Abstract
Two-component signal-transduction systems, composed of a histidine-sensor kinase and a DNA-binding response regulator, allow bacteria to detect environmental changes and adjust cellular physiology to live more efficiently in a broad distribution of niches. Although many two-component signal-transduction systems are known, a limited number of signals that stimulate these systems have been discovered. This chapter describes the purification and characterization of the predominant two-component signal-transduction system utilized by Rhodobacter capsulatus, a nonsulfur purple photosynthetic bacterium. Specifically, we explain the overexpression, detergent solubilization, and purification of the full-length membrane-spanning histidine-sensor kinase RegB. We also provide a method to measure autophosphorylation of RegB and discern the effect of its signal molecule, ubiquinone, on autophosphorylation levels. In addition we describe the overexpression and purification of the cognate response regulator RegA and a technique used to visualize the phosphotransfer reaction from RegB to RegA.
Introduction
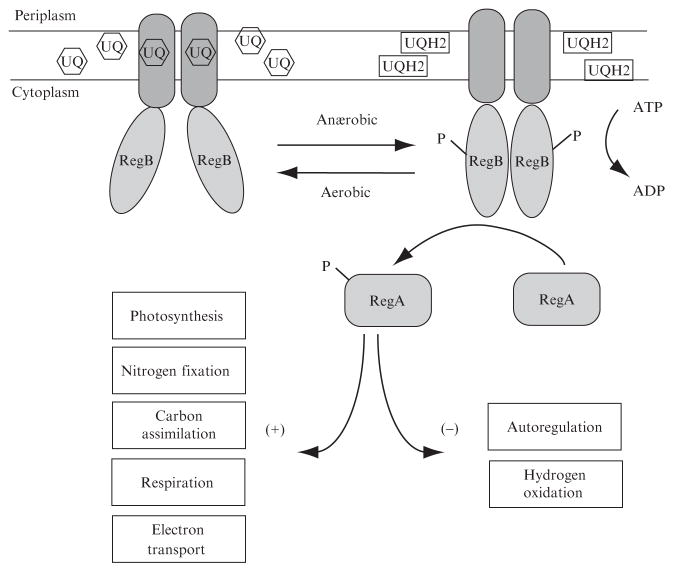
The RegB–RegA two-component signal-transduction system is highly conserved among 28 species of proteobacteria (Elsen et al., 2004). Extensive genetic analysis of RegB and RegA deletion mutants in Rhodobacter capsulatus has demonstrated that many energy-generating and energy-utilizing processes are controlled by the RegB–RegA system (Bird et al., 1999; Du et al., 1999; Dubbs and Tabita, 2003; Elsen et al., 2000, 2004; Swem and Bauer, 2002; Swem et al., 2001). Also, biochemical analysis has provided evidence that phosphorylated RegA binds DNA and directly controls the transcription of genes involved in these processes (Bird et al., 1999; Du et al., 1998). Operons within the RegB–RegA regulon include components required for photosynthesis, hydrogen utilization, cytochrome biogenesis, tetrapyrole biosynthesis, carbon fixation, nitrogen fixation, and dehydrogenase synthesis (Fig. 1) (Elsen et al., 2004). The common theme among all members of this regulon is that they either generate reducing power in the form of reduced ubiquinol or utilize reducing power either directly or indirectly from the ubiquinone pool (Fig. 1). Not coincidentally it was discovered that ubiquinone serves as the signal that regulates the RegB–RegA system (Swem et al., 2006).
Fig. 1.
Schematic of the RegB×RegA regulon. RegB is depicted as a membrane-bound histidine sensor kinase, and autophosphorylation is inhibited by ubiquinone (UQ) under aerobic conditions. Under anaerobic conditions the ubiquinone pool becomes reduced to the ubiquinol state (UQH2) and RegB is able to phosphorylate. The phosphate is transferred to RegA, which regulates multiple cellular processes.
