. TCRα was retrieved from SDS-lysates by immunoprecipitation with anti-TCRα antibodies and visualized by autoradiography.
(D) Quantification of TCRα levels. Plotted is the mean value of two independent experiments with error bars.
Since substrates with different topologies may use distinct, yet overlapping, molecular machineries involved in ER-dislocation (Carvalho et al., 2006; Ravid et al., 2006; Vashist and Ng, 2004), we examined the fate of an unpaired TCRα chain. TCRα is a type I transmembrane protein that is promptly dislocated and degraded by the UPS when expressed in the absence of its partner, the TCRβ chain (Huppa and Ploegh, 1997; Yu et al., 1997). As seen for luminal proteins, the TCRα chain was strongly stabilized by YOD1 C160S (Fig. 5 C and D). Therefore, although distinct topologies may require different mechanisms to achieve dislocation, YOD1 C160S imposed a strong dominant negative effect on the processing of all tested substrates.
Overexpression of YOD1 C160S results in the accumulation of polyubiquitinated ER-dislocation substrates
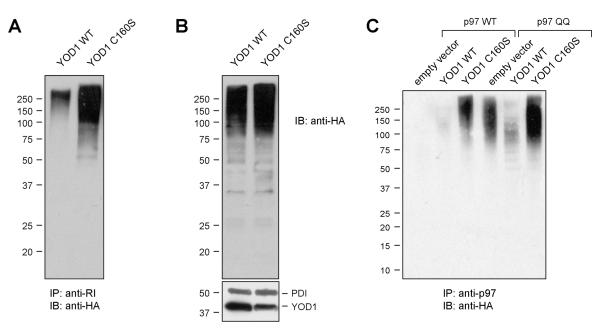
Having shown that YOD1 is involved in protein dislocation and given the role of substrate ubiquitination in ER dislocation (Ye et al., 2001), we asked if dislocation substrates could be a target for the deubiquitinating activity of YOD1. Therefore, RI332 was co-transfected with either YOD1 WT or YOD1 C160S, together with HA-tagged Ub. After retrieval of RI332 by immunoprecipitation, the extent of RI332 ubiquitination was assessed by immunoblotting using an anti-HA antibody. We observed a profound increase in high molecular weight (HMW), HA-reactive material in YOD1 C160S expressing cells relative to those expressing YOD1 WT, indicative of an accumulation of polyubiquitinated RI332 species (Fig. 6 A). Overall, the levels of HMW Ub-conjugates are somewhat elevated in the presence of catalytically inactive YOD1 in the input samples (Fig. 6 B). Accordingly, the steady state level of RI332 is increased in YOD1 C160S expressing cells (Fig. S5A). However, the amount of ubiquitinated RI332 recovered from cells expressing YOD1 C160S as opposed to YOD1 WT is much larger, and therefore cannot be attributed solely to an increase in total HMW Ub species or RI332.
Figure 6. Polyubiquitin chains accumulate on the dislocation substrate RI332 and on p97-associated substrates in the presence of catalytically inactive YOD1 C160S.
(A) 293T cells transiently transfected with RI332, HA-ubiquitin and either YOD1 WT or C160S were lysed in NP40 lysis buffer, and immunoprecipitated with anti-RI antibodies. The eluates were visualized by immunoblotting with anti-HA antibodies.
(B) The inpute lysates from A were directly immunoblotted and probed with anti-HA antibodies for ubiquitin levels, anti-FLAG antibodies for YOD1 levels and anti-PDI antibodies as loading control. See Fig. S5A for additional loading controls.
(C) 293T cell were transiently transfected with HA-ubiquitin, p97 WT or p97 QQ, and either empty vector (pcDNA), YOD1 WT or YOD1 C160S as indicated. After NP40 lysis and immunoprecipitation with anti-p97 antibodies the eluates were immunoblotted and probed with anti-HA antibodies. The corresponding input lysates are shown in the supplementary materials (Fig. S5 B).
