Abstract
Haem d1 is a unique tetrapyrrole molecule that serves as a prosthetic group of cytochrome cd1, which reduces nitrite to nitric oxide during the process of denitrification. Very little information is available regarding the biosynthesis of haem d1. The extreme difficulty in studying the haem d1 biosynthetic pathway can be partly attributed to the lack of a theoretical basis for experimental investigation. We report here a gene cluster encoding enzymes involved in the biosynthesis of haem d1 in two heliobacterial species, Heliobacillus mobilis and Heliophilum fasciatum. The gene organization of the cluster is conserved between the two species, and contains a complete set of genes that lead to the biosynthesis of uroporphyrinogen III and genes thought to be involved in the late steps of haem d1 biosynthesis. Detailed bioinformatics analysis of some of the proteins encoded in the gene cluster revealed important clues to the precise biochemical roles of the proteins in the biosynthesis of haem d1, as well as the membrane transport and insertion of haem d1 into an apocytochrome during the maturation of cytochrome cd1.
INTRODUCTION
Tetrapyrrole derivatives such as haems, chlorophylls, cobalamin and sirohaem are essential components in many metabolic processes in living organisms. The early steps of biosynthesis of the tetrapyrroles are universally similar in that there are a number of common intermediates produced from 5-aminolevulinic acid to uroporphyrinogen III (e.g. Beale, 1995, 2000; Frankenberg et al., 2004). Uroporphyrinogen III serves as a key branching point to synthesize different end products such as haems, chlorophylls, sirohaem, cobalamin and haem d1. Biochemical details of the synthetic pathways for most of the tetrapyrroles except haem d1 have been elucidated, with haem d1 remaining as one of the most enigmatic tetrapyrroles in terms of biosynthesis.
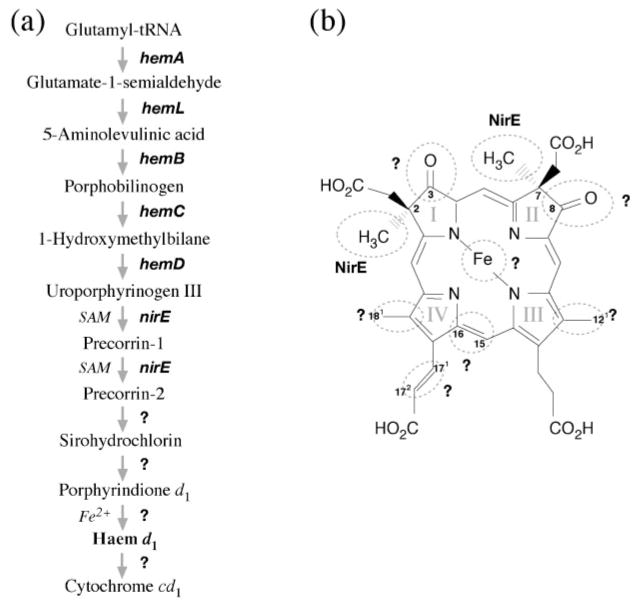
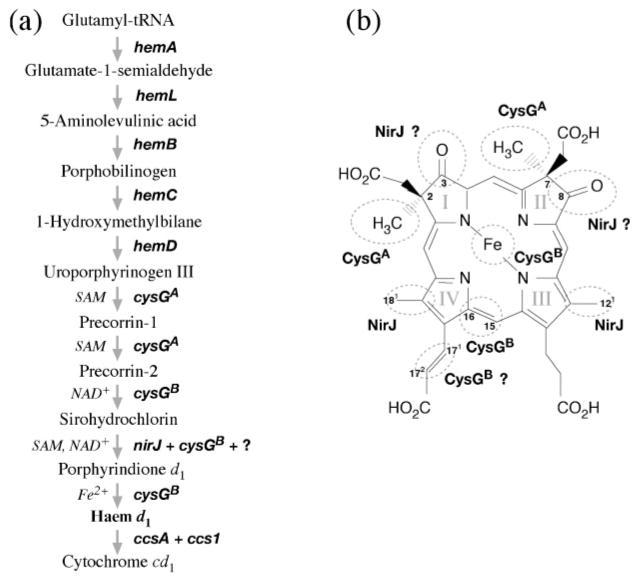
Haem d1 is related to the denitrification process that converts nitrate to gaseous nitrogen as part of the anaerobic respiration of bacteria and archaea. Among the denitrifying enzymes is nitrite reductase, which converts nitrite to nitric oxide as an intermediate step of denitrification. Two types of nitrite reductase are known, copper-containing nitrite reductase and cytochrome cd1. The latter contains a unique tetrapyrrole, haem d1, as one of the prosthetic groups (e.g. Timkovich, 2003). Little is known regarding the biosynthesis of haem d1 except that it may utilize uroporphyrinogen III, precorrins, sirohydrochlorin and porphyrindione d1 as intermediates (Yap-Bondoc et al., 1990; Youn et al., 2004; von Mering et al., 2005) (Fig. 1a).
Fig. 1.
(a) Outline of the putative biosynthetic pathway of haem d1 in bacteria in which 5-aminolevulinic acid is synthesized through the C-5 pathway. The C-4 pathway, through condensation of succinyl-CoA and glycine, found only in proteobacteria is omitted in this figure. The catalysis of the second half of haem d1 biosynthesis as well as incorporation of haem d1 into cytochrome cd1 are still largely unknown and thus labelled with question marks. (b) Structure of haem d1. Based on current knowledge, modifications of most of the moieties of uroporphyrinogen III to produce haem d1 are performed by unknown enzymes (circled and labelled with ?).
The unique features of the structure of haem d1 compared to its precursor uroporphyrinogen III include methyl groups at C2 and C7, methyl groups in place of the acetate groups at C12 and C18, oxo groups in place of propionate groups at C3 and C8, and an acrylate group oxidized from a propionate group at C171,2 (Fig. 1b). Therefore, to synthesize haem d1 from uroporphyrinogen III requires methylation at rings I and II, decarboxylation at rings III and IV, introduction of the oxo groups at rings I and II, and dehydrogenation of the propionate sidechain on ring IV. Although insertion of a ferrous iron to the centre of the porphyrin is itself not unique, it is considered the last step in the haem d1 synthesis (Youn et al., 2004). The fate of the synthesized haem d1 includes transport across the membrane so that it can be inserted into an apocytochrome, along with haem c, to complete the maturation of cytochrome cd1. Enzymes responsible for each of these modification steps as well as subsequent haem transport and cytochrome maturation, however, remain largely unknown.
Insertional mutagenesis analysis of Pseudomonas stutzeri has identified a nir locus that is necessary for haem d1 biosynthesis (de Boer et al., 1994; Palmedo et al., 1995; Glockner & Zumft, 1996; Kawasaki et al., 1997). In this locus, there are two nir operons, one containing nirJ, nirE and nirN genes, and the other nirC, nirF, nirD, nirL, nirG and nirH genes. NirN is homologous to NirS, the known structural polypeptide of cytochrome cd1, and shares regional homology with NirC and NirF (Timkovich, 2003). The nirD, nirL, nirG and nirH genes are all strongly similar to each other at the sequence level and are proposed to have arisen from gene duplication events, although they do not have clearly defined functions. NirJ is a member of the radical S-adenosylmethionine (SAM) protein family, and does not have a clearly defined function in haem d1 biosynthesis. NirE is a SAM-dependent uroporphyrinogen methylase homologous to sirohaem synthase CysGA. This is the only enzyme that has clearly been suggested to catalyse the sequential methylation at C2 and C7 of the porphyrin to produce precorrin-1 and precorrin-2 during haem d1 biosynthesis (Kawasaki et al., 1997). Except for NirE, the precise roles of other Nir proteins in the haem d1 biosynthetic pathway remain undefined.