The sensor kinase, RegB, contains six membrane-spanning helices at the amino terminus followed by a cytosolic domain containing a conserved H-box site of phosphorylation, a redox box containing a redox-active cyteine residue, followed by an adenine triphosphate (ATP) kinase domain at the carboxyl terminus (Potter et al., 2002; Swem et al., 2003). The primary amino acid sequence of the membrane-spanning domain of RegB is not highly conserved with the exception of the second periplasmic loop that contains a universally conserved GGXXNPF motif (Swem et al., 2006). Mutational and biochemical analyses have demonstrated that this region interacts with the ubiquinone pool and that the presence of oxidized ubiquinone inhibits the autophosphorylation activity of isolated full-length RegB in vitro (Swem et al., 2006). Interestingly, the reduced form of ubiquinone (ubiquinol) does not inhibit RegB autophosphorylation. The ubiquinone specificity of RegB allows this system to sense the cellular redox state of R. capsulatus and elicit physiological changes in response to changes in environmental oxygen tension. The autophosphorylation level of RegB directly controls the phosphorylation state of RegA, which affects its ability to bind DNA and regulate diverse oxygen-dependent cellular processes (Bird et al., 1999; Du et al., 1998).
This chapter describes the methods used to overexpress and isolate full-length RegB and its cognate response regulator, RegA. We also offer an assay to measure the signaling properties that ubiquinone exerts on RegB. Finally, we provide an assay for measuring RegB autophosphorylation and phosphate transfer to RegA.
Expression and Purification of RegB
Culture Media
Luria-Bertani (LB) broth: 10 g tryptone, 5 g yeast extract, and 10 g NaCl in 1 liter dH2O
Terrific broth: 12 g tryptone, 24 g yeast extract, and 4 ml glycerol in 900 ml dH2O. After autoclaving, add 100 ml of sterile 0.17 M KH2PO4 and 0.72 M K2HPO4 to bring the final volume to 1 liter.
Buffers
RegB lysis buffer: 10 mM Tris, pH 8, and 100 mM NaCl
Solubilization buffer: 20 mM Tris, pH 8.0, 20 mM imidazole, 300 mM NaCl, 20% glycerol, and 1% n-dodecyl-β-D-maltoside
Wash buffer: 20 mM Tris, pH 8.0, 20 mM imidazole, 150 mM NaCl, and 10% glycerol
Elution buffer: 20 mM Tris, pH 8.0, 200 mM imidazole, 150 mM NaCl, and 10% glycerol
Storage buffer: 10 mM Tris, pH 8.0, 150 mM NaCl, and 50% glycerol
4× SDS-PAGE loading buffer: 200 mM Tris, pH 6.8, 8% SDS, 0.4% bromophenol blue, 40% glycerol, and 4% β-mercaptoethanol
Western transfer buffer 1×: 25 mM Tris-base, 192 mM glycine, and 20% (v/v) methanol
Selection of RegB Overexpression Strains
Full-length regB is polymerase chain reaction (PCR) amplified from R. capsulatus genomic DNA using primers RegBFullNcoI (5′-TACCATGGTGAGGGCTGTCGACC) and RegBFullXhoI (5′-ATCTCGAGGGCGGTGATCGGAACATTC). The PCR product is cloned into the NcoI and XhoI sites of pET28 (Novagen), which places RegB in-frame with a carboxyl terminus 6 His tag. The resulting plasmid, pET28RegBfull, is then transformed into the T7-based isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible Escherichia coli expression strain, BL21(DE3). The transformants are selected for on LB agar plates containing 50 μg/ml kanamycin and grown at 37° overnight. Six to eight independent colonies are streaked for isolation on fresh LB-agar medium and grown overnight at 37°. Five-milliliter liquid Terrific broth cultures are started from each independent isolate and grown overnight with shaking (250 rpm) at 37°. The next day the cultures are diluted 1:50 in fresh Terrific broth and grown to an OD600 of 0.4 to 0.6, at which point IPTG is added to a final concentration of 500 μM. The cultures are induced for an additional 4 h of shaking (250 rpm) at 37°, and then 16 μl of each cell culture is mixed with 4 μl of 4× SDS-PAGE loading buffer, heated to 95° for 5 min, and separated on a 7.5% SDS-polyacrylamide gel. The SDS-polyacrylamide gel-separated protein is then transferred to a nitrocellulose membrane in 1× Western transfer buffer overnight at 30 V at 4°. The nitrocellulose membrane is probed by Western blot analysis with a horseradish peroxidase-conjugated His-tag antibody according to the manufacturer’s instructions (Santa Cruz Biotech). Overexpressed RegB protein is visualized using the WestDura Super Signal substrate (Pierce), and a 50-kDa protein band is identified that is absent from a control strain containing the pET28 vector without the RegB open reading frame. Isolates judged to contain maximal expression of RegB are grown overnight in LB broth supplemented with 50 μg/ml kanamycin in the absence of IPTG and stored at −80° in 20% glycerol.