Are ubiquitinated species that are stabilized by YOD1 C160S associated with p97? We co-transfected HA-Ub, YOD1 WT or YOD1 C160S, and p97 WT. P97 was retrieved by immunoprecipitation, and the retrieval of associated ubiquitinated species was assayed by immunoblotting with anti-HA antibodies. Ubiquitinated species were readily detectable in presence of YOD1 C160S, but were virtually undetectable in the presence of YOD1 WT or a vector control (Fig. 6 C; Fig S5 B). This result we attribute to the tight coupling of ER-dislocation and proteasomal turnover in absence of YOD1 C160S. We also monitored the ubiquitination status of substrates that are trapped in association with p97 by employing an ATPase-deficient p97 E305Q/E578Q mutant (p97 QQ). P97 QQ associates with ER dislocation substrates in stable fashion, and thus blocks their processive deubiquitination and degradation (Ye et al., 2003). Ubiquitinated species are already detectable in absence of YOD1 C160S, but co-expression of YOD1 C160S has an additive effect, as judged by the increase in HA-reactive material (Fig. 6 C). In contrast, a clear reduction of HA-reactivity is seen in presence of YOD1 WT, suggesting that YOD1 WT deconjugates Ub from substrates that are stably associated with p97 QQ. p97 QQ associated polyubiquitinated proteins can be deubiquitinated in vitro by purified YOD1 (Fig. S6). We may thus attribute the inhibitory effect of YOD1 C160S on dislocation to a failure to deubiquitinate p97-associated dislocation substrates.
YOD1 associates with the ER-dislocation machinery
As the expression of YOD1 C160S dramatically stabilized several dislocation substrates, we wondered whether YOD1 physically associates with known components of the ER-dislocation machinery such as SEL1L, Derlin-1 and the UBX-domain containing proteins UBXD8 and UBXD2. We immunoprecipitated FLAG-tagged YOD1 variants from cell lysates and probed for co-precipitated, endogenous proteins with respective antibodies. Using this approach, we identified Derlin-1 and UBXD8 as novel interaction partners of YOD1, while SEL1 and UBXD2 apparently do not interact with YOD1 (Fig 7 A, B and data not shown). The UBX domain of YOD1 was essential for Derlin-1 binding (Fig. 7 B). Thus, binding of Derlin-1, a known interactor of p97, may be indirect and mediated via UBX domain-dependent retrieval of p97. This scenario cannot apply to UBXD8, as the UBX domain of YOD1 is not essential for UBXD8 binding. In fact, neither the UBX domain nor the Znf of YOD1 is essential for UBXD8 binding, but both do stabilize the interaction in synergistic fashion (Fig. 7 B). In agreement with a catalytic role of YOD1 in protein dislocation, the catalytically inactive forms of YOD1 retrieved Derlin-1 and UBXD8 more efficiently than the WT counterpart.
Figure 7. YOD1 associates with the ER-dislocation machinery.
(A) 293T cells were transfected with the indicated YOD1 constructs or empty vector as control and lysed in NP40. After immunoprecipitation with anti-FLAG antibodies the eluates were subjected to immunoblotting with anti-Derlin-1 antibodies.
(B) Cell lysates were prepared as in (A). Eluates were subjected to immunoblotting with anti-UBXD8 and anti-UBXD2 antibodies.
In aggregate, YOD1 interacts directly with p97 and is a novel constituent of the ER dislocation machinery, thereby placing it in a perfect position to process ubiquitinated dislocation substrates in association with p97.
Discussion
Ubiquitination is a highly dynamic process carefully controlled by opposing Ub-conjugating and deconjugating activities. The p97 complex and the 19S cap of the proteasome, the two major Ub-dependent protein unfolding machines in the eukaryotic cytosol, employ both types of activity to regulate the fate of their substrates (Elsasser and Finley, 2005; Jentsch and Rumpf, 2007). In the case of the 26S proteasome, deubiquitination removes the impediment of attached Ub chains to allow the passage of substrates to the interior of the proteolytic core particle, accessible only through a narrow pore (Pickart and Cohen, 2004). Given the similarity between the p97 complex and the 19S cap of the proteasome, we hypothesized that p97 might make use of deubiquitinating activities in most, if not all pathways in which ubiquitinated substrates are engaged by this complex. The involvement of p97 in ER dislocation (Ye et al., 2001) inspired us to look for Ub-specific proteases that might directly interact with it, thus prompting our examination of YOD1, a UBX-domain-containing member of the otubain family of unknown function.