The photosynthetic bacteria heliobacteria (Heliobacteriaceae) were first discovered in the early 1980s (Gest & Favinger, 1983; Gest, 1994) and have now been expanded to about a dozen strains encompassing five different genera (NCBI Taxonomy Database; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Taxonomy). Heliobacteria, which belong phylogenetically to the low-GC Gram-positive group, are a unique group of photosynthetic bacteria in that they contain a bacteriochlorophyll g pigment and a simplified type I photosynthetic reaction centre (Madigan & Ormerod, 1995). They are also known to be able to fix nitrogen and perform ammonia assimilation (Kimble & Madigan, 1992). No other aspects of nitrogen metabolism are known for heliobacteria nor is there any indication that they may catalyse haem d1 biosynthesis.
We report here the discovery of a gene cluster related to haem d1 biosynthesis in two heliobacterial species, Heliobacillus mobilis and Heliophilum fasciatum. Subsequent bioinformatics analysis of the genes encoding the haem d1 biosynthesis enzymes yielded a significant insight into the biochemical pathway for the synthesis of this unique tetrapyrrole molecule.
METHODS
Bacterial culture and DNA isolation
Hb. mobilis was grown in a PYE liquid medium (Beer-Romero & Gest, 1987) at 25 °C under anaerobic conditions with tungsten-light illumination. The anaerobic conditions were created using an anaerobic chamber (Coy Laboratory). The bacterial cells were harvested after culturing for 2 days by centrifugation (5000 g). Genomic DNA was isolated according to Pospiech & Neumann (1995). Hp. fasciatum was purchased from ATCC, but was found to be non-viable. The lyophilized bacterial stock was used directly for DNA isolation and subsequent downstream analysis.
General DNA manipulation
The analysis with Hb. mobilis began by first identifying an evolutionarily conserved segment of the hemB gene sequence among a group of Gram-positive bacteria through database searching using BLAST (Altschul et al., 1997) and sequence alignment using CLUSTAL (Thompson et al., 1994) and T-Coffee (Notredame et al., 2000). The conserved region allowed the design of a pair of degenerate PCR primers with the aid of Oligo software (National Biosciences). The forward primer (TCKGCYTTYTAY-GGACCHTTYC) and reverse primer (AYTCACCGSASACATTATA) used in degenerate PCR were synthesized by Integrated DNA Technologies. The analysis with Hp. fasciatum, which began after the entire Hb. mobilis sequence was obtained, was facilitated by the availability of the Hb. mobilis sequence information. It began by obtaining partial sequences from hemB, hemA2, hemD, hemL and hep2 using degenerate PCR (for hemA2, forward primer TCMAC-RTGCAAYCGDACGGA and reverse primer CACCTGYCCRAGAA-TTTGBGT; for hemL, forward primer TGGGGYCCICTKATYYTRGG and reverse primer GGTYAGIGCKCCIGAACC; for hep2, forward primer GGAAAAMGWYTVMGICCGGC and reverse primer ARWA-RRRRGCKGTYTTICG; and for hemD, forward primer AARGGMGG-VGAYCCCTTYGT and reverse primer TSCCBGGHATCACYTCRGC).
The PCR products were cloned into the pUC19 vector with the PCR-Script Cloning kit (Stratagene). Small-scale plasmid DNA preparations were made by using the Qiaprep Spin Miniprep kit (Qiagen). DNA sequencing of the clones was performed with the universal primers for the pUC19 plasmid (forward primer CGCCAGGGTTT-TCCCAGTCACGAC and reverse primer TCACACAGGAAACAG-CTATGAC). Nucleotide sequences were determined by the dideoxy chain-termination method (Sanger et al., 1977) using the BigDye Sequencing kit v3.1 (Applied Biosystems).
Once the partial hemB gene of Hb. mobilis was sequenced, the upstream and downstream flanking DNA was obtained by using the inverse PCR technique (Ochman et al., 1988) repeatedly. For Hp. fasciatum, the partial gene fragments resulting from degenerate PCR were first joined using regular PCR and subsequently sequenced. Further upstream and downstream sequences were obtained using a novel genome-walking technique developed by Guo & Xiong (2006). The novel technique was necessary in this case because inverse PCR required a substantial quantity of genomic DNA that was not available for Hp. fasciatum. The novel method had the advantage of consuming only minute amounts of starting DNA.
Sequence analysis
Sequencing was performed on both strands of DNA for cross-verification. The final sequence contigs were assembled by matching and removing overlapping regions of individual fragments and joining the remainder of the fragments. ORFs of the final sequences were determined using multiple hidden Markov model (HMM)-based gene-prediction programs: GeneMark.hmm (Lukashin & Borodovsky, 1998), GeneMark frame-by-frame (Shmatkov et al., 1999), AMIgene (Bocs et al., 2003) and FrameD (Schiex et al., 2003). The predictions were made with the HMMs of each program trained for a closely related low-GC Gram-positive bacterium such as Bacillus subtilis. To confirm the gene prediction, the putative ORFs were checked for the presence of RBSs immediately upstream of the start codons. Only the predicted frames that were preceded by the canonical RBS were accepted.
Once the genes and gene boundaries were determined, sets of genes that might be transcriptionally linked to form operons were predicted using the rule developed by Wang et al. (2004). The method, which has been shown to be 91 % accurate, required three pieces of information: gene orientation, intergenic distance and gene linkage conservation. To obtain the gene linkage information in other genomes, cross-genome comparison was performed with the aid of the STRING server (http://string.embl.de/), which compiled gene neighbourhood information of 179 completely sequenced genomes (von Mering et al., 2005). To determine whether a pair of adjacent genes belonged to a common operon, a scoring scheme was used with the operon assignment threshold set at 2.
Gene functional annotation was based on a combined approach: (1) direct BLAST searches against the non-redundant GenBank database for translated proteins (Altschul et al., 1997); (2) searches against the protein classification database Protonet (Sasson et al., 2003), which annotates protein functions using a hierarchical tree-based approach with the aid of gene ontology, and provides information on the biological process, molecular function and cellular localization of each protein (e.g. Azuaje et al., 2006; Thomas et al., 2007); and (3) structural and functional feature prediction using Phylofacts (Krishnamurthy et al., 2006) and Phobius (Kall et al., 2004).
The statistical significance of pairwise sequence similarities was evaluated using the probability of random shuffles (PRSS) test (Pearson & Lipman, 1988), which calculates the probability of similarities of randomly shuffled and unshuffled sequences using a distance matrix Monte Carlo procedure. The test was performed with 1000 global shuffles with the gap-opening penalty set at 12 and the gap-extending penalty at 2 by using the BLOSUM50 scoring matrix.
Phylogenetic analysis
Phylogenetic analysis was carried out for several of the proteins encoded in the gene cluster. The sequence homologues of the heliobacterial proteins were retrieved from searching sequence databases using BLAST (Altschul et al., 1997) with an E value cutoff of 10−20. After removing redundant and nearly redundant homologues, the sequences were aligned using a profile-based approach (Simossis et al., 2005), followed by manual refinement. The final sequence alignments were used to construct phylogenetic trees based on maximum-likelihood with the aid of the PHYML program (Guindon & Gascuel, 2003) under the Whelan and Goldman (WAG) substitution model (Whelan and Goldman, 2001) with four substitution rate categories. Nonparametric bootstrapping was subsequently performed with 100 replicates of the datasets.