Optimization of RegB Induction Conditions
Overproduction of large amounts of full-length RegB in E. coli is determined to be lethal, leading to a very low yield of recombinant protein. To overcome lethality, a minimal amount of IPTG is utilized to overexpress RegB from the pET28 expression system. Optimum expression is determined by performing an IPTG killing experiment involving the plating of BL21(DE3)/pET28RegBfull onto LB plates containing IPTG at concentrations ranging from 10 μM to 1 mM. All IPTG concentrations above 200 μM resulted in nearly 100% lethality as judged by low plating efficiency. There is no observable inhibition of growth at IPTG concentrations of 20 μM or below. However, LB plates containing 75 μM IPTG give rise to very thin colonies that are translucent in appearance, presumably because of overproduction of RegB. A concentration of 75 μM IPTG is thus used for overexpressing full-length RegB in liquid cultures.
Purification of Full-Length RegB
An overnight culture of BL21(DE3) containing plasmid pET28/RegBfull is grown for 16 h with shaking (250 rpm) at 37° in LB supplemented with 50 μg/ml kanamycin. The culture is diluted 1:50 into Terrific broth with 50 μg/ml kanamycin (1-liter batches in 2-liter flasks) and grown at 37° shaking (250 rpm) until an OD600 of 0.4–0.6 is reached, at which time IPTG is added to a final concentration of 75 μM. Protein expression is allowed to proceed for 4 h, shaking at 37° before the cells are harvested by centrifugation (7600g, 4°). The cell pellet is then stored at −80° until protein purification is initiated.
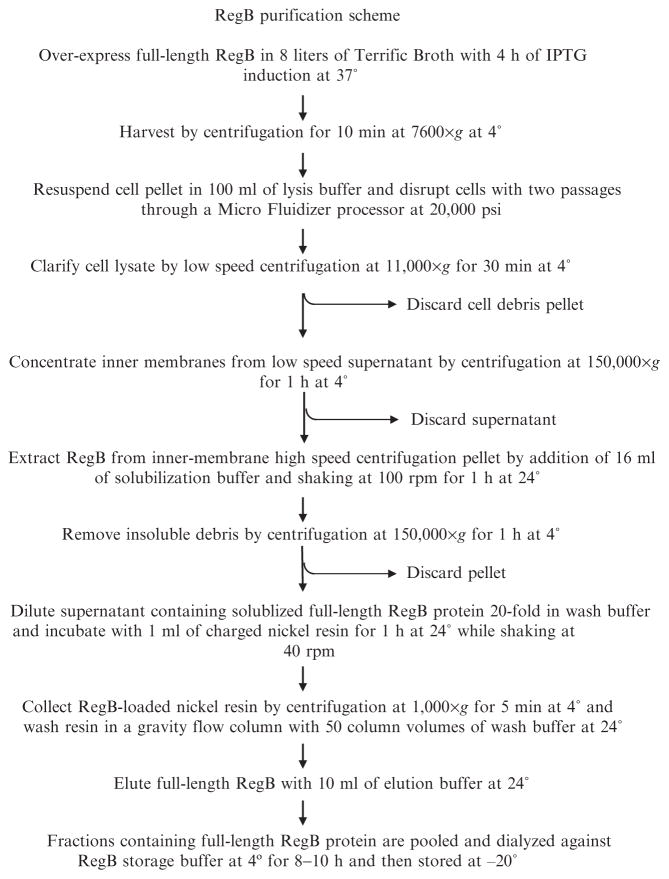
For purification (Fig. 2), cell pellets obtained from 8 liters RegB overexpressed culture are gently thawed on ice in the presence of 100 ml lysis buffer until a homogeneous solution is obtained. Cells are lysed by two passages through the M-110L Micro Fluidizer Processor at 20,000 psi (Microfluidics). The cell lysate is clarified by centrifugation at 11,000g for 30 min at 4° to remove the outer membranes and any nonruptured E. coli. The supernatant, which contains RegB-loaded inner membranes, are pelleted by centrifugation at 150,000g for 1 h at 4°. The inner membrane pellet is solubilized by the addition of 2 ml solubilization buffer per liter of lysed E. coli cells (typically 16 ml solubilization buffer). A Pyrex tissue grinder is used to homogenize the inner membranes in the presence of solubilization buffer. The solubilized membranes are shaken in a 50-ml conical tube attached to an orbital shaker, rotating at 100 rpm for 1 h at 24°. Any insoluble debris is then separated from the solubilized RegB protein by centrifugation at 150,000g for 1 h at 4°. The supernatant containing soluble full-length RegB protein is diluted 