We showed that p97 associates with YOD1 by virtue of its UBX domain and that YOD1 participates in a p97 complex that also contains NPL4 and UFD1 (Fig. 1 and Fig. S1), involved in the dislocation of misfolded proteins from the ER (Ye et al., 2001, 2003). This complex also contains Derlin-1 and UBXD8 (Fig. 7), ER-resident components of the dislocation machinery (Lilley and Ploegh, 2004; Mueller et al., 2008; Ye et al., 2004). Importantly, a deletion variant of YOD1 (ΔUBX-GFP YOD1) that does not bind p97 still co-precipitated UBXD8 (Fig. 7 B). Thus, binding of UBXD8 is not mediated indirectly via p97 alone and therefore cannot be a mere consequence of YOD1 overexpression and unspecific retrieval of p97 interactors. The specificity of the UBXD8 binding is further supported by the fact that UBXD2, another UBX-domain containing interactor of p97 (Liang et al., 2006), does not interact with YOD1 (Fig. 7 B).
The catalytic role of YOD1 in dislocation is demonstrated by overexpression of a catalytically inactive form (YOD1 C160S), which almost quantitatively stabilized RI332, a model substrate for ER dislocation. The magnitude of this dominant negative effect (Fig. 3 A) was comparable to that commonly seen upon complete proteasomal inhibition (Fig. S4). Similarly to RI332, NHK and unpaired TCRα-chain, were profoundly stabilized by YOD1 C160S. The dominant negative effect was entirely dependent on the ER localization of the dislocation substrate: we saw no effect on protein stability once the ribophorin substrate was topologically uncoupled from the dislocation pathway.
We may thus place YOD1 in an ER-dislocation pathway, and suggest that the correct positioning of the substrate and of YOD1 itself relative to p97 are important for p97 function. Given its effect on both luminal and membrane-bound substrates, YOD1 must lie at or beyond the point where the relevant dislocation pathways converge, most likely between the dislocon and the proteasome, in agreement with its ability to associate with p97. It is still not known how many distinct dislocation pathways exist in the mammalian cell and whether or not they are assigned to particular substrate classes. Knockdown experiments with YOD1 or ataxin-3, another DUB implicated in protein dislocation from the ER, did not affect the degradation of several dislocation model substrates (Wang et al., 2004; Zhong and Pittman, 2006). At this point, we cannot assess whether the remaining levels of YOD1 that escape RNA silencing are sufficient to promote dislocation, or if p97-associated DUBs have overlapping or redundant functions. Several DUBs contain an UBX domain that could facilitate their binding to p97. The extent to which these other UBX-containing DUBs participate in dislocation remains to be established.
Is the arrest in dislocation imposed by YOD1 C160S indeed attributable to a deubiquitination defect? Cells expressing catalytically inactive YOD1 C160S accumulated polyubiquitinated RI332 to a much higher degree than YOD1 WT cells (Fig. 6 A), suggesting that the loss of catalytic activity of YOD1 is responsible for the accumulation of p97-associated RI332 in ubiquitinated form. In fact, this interpretation was substantiated by the observation that YOD1 modulates the ubiquitination status of not only RI332, but also of other p97-associated substrates (Fig. 6 C). The identity of these p97-associated Ub conjugated adducts was not determined, but likely includes a multitude of different dislocation substrates. YOD1 WT reversed the ubiquitination of dislocation substrates trapped by stable association with an ATPase-deficient p97 mutant (p97 QQ), whereas expression of YOD1 C160S in the same cells stabilized the Ub-adducts even more (Fig. 6 C and Fig. S5 B). We conclude that YOD1 activity is required for the p97-driven dislocation of misfolded proteins, and we attribute the inhibitory effect exerted by YOD1 C160S to a failure to deubiquitinate p97-associated dislocation intermediates en route to the cytosol (Fig. S7).
In our model, p97 engages dislocation substrates by means of the UFD1/NPL4 adaptor as soon as they emerge in the cytosol and undergo E3-catalyzed ubiquitination (Fig. S7 I). We speculate that once the substrate is transferred to the p97 pore and threaded through p97’s axial channel, YOD1 trims the Ub chain on the associated substrate to remove Ub moieties that present a steric impediment to the threading process (Fig. S7 II). Based on the data presented, we do not know whether this mechanism requires the en bloc removal of the entire Ub chain, akin to Rpn11 (Verma et al., 2002), or if a processive trimming-type activity like Ubp6 (Crosas et al., 2006) is sufficient to allow such threading. In the presence of YOD1 C160S, the Ub chain on the dislocation intermediate persists, thus preventing the processive movement of the substrate towards the cytosol (Fig. S7 IIb). In this scenario, a stoichiometric quantity of arrested intermediates is sufficient to titrate a limited number of dislocons, resulting in a pronounced accumulation of misfolded proteins within the ER (Fig. S7 IIIb).