Molecular modelling
3D protein structures of a number of proteins encoded by the gene cluster were constructed based on the principle of homology modelling. The homology models could be built because of the extremely conserved nature of protein structures given the small number of protein folds available (<800) against the huge number of protein sequences in nature (>1 × 106 individual sequences). The practical boundaries of sequence identity for proteins adopting the same structures were defined by Rost (1999) as a function of sequence length in pairwise alignment, e.g. a sequence identity of 20 % for an alignment of 150 aa can fall within the ‘safe’ zone for protein homology modelling. Below the safe zone is the ‘twilight’ zone, where identical structure can still be found (sometimes as low as 12–15 %), although statistical tests such as the PRSS test have to be used to differentiate random matching from truly related sequences. The sequence alignments used in this study were well within the range suitable for homology model building.
The structural templates for the modelling were chosen from the Protein Data Bank (PDB) using an HMM-based approach, HHPred (Soding et al., 2005). The resulting statistically most significant alignment was used as a basis for manual refinement. The refined alignment was used as input for the modelling software Modeller (Sali et al., 1995), which was able to model both conserved regions and loops to generate a raw model that was subsequently refined with built-in energy-minimization features. The quality of the protein model was evaluated using Verify3D (Eisenberg et al., 1997). The protein cofactors were subsequently modelled by transferring the coordinates directly from the template to the protein model. For NirL, quaternary modelling involving a complex structure of a NirL dimer and dsDNA was also performed. The NirL dimer was modelled by superimposing two monomers upon an Lrp dimer unit from the octameric structure generated by Ren et al. (2007). The dimer was then manually docked onto a 22 bp DNA structure (PDB code 1CGP) in the Quanta (Accelrys) molecular-modelling environment. The final modelling result was rendered using Pymol (DeLano Scientific LLC).
NirL expression and purification
To test the hypothesis that NirL is a transcription factor, the protein was purified to homogeneity and its DNA-binding activity characterized. Briefly, the nirL gene was amplified using PCR with the primers CGCATATGTGGACTGAAAAAGACAAAGAG and CGGAATTCCGCTTCTTTTTCCATGAAG. The PCR product was subsequently cloned into an expression construct pTYB1 (New England Biolabs) between the NdeI and EcoRI restriction sites. The cloned gene was resequenced to verify the absence of mutations and was subsequently used for heterologous expression in Escherichia coli ER2566.
NirL was expressed as a C-terminal fusion protein to an intein (an inducible protein self-splicing element) and a chitin-binding domain. The strain with the NirL expression construct (pTYB1:: nirL) was grown at 37 °C in Terrific Broth (TB) medium containing ampicillin (100 μg ml−1) to OD600 0.6, when IPTG was added to a final concentration of 0.5 mM. The cells were incubated at room temperature (22 °C) overnight before being harvested.
The cells were harvested by centrifugation at 5000 g for 10 min at 4 °C. The cell pellet was resuspended in 5 ml cell lysis buffer (20 mM Tris/HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA, 0.1 % Triton X-100, 20 μM PMSF) and lysed by agitation in fine glass beads (0.1 mm diameter) using a mini-BeadBeater (Glen Mills). The lysed cell suspension was centrifuged at 1500 g for 10 min to remove the cell debris and glass beads. The cell lysate was centrifuged at 20 000 g for 30 min at 4 °C. The supernatant was subsequently loaded onto a chitin column equilibrated with column buffer (20 mM Tris/HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA). The column was washed with 5 vols column buffer followed by 1 vol. cleavage buffer (20 mM Tris/HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA, 20 μM PMSF, 50 mM DTT). The on-column protein cleavage was performed by incubating the fusion protein in the cleavage buffer at room temperature in an anaerobic chamber (Coy Laboratory) overnight (18 h). The column was then eluted with 2 vols of elution buffer (50 mM Tris, pH 8.0, 150 mM KCl, 5 mM DTT, 5 %, v/v, glycerol). The eluate was collected and concentrated using a Centricon-10 concentrator (Millipore). Protein samples were taken and analysed by SDS-PAGE on 12.5 % gels that were subsequently stained with Coomassie brilliant blue (R-250) dye.
DNA mobility shift assay
The DNA fragment used for the mobility shift assay was a PCR-amplifed 200 bp region immediately upstream of nirJ2 in Hp. fasciatum, and contains the putative promoter for the nir operon. The PCR product was purified using the Qiaquick Gel Extraction kit (Qiagen). For the DNA-binding assay, 50 ng DNA was added to the binding buffer (10 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT, 2.5 %, v/v, glycerol, 5 mM MgCl2, 0.05 % Nonidet P-40) in a final volume of 20 μl, either with or without 10 μg purified NirL protein. The reaction was carried out at room temperature for 30 min. The reaction mixture was subjected to electrophoresis in 5 % polyacrylamide gels in native TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.3) at 100 V for 1 h. Following electrophoresis, the gel was stained with 50 ml 0.001 % SYBR-Gold (Invitrogen) for 30 min, and visualized using the EpiChemi3 Imaging System (UVP).
RESULTS AND DISCUSSION
Overall organization of the gene cluster and gene annotation
In this study, a gene cluster related to haem biosynthesis was obtained from two different heliobacterial species. Both were isolated from rice fields, contained bacteriochlorophyll g, produced endospores, and were capable of nixtrogen fixation and photoheterotrophic growth (Beer-Romero & Gest, 1987; Ormerod et al., 1996). Their phylo-genetic distance was relatively divergent within the family Heliobacteriaceae (Ormerod et al., 1996).
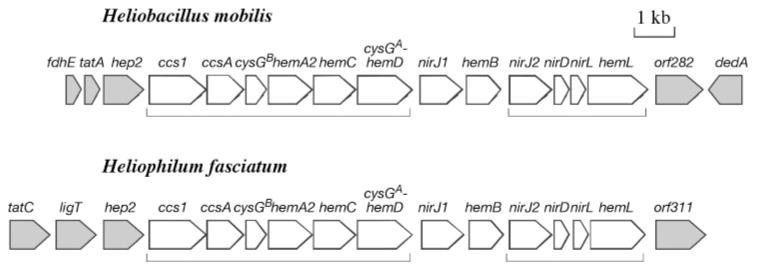
The sequencing of the gene cluster was initiated by obtaining a number of conserved gene fragments through degenerate PCR. The flanking sequences of the segments were subsequently obtained by using two different genome-walking techniques: inverse PCR (Ochman et al., 1988) and a method newly developed by Guo & Xiong (2006). The final nucleotide sequence length for the Hb. mobilis gene cluster was 16 361 bp, and for Hp. fasciatum, 17 398 bp. The locations and boundaries of the ORFs were determined based on a combination of de novo gene prediction programs and the presence of RBS in the immediate vicinity of the predicted ORFs to minimize errors. We found 17 protein-encoding genes, including partial ones at both ends, in the Hb. mobilis sequence and 16 genes in the Hp. fasciatum sequence (Fig. 2).
Fig. 2.
Physical maps of the DNA fragments sequenced from Hb. mobilis and Hp. fasciatum. The arrowed boxes indicate predicted ORFs and direction of transcription. White boxes, genes related to haem biosynthesis and transport; grey boxes, genes presumably irrelevant to haem biosynthesis and transport. Groups of genes predicted to be operons are indicated with brackets.