20-fold in wash buffer and allowed to incubate with 1 ml of settled charged nickel resin (Novagen) for 1 h at room temperature shaking at 40 rpm on an orbital shaker. The dilution of solubilized RegB with wash buffer is critical for the nickel affinity purification, as the high n-dodecyl-β-D-maltoside concentrations used for RegB solublization will inhibit His-tag/nickel affinity chromotagraphy. The RegB-loaded nickel resin is then pelleted by centrifugation at 1000g for 5 min at 4°. The supernatant is retained for SDS-PAGE analysis, and the resin is applied to a disposable gravity flow column and washed with 50 column volumes of wash buffer at 24°. Full-length RegB is eluted with 10 ml of elution buffer at 24°, collected in 1-ml fractions, and analyzed for protein content using a Bio-Rad protein assay (Bio-Rad). The RegB protein concentration in milligrams per milliliter is calculated using a bovine serum albumin standard curve generated in the presence of the Bio-Rad protein assay solution. The molecular mass of RegB (50 kDa) is then used to determine the molar concentration of protein. Fractions containing protein are separated on a 7.5% SDS-polyacrylamide gel to confirm the presence of RegB. Fractions containing RegB protein are pooled and dialyzed against storage buffer at 4° for 8–10 h and then stored at −20° until kinase assays are completed.
Fig. 2.
RegB purification flowchart. A simple flowchart illustrating inner membrane isolation and RegB solubilization and purification.
Expression and Purification of RegA
Buffers
Lysis/wash buffer: 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 0.1 mM EDTA, and 0.1% Triton X-100
Cleavage buffer: 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 0.1 mM EDTA, and 30 mM dithiothreitol
Elution buffer: 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 0.1 mM EDTA
Dialysis buffer: 50 mM Tris-HCl (pH 8.0), 200 mM KCl, 10 mM MgCl2, and 50% glycerol
Overexpression of RegA
The expression vector used to fuse RegA to the chitin-binding domain was constructed as described by Du et al. (1998). The RegA expression plasmid, pET29CBD::regA, is transformed into the T7-based IPTG-inducible expression strain, BL21(DE3). The RegA overexpression strain is grown for 14 to 16 h with shaking (250 rpm) at 37° in LB containing 50 μg/ml kanamycin. The culture is then diluted 1:50 in Terrific broth containing 50 μg/ml kanamycin (1-liter batches in 2-liter flasks). The cells are grown to an OD600 of 0.4 to 0.6, at which time RegA expression is induced by the addition of 500 μM IPTG. RegA expression is allowed to proceed for 4 h, shaking (250 rpm) at 37°, and is then harvested by centrifugation at 7600g at 4° for 10 min. The cell pellet is stored at −80° until purification is initiated.