Given that AAA ATPases of the 19S cap can accommodate looped polypeptides (Lee et al., 2002; Liu et al., 2003), and that this capability is conserved even in distantly related prokaryotic AAA+ ATPases endowed with threading activity (Haslberger et al., 2008), single Ub modifications may well be tolerated by the p97 assembly. A mono- or oligo-ubiquitinated substrate that emerges from p97/cdc48 at the other end may then be subjected to Ufd2-like E4 activities known to associate with Cdc48 (Richly et al., 2005). This would eventually extend the Ub chain to a length that is optimal for the transfer to proteasomal shuttling factors, such as Rad23/Dsk2 (Raasi et al., 2004; Richly et al., 2005). In this scenario, substrate dislocation is processively coupled to proteasomal degradation. Genetic evidence in S. cerevisiae established that Cdc48 cooperates with numerous cofactors in the following order: NPL4/UFD1→Cdc48→Ufd2→Rad23/Dsk2 (Medicherla et al., 2004; Richly et al., 2005). The proposed threading mechanism is reminiscent of that used by many other pore-forming hexameric AAA+ ATPases (Hinnerwisch et al., 2005; Martin et al., 2008; Schlieker et al., 2004; Weibezahn et al., 2004). The presence of a central channel in p97 (DeLaBarre and Brunger, 2003; Huyton et al., 2003), as well as the observation that mutations in pore-located residues in both AAA domains of p97 severely compromise ER dislocation activity (DeLaBarre et al., 2006) is in excellent agreement with our model. The functional assignment of the Otu1, VCIP135, Ataxin 3, and YOD1 to p97-dependent cellular processes in yeast (Rumpf and Jentsch, 2006) and mammalian cells (Wang et al., 2006; Wang et al., 2004; Zhong and Pittman, 2006) suggests an evolutionary conserved role for DUBs in the context of p97/Cdc48 activity. Future work will be required to probe the proposed mechanism further and to address whether other biological activities that rely on p97 function employ deubiquitinating activities as well.
In conclusion, we have firmly placed YOD1 in the dislocation pathway at a point where several of the distinct mammalian dislocation pathways converge. Given the multiplicity of exit strategies, combined with the near-universal involvement of the UPS in targeting the extracted proteins for degradation, it will be interesting to see how many other deubiquitinating enzymes are involved. While the ER is in sharp focus as a compartment where dislocation occurs or is initiated, we should remain open to the possibility that other intracellular locations might participate as well, each with unique machinery dedicated to the task, including perhaps novel components of the UPS.
Experimental procedures
Antibodies, Cell lines, Constructs
Antibodies against the HA-epitope were purchased from Roche (3F10), anti-FLAG, anti-TCRα and anti-ubiquitin antibodies were purchased from Sigma-Aldrich. Anti-α1 AT was purchased from Novus Biologicals, monoclonal anti-p97 from Fitzgerald Industries. The anti-Ribophorin antibody was a generous gift from N. Erwin Ivessa (Vienna Biocenter, Austria), anti-UBXD2 antibody was kindly provided by Mervyn J. Monteiro (University of Maryland Biotechnology Institute, USA). Recombinant, purified YOD1 was sent to Covance Research Products to generate rabbit polyclonal antibodies. The polyclonal p97 antibody was described previously (Lilley and Ploegh, 2005). 293T cells were cultured and transfected as previously described (Lilley et al., 2003). A plasmid coding for untagged RI332 was a generous gift from N. Erwin Ivessa (Vienna Biocenter, Austria) (Kitzmuller et al., 2003). YOD1 variants were cloned into the pcDNA 3.1 vector via HindIII and XbaI with an N-terminal FLAG-tag. The ΔUBX YOD1 expression construct has an N-terminal FLAG-tagged EGFP followed by aa129-348 of YOD1. SS RI332, with a C-terminal HA-tag, deprived of its signal sequence, was cloned into the pcDNA 3.1 vector. The TCRα and the α1 AT expression construct were described previously (Hosokawa et al., 2003; Huppa and Ploegh, 1997). Site-directed mutagenesis of p97, YOD1 and RI332 was performed with the QuickChange II mutagenesis kit (Stratagene).