The functional annotation of the gene products (Table 1) was derived from the combined information of sequence similarity matches using BLAST, a tree-based protein classification with the aid of gene ontology (Sasson et al., 2003) and protein structural feature prediction. In the centre of the sequence is a cluster of 12 genes with an identical gene organization in both heliobacterial species. These genes share a common functional theme, which is haem biosynthesis and transport. The cluster begins with the ccs1 and ccsA genes, which are ATP-binding cassette (ABC)-type transmembrane proteins whose homologues are involved in haem transport across the membrane for cytochrome c biosynthesis. Downstream of the ccs genes are a number of hem genes, namely hemA, hemL, hemB, hemC and hemD, which encode enzymes for each step of biosynthesis leading to uroporphyrinogen III. In addition, there are nirJ1, nirJ2, nirD and nirL, which are annotated as haem d1 biosynthesis proteins. In addition, hemD is found to be fused with cysGA upstream; the latter, together with cysGB, is known to be involved in the biosynthesis of sirohaem, the structure of which is closely related to that of haem d1. This 12-gene cluster is loosely termed ‘haem biosynthesis gene cluster’ in this communication.
Table 1. Location and functional annotation of the ORFs identified in this study.
Major motifs and transmembrane domains of translated proteins were predicted using a number of protein domain search methods (see Methods for details). Genes of particular interest to haem d1 biosynthesis and transport as well as important motifs of the gene products are highlighted in bold type.
| Position (Hb. mobilis) | Position (Hp. fasciatum) | Gene | Best database match | Major motifs/features | Brief functional annotation |
|---|---|---|---|---|---|
| 425–1195 | tatC | Protein translocase | Six transmembrane helices | Sec-independent protein secretion pathway | |
| 1322–1942 | ligT | 2′-5′ RNA ligase | RNA metabolism | ||
| 1–574 | fdhE (partial) | Formate dehydrogenase formation protein | C3HC4-type zinc finger (RING finger) | Lipid transport | |
| 595–828 | tatA | Protein translocase | One transmembrane helix | Sec-independent protein secretion pathway | |
| 926–1888 | 2097–3059 | hep2 | Heptaprenyl diphosphate synthase component II | Polyprenyl synthetase domain | Isoprenoid biosynthesis |
| 1967–3358 | 3133–4488 | ccs1 | Cytochrome c biogenesis protein | Four transmembrane helices | Haem transport |
| 3381–4229 | 4475–5320 | ccsA | Cytochrome c biogenesis protein | Eight transmembrane helices | ABC-type transport system involved in cytochrome c biogenesis |
| 4274–4900 | 5336–5968 | cysGB | Sirohaem synthase | Dehydrogenase domain, NAD-binding motif, ferrochelatase domain | Haem biosynthesis |
| 4882–6189 | 5944–7239 | hemA2 | Glutamyl-tRNA reductase | Haem biosynthesis | |
| 6202–7146 | 7265–8212 | hemC | Porphobilinogen deaminase | Dipyromethane cofactor- binding domain | Haem biosynthesis |
| 7143–8690 | 8212–9750 | cysGA– hemD | Uroporphyrinogen III methyltransferase–synthase | Methylase domain, SAM- binding site | Haem biosynthesis |
| 8835–10 010 | 9928–11 106 | nirJ1 | Haem d1 biosynthesis protein | SAM-binding site, Fe–S- binding sites | High potential Fe–S protein |
| 10 065–11 054 | 11 332–12 321 | hemB | Delta-aminolevulinic acid dehydratase (porphobilinogen synthase) | Haem biosynthesis | |
| 11 374–12 369 | 12 424–13 416 | nirJ2 | Haem d1 biosynthesis protein | SAM-binding site, Fe–S- binding sites | High potential Fe–S protein |
| 12 373–12 858 | 13 421–13 918 | nirD | Haem d1 biosynthesis protein | HTH motif | Transcription regulation |
| 12 884–13 357 | 14 060–14 539 | nirL | Haem d1 biosynthesis protein | HTH motif | Transcription regulation |
| 13 369–14 676 | 14 552–15 826 | hemL | Glutamate-1-semialdehyde 2,1- aminotransferase | Haem biosynthesis | |
| 14 871–15 719 | orf282 | Unknown integral transmembrane protein | Ten transmembrane helices, two internal repeats, ion channel signature | Permease/transporter | |
| 15 721–16 361 | dedA | Uncharacterized membrane protein | Two transmembrane helices | ||
| 16 014–16 949 | orf311 | Hypothetical protein |
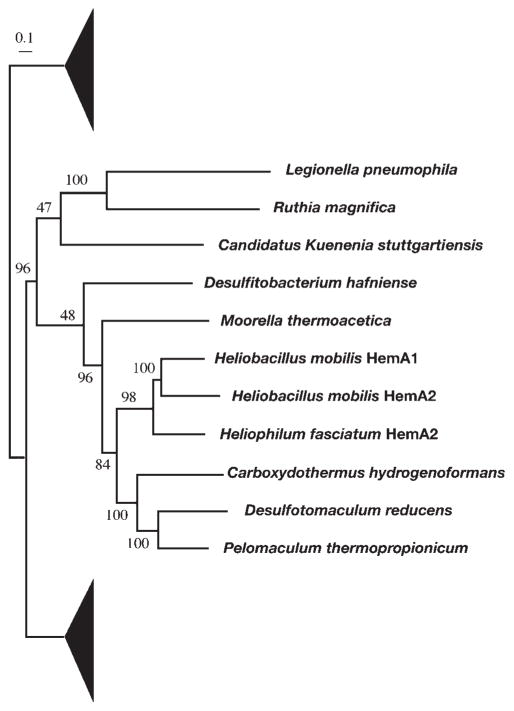
Present among the hem genes is a hemA homologue, termed hemA2, because it is the second hemA gene discovered in heliobacteria after the first one found in the photosynthesis gene cluster (Xiong et al., 1998). The translated products of the two genes share 54 % sequence identity, indicating that they are the result of gene duplication. Our phylogenetic analysis further indicated that the duplication event was very recent and may have occurred only after the speciation of the individual heliobacterial strains (Fig. 3). Since HemA is widely distributed in the bacterial domain, only a portion of the tree surrounding the positions of the heliobacterial taxa is shown.
Fig. 3.
Maximum-likelihood tree of the HemA family, showing a recent duplication event that gave rise to HemA1 and HemA2 in heliobacteria. Due to the large size of this sequence family, only a portion of the tree is shown, with filled triangles representing omitted taxa. The numbers on the branches indicate bootstrap values. The scale bar corresponds to 0.1 amino acid substitutions per site.
The genes outside this haem biosynthesis cluster are, however, not conserved in linkage. They include genes involved in the sec-independent protein secretion pathway (tatA and tatC) and in RNA metabolism (ligT), in addition to a number of ORFs with unknown functions.
As part of the sequence annotation, we performed operon prediction with the newly predicted genes using the Wang et al. (2004) method, which determines operons by the combined information of inter-gene distances and gene linkage conservation among genomes, and has been shown to be highly accurate (~91 % accuracy). Two operons are predicted in the given sequences (Fig. 2), with ccs1, ccsA, cysGB, hemA2, hemC and cysGA–hemD constituting the first operon, and nirJ2, nirD, nirL and hemL forming the second operon. The operon structure for the two heliobacterial strains is well conserved. The first transcriptional unit appears to be mainly involved in the early stage of tetrapyrrole biosynthesis and haem transport, with the exception of cysGA and cysGB. The second operon may be more specific for haem d1 biosynthesis, with the exception of hemL. In between the two operons are nirJ1 and hemB, which appear to be monocistronic.
Of particular interest is the presence of cysGB and cysGA in the first operon along with most of the hem and ccs genes, and of hemL in the second operon along with the nir genes. The hemL gene (encoding glutamate semialdehyde aminotransferase) is involved in the early steps of uroporphyrinogen III biosynthesis, whereas cysGA and cysGB, as illustrated below, may be involved in the late steps of haem d1 biosynthesis. The mixed arrangement of these genes in two different operons appears to indicate that the two stages of the haem d1 biosynthesis pathway as well as the final assembly of cytochrome cd1 are tightly co-regulated at the functional level.