Purification of RegA
The cell pellet obtained from 4 liters of overexpressed RegA culture is resuspended in 50 ml lysis buffer. Cells are lysed by two passages through the M-110L Micro Fluidizer Processor at 20,000 psi (Microfluidics), and the cell lysate is clarified by centrifugation at 11,000g for 30 min at 4°. A 10-ml chitin affinity column is prepared by applying 15 ml of chitin bead slurry (New England Biolabs) to a disposable column and allowed to pack by gravity flow. The column is equilibrated with 5 column volumes of lysis/wash buffer. The clarified RegA cell lysate is then applied to the column at a flow rate of 1 ml/min and subsequently washed with 10 column volumes of lysis/wash buffer. The final 1-ml volume of wash buffer is collected and analyzed for protein content using the Bio-Rad protein assay (Bio-Rad). If the wash buffer still contains protein contamination, the column is washed with an additional 10 column volumes or until no more contaminating protein is detected in the column flow through. The chitin column is then quickly purged with 3 column volumes of cleavage buffer and immediately capped off and stored for 10 h at 4°. The RegA protein is then eluted by flushing the column with 20 ml elution buffer and collected in 1-ml fractions. Each fraction is assayed for protein content using the Bio-Rad protein assay (Bio-Rad). Fractions containing protein (typically fractions 3–15) are separated by SDS-PAGE and stained with Coomassie blue to confirm the presence of a 20-kDa protein band corresponding to RegA. Fractions containing RegA protein are pooled and dialyzed overnight at 4° against dialysis buffer and stored at −20°. The RegA protein concentration in milligrams per milliliter is calculated using a bovine serum albumin standard curve generated in the presence of the Bio-Rad protein assay solution. The molecular mass of RegA (20 kDa) is then used to determine the molar concentration of protein.
RegB Kinase and Phosphotransfer Assays
Buffers
10× protein kinase buffer: 200 mM Tris, pH 8.0, 500 mM NaCl, 60 mM MgCl2, 1 mM CaCl2, and 1 M KCl
10× ATP cocktail: 10 mM unlabeled ATP and 0.5 μCi γ-32P-labeled ATP (ICN Biomedical, 5000 Ci/mmol)
RegB Kinase Assays
RegB is diluted to a final concentration of 5–10 μM in 1× protein kinase buffer and then incubated at 37° for 20 min. The kinase reaction is initiated by the addition of a 1/10 volume of a 10× ATP cocktail that contains trace amounts of γ-32P ATP. Aliquots are removed at times ranging between 0.5 and 16 min and quenched by the addition of 4× SDS-PAGE loading buffer. The samples are then separated by SDS-PAGE, analyzed by autoradiographic film, and quantified using a phosphor-imaging system (Typhoon 9200, Amersham Biosciences). It is extremely important to avoid boiling the quenched reaction prior to separation on SDS-PAGE, as the phosphorylated protein is heat labile.
Ubiquinone Inhibition of RegB
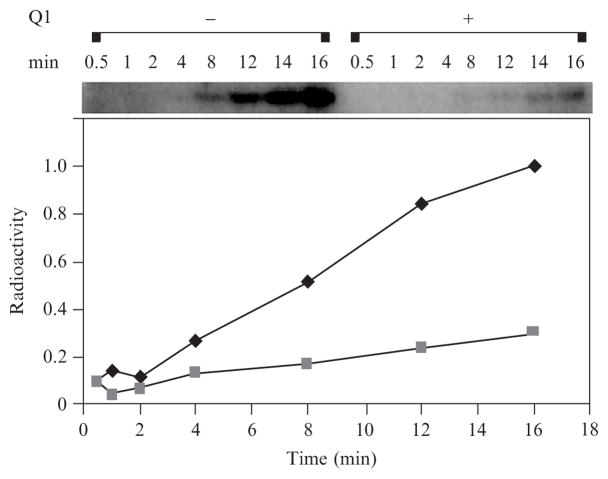
For ubiquinone inhibition experiments, benzoquinone (Sigma) or coenzyme Q1 (Sigma) is solubilized in 95 % ethanol at a concentration of 167 mM and then diluted in water to a final concentration of 16.7 mM. Either benzoquinone or coenzyme Q1 is then added to the RegB kinase reaction at a 20- to 200-fold molar excess over that of RegB. Maximal inhibition is observed at a concentration of 50:1 coenzyme Q1 to RegB. An appropriate concentration (<1%) of ethanol is added to a RegB kinase control reaction that does not contain ubiquinone. Both reactions are incubated at 37° for 20 min prior to the addition of 10× ATP cocktail to initiate the phosphorylation reaction (Fig. 3). The kinase reactions are completed as described earlier, and autophosphorylation inhibition is quantified using a phosphor-imaging system (Typhoon 9200, Amersham Biosciences).