Pulse-chase experiments, immunoprecipitations, PNGase F digestion, Endo H digestion, gel electrophoresis, immunoblotting, transient transfections, and enzmatic assays
Pulse chase experiments were performed as previously described (Wiertz et al., 1996a). For pulse-labeling experiments, cells were starved for 30-45 min in methionine/cysteine-free DMEM at 37°C, and labeled for 10 min at 37°C with 250 μCi of [35S] methionine/cysteine.
Cell lysis, immunoprecipitation, transfection of cells with RI332 and its variants, SDS-PAGE, and fluorography were performed as described earlier (Mueller et al., 2006). Quantification of radioactivity was performed on a phosphoimager. PNGase F and Endo H digestions of radiolabeled RI332 were performed before or after immunoprecipitation according to the recommendations of the manufacturer (New England Biolabs).
For the protease protection assay cells were homogenized by passing through a 23G needle in hypotonic buffer (20 mM Hepes pH 7.5, 5 mM KCl, 5 mM MgCl2, 1 mM DTT, and a protease inhibitor cocktail (Roche)). Proteinase K was added to a final concentration of 100 μg/ml in presence and absence of 0.5% NP40. After 20 min on ice, the proteinase K was inactivated by inclusion of PMSF (5 mM). All samples were adjusted to 1% SDS and analyzed by SDS-PAGE. Deubiquitination assays are described in the supplementary materials.
Supplementary Material
Acknowledgements
We thank Eric Spooner for mass spectrometry support and Elizabeth Klemm for providing ΔSS RI332. R.E. is supported by an EMBO long term Fellowship, 2008-379.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Edelmann MJ, Iphofer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierman D, Ye Y, Dai M, Chau V, Rapoport TA. Polyubiquitin serves as a recognition signal, rather than a ratcheting molecule, during retrotranslocation of proteins across the endoplasmic reticulum membrane. J Biol Chem. 2003;278:34774–34782. doi: 10.1074/jbc.M303360200. [DOI] [PubMed] [Google Scholar]
- Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, Bukau B. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol. 2008;15:641–650. doi: 10.1038/nsmb.1425. [DOI] [PubMed] [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- Huyton T, Pye VE, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS. The crystal structure of murine p97/VCP at 3.6A. J Struct Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Kalies KU, Allan S, Sergeyenko T, Kroger H, Romisch K. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. Embo J. 2005;24:2284–2293. doi: 10.1038/sj.emboj.7600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmuller C, Caprini A, Moore SE, Frenoy JP, Schwaiger E, Kellermann O, Ivessa NE, Ermonval M. Processing of N-linked glycans during endoplasmic-reticulum-associated degradation of a short-lived variant of ribophorin I. Biochem J. 2003;376:687–696. doi: 10.1042/BJ20030887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Prakash S, Matouschek A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J Biol Chem. 2002;277:34760–34765. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- Lee RJ, Liu CW, Harty C, McCracken AA, Latterich M, Romisch K, DeMartino GN, Thomas PJ, Brodsky JL. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. Embo J. 2004;23:2206–2215. doi: 10.1038/sj.emboj.7600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Tortorella D, Ploegh HL. Dislocation of a type I membrane protein requires interactions between membrane-spanning segments within the lipid bilayer. Mol Biol Cell. 2003;14:3690–3698. doi: 10.1091/mbc.E03-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. Embo J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci U S A. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Ng W, Sergeyenko T, Zeng N, Brown JD, Romisch K. Characterization of the proteasome interaction with the Sec61 channel in the endoplasmic reticulum. J Cell Sci. 2007;120:682–691. doi: 10.1242/jcs.03351. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Raasi S, Wolf DH. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. Embo J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell. 2007;25:677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Schekman R. Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J Cell Biol. 2008;181:1095–1105. doi: 10.1083/jcb.200804053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996a;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996b;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.