The linkage of the hem genes responsible for the biosynthesis of uroporphyrinogen III appears to be consistent among Gram-positive bacteria such as B. subtilis, Staphylococcus aureus and Paenibacillus macerans (Hansson et al., 1991; Kafala & Sasarman, 1997; Johansson & Hederstedt, 1999). The reported linkage patterns are in some ways similar to that in heliobacteria. It remains to be investigated whether the consistent clustering indicates possible physical interactions at the protein level or simply an evolutionary pressure for coexpression of the functionally related genes.
The discovery of the haem d1 biosynthesis genes was in fact a matter of serendipity as a result of genome walking. The analysis of the haem d1 biosynthesis genes turned out to be most interesting in filling the knowledge gaps for the enzymic involvement in the haem d1 biosynthesis pathway. The following sections concentrate on the proteins encoded by the cluster that are specifically related to haem d1 biosynthesis and its transport for cytochrome maturation.
CysGA
The database search analysis for the translated ORF downstream of hemC revealed a fusion gene of cysGA and hemD (Fig. 2) (BLAST E value 0). The CysGA domain of the fusion product is on the N terminus (amino acids 1–251). Its homologues in other species have been annotated as sirohaem synthase, which is a SAM-dependent uroporphyrinogen III methylase catalysing the first two steps of sirohaem synthesis, namely methylation at rings I and II of uroporphyrinogen III to produce precorrin-1 and pre-corrin-2. The HemD domain on the C terminus (amino acids 252–512) is a uroporphyringen III synthase known to catalyse the cyclization of the linear tetrapyrrole 1-hydroxymethylbilane to produce the macrocyclic uroporphyrinogen III. The fusion of CysGA and HemD appears to be rather common in Gram-positive bacteria, as observed in Bacillus, Paenibacillus and Clostridium species (Johansson & Hederstedt, 1999; Fujino et al., 1995). The genetic fusion apparently generates an efficient mechanism to produce precorrin-2 from 1-hydroxy-methylbilane, with three consecutive steps of catalysis being carried out by the same polypeptide.
Sirohaem is a similar compound to haem d1. It has been suggested that the initial methylation steps leading to the synthesis of precorrin-2 should be shared between sirohaem biosynthesis and haem d1 biosynthesis (Zumft, 1997). In Pseudomonas, NirE has been shown by genetic analysis to be necessary to catalyse the conversion of uroporphyrinogen III to precorrin-2 (de Boer et al., 1994; Kawasaki et al., 1997) during haem d1 biosynthesis. NirE in fact shares 60 % sequence identity with CysGA from E. coli (Warren et al., 1994), which confirms that NirE can essentially be treated as CysGA and that the latter can be directly involved in these reactions. In addition, it has been shown that there is an absolute requirement for SAM in the initial steps of haem d1 biosynthesis (Yap-Bondoc et al., 1990).
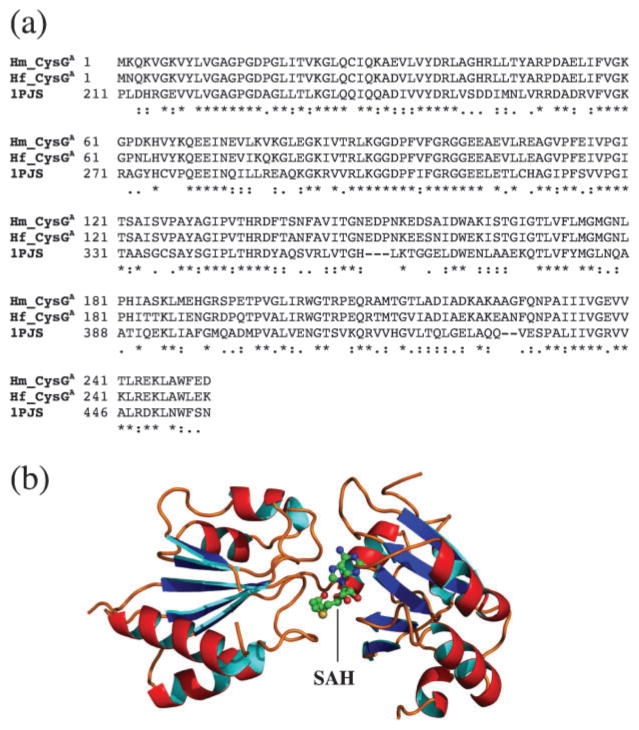
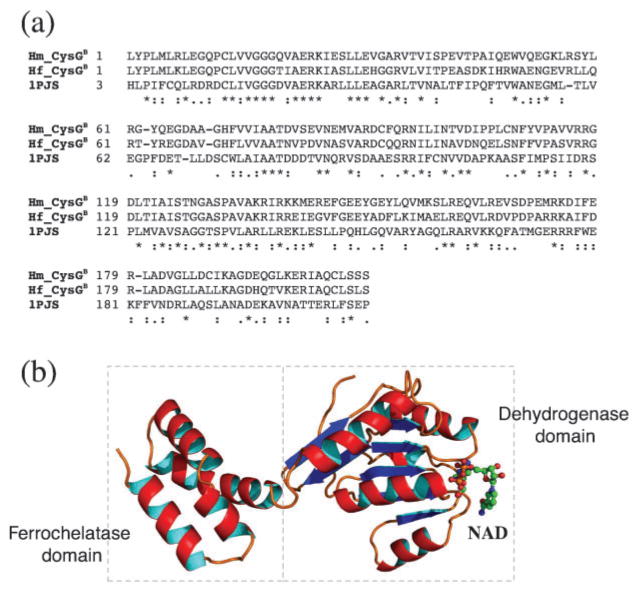
To provide a structural basis for CysGA, we applied a comparative modelling approach. The CysGA template used for the modelling was obtained by searching PDB using an HMM-based approach to produce a high-quality alignment with a significantly related homologous sequence in the database (Soding et al., 2005). The search identified the CysGA domain of CysG from Salmonella enterica as the closest homologue (1PJS) (Stroupe et al., 2003). The full-length match between the CysGA domain of Hb. mobilis and that of S. enterica was 49 % in sequence identity (Fig. 4a). A homology model was subsequently built based on a refined alignment with a bound cofactor S-adenosyl homocysteine (SAH) (Fig. 4b), which is demethylated SAM. The model was evaluated using a statistical profile-based approach (Eisenberg et al., 1997) and was shown to be of high quality (results not shown). There are two structural domains in the modelled structure, domain I (Fig. 4b, left) and domain II (Fig. 4b, right), both consisting of a β-sheet surrounded by α-helices. The two domains are arranged in a V shape with the SAH/SAM cofactor bound to domain II near the centre. CysGA is thought to be able to transfer a methyl group from SAM to C2 or C7 of the macrocyclic ring via a stereochemical inversion of the reactive carbon on the porphyrin substrate (Stroupe et al., 2003). Since the closely related Salmonella methylase carries out the catalysis with a homodimeric quaternary structure, it is reasonable to postulate that heliobacterial CysGA achieves the same functionality through a similar architecture.
Fig. 4.
(a) Sequence alignment of the CysGA domain of the CysGA–HemD fusion protein from Hb. mobilis and Hp. fasciatum (Hm_CysGA and Hf_CysGA, respectively) with the CysGA domain of the CysG protein of S. enterica for which a crystal structure is available (1PJS). Identical sequence matches in the alignment are indicated by ‘*’, strongly similar matches by ‘:’, and weakly similar matches by ‘.’. (b) 3D model of the CysGA domain for Hb. mobilis based on the above alignment. The position of the bound cofactor SAH (demethylated SAM) is also shown.