Fig. 3.
Inhibitory effect of ubiquinone on RegB autophosphorylation. RegB autophosphorylation is assayed in the presence and absence of ubiquinone (Q1). The concentration of ubiquinone (Q1) used in the reaction is 200-fold molar excess to RegB. Reaction aliquots are removed at 0.5, 1, 2, 4, 8, 12, 14, and 16 min, quenched with 4× SDS-loading buffer, and separated by SDS-PAGE. RegB autophosphorylation is visualized and quantified using the Typhoon phosphor-imaging system (Amersham Biosciences). ■ represents the radioactivity of the reaction aliquots in the presence of ubiquinone. ◆ represents the radioactivity of the reaction aliquots in the absence of ubiquinone.
RegB Phosphorylation of RegA
To measure RegB phosphotransfer, a RegB kinase reaction is assembled as described earlier and the reaction is started by the addition of 10 × ATP cocktail. The RegB protein is allowed to autophosphorylate for 15 min, before the addition of a 1 M equivalent of RegA is added to the reaction. Aliquots are removed at 0.5, 1, 1.5, 2, 3, and 5 min following RegA addition and are quenched by the addition of 4× SDS-PAGE loading buffer. The reactions are placed on ice until being separated on a 10% SDS-polyacrylamide gel. The gel is then visualized using the Typhoon phosphor-imaging system (Amersham Biosciences).
Acknowledgments
This study is supported by National Institutes of Health Grant GM040941 awarded to C. E. Bauer.
References
- Bird TH, Du S, Bauer CE. Autophosphorylation, phosphotransfer, and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J Biol Chem. 1999;274:16343–16348. doi: 10.1074/jbc.274.23.16343. [DOI] [PubMed] [Google Scholar]
- Du S, Bird TH, Bauer CE. DNA binding characteristics of RegA: A constitutively active anaerobic activator of photosynthesis gene expression in Rhodobacter capsulatus. J Biol Chem. 1998;273:18509–18513. doi: 10.1074/jbc.273.29.18509. [DOI] [PubMed] [Google Scholar]
- Du S, Kouadio JL, Bauer CE. Regulated expression of a highly conserved regulatory gene cluster is necessary for controlling photosynthesis gene expression in response to anaerobiosis in Rhodobacter capsulatus. J Bacteriol. 1999;181:4334–4341. doi: 10.1128/jb.181.14.4334-4341.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbs JM, Tabita FR. Interactions of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides. J Biol Chem. 2003;278:16443–16450. doi: 10.1074/jbc.M211267200. [DOI] [PubMed] [Google Scholar]
- Elsen S, Dischert W, Colbeau A, Bauer CE. Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J Bacteriol. 2000;182:2831–2837. doi: 10.1128/jb.182.10.2831-2837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen S, Swem LR, Swem DL, Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev. 2004;68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CA, Ward A, Laguri C, Williamson MP, Henderson PJ, Phillips-Jones MK. Expression, purification and characterisation of full-length histidine protein kinase RegB from Rhodobacter sphaeroides. J Mol Biol. 2002;320:201–213. doi: 10.1016/S0022-2836(02)00424-2. [DOI] [PubMed] [Google Scholar]
- Swem DL, Bauer CE. Coordination of ubiquinol oxidase and cytochrome cbb (3) oxidase expression by multiple regulators in Rhodobacter capsulatus. J Bacteriol. 2002;184:2815–2820. doi: 10.1128/JB.184.10.2815-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Elsen S, Bird TH, Swem DL, Koch HG, Myllykallio H, Daldal F, Bauer CE. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J Mol Biol. 2001;309:121–138. doi: 10.1006/jmbi.2001.4652. [DOI] [PubMed] [Google Scholar]
- Swem LR, Gong X, Yu CA, Bauer CE. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J Biol Chem. 2006;281:6768–6775. doi: 10.1074/jbc.M509687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Kraft BJ, Swem DL, Setterdahl AT, Masuda S, Knaff DB, Zaleski JM, Bauer CE. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 2003;22:4699–4708. doi: 10.1093/emboj/cdg461. [DOI] [PMC free article] [PubMed] [Google Scholar]