CysGB
The ORF immediately preceding the hemA2 gene was identified as cysGB (Fig. 2) through database similarity searches (BLAST E value 10−49). The CysGB homologues in other species are involved in sirohaem biosynthesis by serving two functions, precorrin dehydrogenation and iron metallation (Stroupe et al., 2003). In sirohaem biosynthesis, CysGB catalyses dehydrogenation at C15 and C16 of the macrocyclic ring to generate sirohydrochlorin with the aid of NAD+, and catalyses the insertion of a ferrous iron into sirohydrochlorin to make sirohaem. In many Gram-negative bacteria that synthesize sirohaem, CysGB is found to be fused with CysGA to form a multi-domain, multifunctional CysG protein (Warren et al., 1994; Stroupe et al., 2003). In E. coli, CysGB has been shown to regulate CysGA activity by preventing it from overmethylation of the porphyrin ring (Woodcock et al., 1998). Since CysGB in heliobacteria exists as a separate protein, while its orthologues exist as a fusion protein with CysGA, it may be reasonable to predict that CysGB in heliobacteria functions through physical interaction with CysGA–HemD on the basis of the ‘Rosetta stone’ principle (Marcotte et al., 1999; Enright et al., 1999) for protein–protein interaction prediction (with ~70 % accuracy).
The structural analysis of CysGB was similarly carried out using homology modelling with the CysGB domain of the same CysG protein from S. enterica serving as template (1PJS) (Stroupe et al., 2003). The full-length alignment between Hb. mobilis CysGB and the CysGB domain of the template protein was 31 % by identity (Fig. 5a). A homology model was subsequently built based on the refined alignment with the bound cofactor NAD (Fig. 4b). From the structural model, it is clear that the dual function of CysGB is realized by two distinct structural domains in the protein, the dehydrogenase domain (Fig. 5b, right) on the N terminus (residues 1–146), in which the cofactor NAD is bound, and the ferrochelatase domain (Fig. 5b, left) on the C terminus (residues 147–208) (the metal ions are not bound to the protein but presumably exist in the aqueous environment).
Fig. 5.
(a) Sequence alignment of CysGB from the two heliobacterial species with the CysGB domain of the CysG protein of S. enterica for which a crystal structure is available (1PJS). (b) 3D model of CysGB for Hb. mobilis based on the above alignment. The bifunctional enzyme has two distinct structural domains, the dehydrogenase domain on the N terminus and the ferrochelatase domain on the C terminus. The bound cofactor NAD for the dehydrogenase domain is also shown.
The transformation of precorrin-2 into sirohydrochlorin has been suggested to be one of the intermediate steps in haem d1 biosynthesis (Zumft, 1997). Based on the knowledge of the function of CysGB in sirohaem biosynthesis, we propose that heliobacterial CysGB is involved in sirohydrochlorin formation during haem d1 biosynthesis. The same protein may also be responsible for the last step of iron insertion into porphyrindione d1 to produce haem d1. Since CysGB in Salmonella functions as a homodimer, it is reasonable to assume that a similar architecture exists in heliobacterial CysGB as well.
NirJ
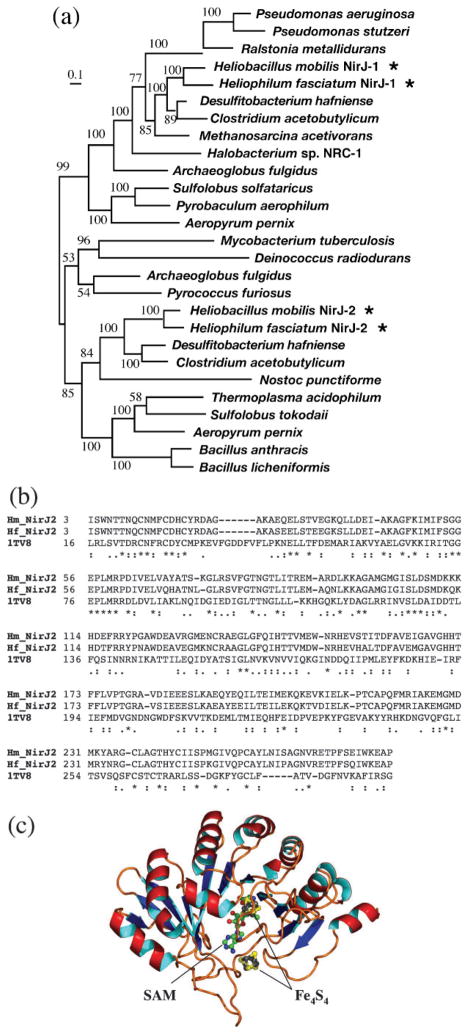
The ORFs immediately upstream and downstream of the hemB frame were both identified as nirJ, encoding a haem d1 biosynthesis protein (Fig. 2). They were differentiated as nirJ1 and nirJ2 (BLAST E values 8×10−147 for NirJ1 and 7×10−125 for NirJ2). The two gene products were 32 % identical to one other at the amino acid level and were apparently the result of gene duplication. Our phylogenetic analysis of the NirJ family indicated that the duplication event was quite ancient, with the two versions of NirJ branching before the separation of bacteria and archaea (Fig. 6a).
Fig. 6.
(a) Maximum-likelihood tree of the NirJ family, showing ancient divergence of NirJ1 from NirJ2 (indicated by asterisks). The numbers on the branches indicate bootstrap values. The scale bar corresponds to 0.1 amino acid substitutions per site. (b) Sequence alignment of NirJ2 from the two heliobacterial species with MoaA of Staph. aureus for which a crystal structure is available (1TV8). (c) 3D model of NirJ2 for Hb. mobilis based on the above alignment. The positions of the bound cofactors SAM and iron–sulfur centres (Fe4S4) are also shown.
The motif analysis of NirJ1 and NirJ2 showed that they contain a SAM-binding site and two Fe4S4-binding sites, with a consensus CX2–3CX2–5C motif. The sequence similarity search using the BLAST program returned a number of significant hits that belonged to the radical SAM protein family with diverse functions. A more sensitive HMM-based search against the structure database revealed a significant remote homologue in the form of MoaA from Staph. aureus (1TV8) (Fig. 6b, pairwise full-length identity 16 %, P<0.01 from the PRSS test, which is a randomization and realignment test for sequence homology) (Pearson & Lipman, 1988). MoaA catalyses the formation of a precursor for a molybdenum cofactor (Hänzelmann & Schindelin, 2004). The reaction involves structural rearrangement of GTP into 6-alkyl pterin with a cyclic phosphate. The reaction, however, has little resemblance to any of the reactions required for haem d1 biosynthesis. Other members of the same ‘radical SAM family’ with Fe4S4 centres catalyse a diverse range of reactions, including biotin synthase (BioB), which converts dethiobiotin to biotin, lysine aminomutase (LAM), which catalyses the interconversion of L-α-lysine and L-β-lysine, and coproporphyrinogen III oxidase (HemN), which converts coproporphyrinogen III to protoporphyrinogen IX. None of these catalytic reactions, except that of HemN, is obviously related to haem d1 biosynthesis. HemN is the only member of the radical SAM protein family involved in tetrapyrrole biosynthesis, and catalyses two successive oxidative decarboxylation reactions on the propionate sidechains of coproporphyrinogen III with the aid of two SAM cofactors and one Fe4S4 centre (Layer et al., 2003).
In our HMM-based database search using NirJ2 of Hb. mobilis as query, HemN from E. coli indeed turned out to be one of the top hits in the search result. More detailed pairwise comparison between the two proteins showed a regional alignment covering 51 % of the total length with identity 12 %, similarity 57 % and P<0.05 from the PRSS test. Thus, this supports a remote homology between NirJ and HemN, which allowed us to propose that NirJ functions similarly to HemN. Since HemN catalyses decarboxylation of the propionate groups, this may be considered similar to the decarboxylation of the acetate groups that is required for haem d1 biosynthesis.
The homology model of NirJ2 was constructed based on the alignment with MoaA from Staph. aureus with a bound SAM and two Fe4S4 centres (Fig. 6c). The overall protein model resembles a triosphosphate isomerase (TIM) barrel with an eight-stranded β-sheet wrapped around by eight α-helices. One of the bound Fe4S4 centres is thought to be able to transfer an electron to the SAM molecule and induce its cleavage, producing methionine and a 5′-deoxyadenosyl radical. The highly oxidizing radical then abstracts a hydrogen from a carbon atom on the substrate to induce a glycyl radical that catalyses a subsequent bond cleavage reaction on the substrate (Hänzelmann & Schindelin, 2004). This mode of reaction is considered common among SAM radical enzymes with Fe4S4 centres, and may provide a mechanistic clue to the presumed bond breakage reaction of NirJ.
NirD and NirL
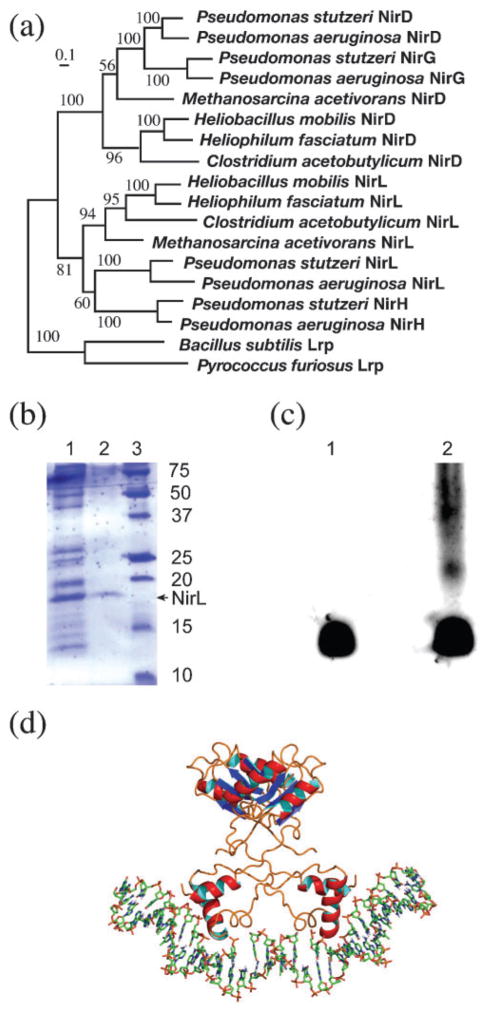
The heliobacterial nir operon contains two other nir genes, nirD and nirL (Fig. 2). These two gene products share 24 % identity with each other at the translated amino acid level. They can be considered to be the result of gene duplication from a common ancestor. As shown in Fig. 7(a), the gene duplication appears to be very ancient, and may have occurred before the separation of bacteria and archaea. In fact, in the Pseudomonas lineage, an additional gene-duplication event appears to have occurred with this pair of gene homologues, giving rise to the four similar genes nirD, nirL, nirG and nirH. Deletion of any of these genes is able to abolish the production of haem d1 in Pseudomonas (Palmedo et al., 1995; Kawasaki et al., 1997).
Fig. 7.
(a) Maximum-likelihood tree of the NirD/L and Lrp family. With the Lrp sequences forming a natural outgroup, the ancient gene duplication event leading to the separation of NirD and NirL is evident. Further gene duplication from the ancestor of either NirD or NirL gave rise to NirG and NirH in Pseudomonas. The number on each branch represents a bootstrap value. The scale bar corresponds to 0.1 amino acid substitutions per site. (b) The result of expression and purification of NirL from Hp. fasciatum using an intein-mediated approach. The protein samples were fractionated in a 12.5 % SDS-polyacrylamide gel stained with Coomassie brilliant blue R-250. Lane 1, clarified cell lysate applied to the chitin-containing affinity column; lane 2, protein sample eluted from the column after in situ protein splicing, showing NirL (17 kDa) being purified to near homogeity; lane 3, protein molecular mass markers with numbers on the right indicating protein size in kDa. (c) The result of DNA mobility shift assay for NirL in a 5 % native polyacrylamide gel stained with SYBR-Gold. Lane 1, nirJ2 promoter DNA only; lane 2, nirJ2 promoter DNA incubated with NirL. The DNA band shift is clearly visible in lane 2, indicating the formation of the DNA–protein complex. (d) Model of NirL binding to DNA. The homology model of NirL was constructed based on an alignment (not shown) with the most closely related Lrp transcription factor from Neisseria meningitidis (Koike et al., 2004; PDB code 1RI7). NirL was modelled in the dimer form based on a dimer unit of the same octameric structure with a double-stranded DNA ligand modelled based on the suggestions of Koike et al. (2004) and Ren et al. (2007). The DNA coordinates were extracted from the structure of Schultz et al. (1991) (PDB code 1CGP).
Both NirD and NirL from heliobacteria can be annotated as transcription factors that are members of the Lrp family of transcription regulators on the basis of the BLAST search results (E values 10−41 for NirD and 4×10−45 for NirL). The Lrp (leucine-responsive regulatory) transcription factors regulate many specific metabolic functions, such as amino acid biosynthesis and pilus synthesis (Brinkman et al., 2003). The best-studied Lrp proteins have been shown to control gene expression through two distinct structural domains, the DNA-binding and regulatory domains. The DNA-binding domain on the N terminus binds to promoter DNA with a helix–turn–helix (HTH) fold to induce DNA conformational changes for transcription activation or inhibition. The regulatory domain on the C terminus, upon binding to a ligand, facilitates protein–protein interactions by forming a homodimer that in turn becomes a building block for a higher order of structure such as an octameric disc (Thaw et al., 2006).
An HTH motif was identified at the N terminus (residues 3–49) of heliobacterial NirD and NirL, and was strongly similar to the one in most Lrp proteins, supporting their putative role as transcription regulators. No enzymic functions were identified through the bioinformatics analysis. Furthermore, a palindromic sequence TTT(N)AT(N5–7)-AT(N)AAA was found in the upstream region (−47.5 ± 8.5 bp from gene start sites) of both nirJ1 and nirJ2, and matched well with the known DNA-binding motif, which is an AT-rich inverted repeat, of many Lrp proteins (Koike et al., 2004). Thus, we suggest that NirD/L serve as transcription factors that regulate the expression of nirJ1 and the nir operon, including the hemL gene. Therefore, they can be considered to be indirectly involved in the biosynthesis of haem d1.
To verify that NirD/L are indeed DNA-binding proteins, we cloned and expressed the nirL gene from Hp. fasciatum and purified the NirL protein using an intein-mediated approach (Fig. 7b). Its DNA-binding characteristics were determined using a gel mobility shift assay with a DNA probe that included 200 bp upstream from the nirJ2 gene, encompassing the putative promoter for the nir operon. DNA band shifts were clearly observed with the addition of partially purified NirL (Fig. 7c). This result thus supports the above proposal that NirL, and likely NirD as well, plays a role in regulation of expression of the nir operon.
We further constructed a 3D model of NirL based on the strong full-length sequence similarity to a closely related Lrp transcription factor from Pyrococcus sp. (Koike et al., 2004; PDB code 1RI7). The pairwise alignment had an identity level of 23 %. Based on the knowledge that all known Lrp transcription factors form an octamer consisting of four dimer units, a dimer of NirL (Fig. 7d) was modelled along with its DNA ligand according to Koike et al. (2004), showing the N-terminal HTH motif of NirL interacting closely with the major groove of the DNA.
It needs to be pointed out that this proposal is novel and contradictory to the current belief that the NirD/L proteins are directly involved in haem d1 synthesis (Zumft, 1997; Timkovich, 2003). Youn et al. (2004) overexpressed a Pseudomonas nirFDLGH operon and obtained an unusual tetrapyrrole termed ‘compound 800’ that had some features related to haem d1. It is not clear whether the result was due to the expression of the five gene products encoded in the operon or upregulation/down-regulation of other nir genes in Pseudomonas as an indirect result of overexpression of the transcription regulators.
Ccs proteins
Also of interest are the two genes at the beginning of the haem biosynthesis gene cluster. They encode two transmembrane proteins related to cytochrome c biosynthesis. Sequence database searching identified them as Ccs1 and CcsA, responsible for the transmembrane delivery of haem c during the biogenesis of cytochrome c holoproteins (Nakamoto et al., 2000) (BLAST E values 7×10−55 for Ccs1 and 4×10−75 for CcsA). This function could be significant, because cytochrome cd1 is known to carry out its catalysis in the periplasmic space (for Gram-positive bacteria, it is the space between the plasma membrane and the cell wall) (Suharti & de Vries, 2005). The transport of the newly synthesized haem d1 across the membrane is thus a necessary step for the final assembly and maturation of cytochrome cd1 (Zumft, 1997). The very existence of the ccs genes in an operon related to haem d1 biosynthesis gives important hints that they may be involved in the transport of haem d1 in addition to haem c for the generation of cytochrome cd1 in the mature form in the periplasm.
CcsA and Ccs1 of cyanobacteria and algal chloroplasts have been shown to function as a closely associated complex in delivering haem to an apocytochrome, with CcsA binding to haem through its tryptophan-rich domain, and Ccs1 interacting with the apocytochrome and anchoring it for haem insertion (Hamel et al., 2003). The tryptophan-rich domain for haem binding has indeed been identified in heliobacterial CcsA. In addition to transport, the CcsA–Ccs1 complex in cyanobacteria and chloroplasts is also able to perform haem ligation to covalently attach a haem to a c-type apocytochrome (Hamel et al., 2003). The latter function, if conserved in heliobacteria, should be confined to the incorporation of haem c into cytochrome cd1, since haem d1 is non-covalently bound to the cytochrome protein.
Working hypothesis on haem d1 biosynthesis
To summarize the above sequence and structural analysis, we propose a working hypothesis for the enzymes involved in the haem d1 biosynthesis pathway. The strong sequence similarity of heliobacterial CysGA to well-characterized SAM-dependent uroporphyrinogen III methyltransferases gives credence to the idea that the CysGA domain of the CysGA–HemD fusion protein is able to methylate uroporphyrinogen III at C2 and C7 via two consecutive steps to produce precorrin-2. CysGB, which contains a dehydrogenase domain, is proposed to catalyse the oxidation of the single bond between C15 and C16 to produce a double bond, leading to the formation of sirohydrochlorin. NirJ, belonging to the same protein family as HemN, which modifies tetrapyrrole sidechains through decarboxylation, is proposed to decarboxylate the acetate sidechains at C12 and C18 to produce methylated groups at rings III and IV. The final step of haem d1 synthesis, iron insertion of porphyrindione d1, is proposed to be carried out by the ferrochelatase domain of CysGB. The newly synthesized haem d1 may be transported across the membrane and subsequently inserted into an apocytochrome via the combined effects of CcsA and Ccs1 during the biogenesis of cytochrome cd1 (Fig. 8a, b).
Fig. 8.
(a) Working hypothesis of haem d1 biosynthesis as well as incorporation of haem d1 into an apocytochrome to produce cytochrome cd1, based on the bioinformatics analysis result. The CysGA domain of the CysGA–HemD fusion protein is proposed to methylate uroporphyrinogen III at C2 and C7 in two consecutive steps to produce precorrin-2. NirJ is proposed to catalyse the decarboxylation of the acetate sidechains on rings III and IV. CysGB, which is a bifunctional dehydrogenase and ferrochelatase, is proposed to catalyse the oxidation of the single bond between C15 and C16 to produce a double bond and the insertion of a ferrous iron in porphyrindione d1 to complete the haem d1 synthesis. The transport of synthesized haem d1 and its insertion into an apocytochrome are thought to be mediated by CcsA and Ccs1. (b) Structure of haem d1 labelled with enzymes proposed to be involved in converting some of the circled moieties. The acrylate formation in ring IV may be catalysed by CysGB, whereas the conversion of the propionate groups to oxo groups in rings I and II may be catalysed by NirJ, both of which are predicted with a lesser degree of confidence at this stage (labelled with ?).
There are two additional reactions in haem d1 synthesis, acrylate formation at C171,2 and the conversion of propionates to oxo groups at C3 and C8, which have not yet been clearly defined in the above proposal. This is because additional haem d1 biosynthesis enzymes may be involved for these two sets of reactions, since not all nir gene homologues in Pseudomonas have been identified in these two heliobacterial species. On the other hand, if no additional genes for haem d1 biosynthesis are found, the existing enzymes encoded in the cluster could catalyse all of these reactions. For instance, the SAM-binding NirJ1/NirJ2 proteins may be involved in the oxidative replacement of the propionates on rings I and II. It has been proposed that the introduction of the oxo groups may involve enzymes with radical species such as SAM through the removal of the propionates by hydroxylation, followed by a reverse aldol condensation (Frankenberg et al., 2004). The reaction bears a slight resemblance to the oxidative decarboxylation carried out by HemN. Since the two different versions of NirJ may have originated before the separation of bacteria and archaea, and have since evolved independently, it is possible that there is a functional separation in which one of the NirJ proteins is responsible for the oxo group formation while the other is specific to the decarboxylation reaction.
The formation of the acrylate group on ring IV is possibly catalysed by CysGB, since the dehydrogenation reaction is similar to that at neighbouring C15 and C16, resulting in a conjugated double bond with the macrocyclic ring. However, it remains to be seen whether the minimalist point of view can be sustained until the full genome data become available, though in Gram-positive bacteria, and especially heliobacteria, a complete set of genes for a biosynthetic pathway tend to be arranged in one operon or superoperon, as is the case for the photosynthesis gene cluster (Xiong et al., 1998). This form of arrangement may ensure a tight gene regulation that is important for anaerobic metabolism. The working hypothesis for the haem d1 biosynthesis pathway offers many tantalizing clues to be tested by experimental investigation.
Acknowledgments
We thank Lauren Gray for participating in the early stage of data collection. J. X. thanks the Welch Foundation (grant no. A1589) and C. E. B. thanks the National Institutes of Health (grant 53940) for support.
Abbreviations
- ABC
ATP-binding cassette
- HMM
hidden Markov model
- HTH
helix–turn–helix
- Lrp
leucine-responsive regulatory
- PDB
Protein Data Bank
- PRSS
probability of random shuffles
- SAH
S-adenosyl homocysteine
- SAM
S-adenosylmethionine
Footnotes
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuaje F, Al-Shahrour F, Dopazo J. Ontology-driven approaches to analyzing data in functional genomics. Methods Mol Biol. 2006;316:67–86. doi: 10.1385/1-59259-964-8:67. [DOI] [PubMed] [Google Scholar]
- Beale SI. Biosynthesis and structures of porphyrins and hemes. In: Blankenship RE, Madigan MT, Bauer CE, editors. Anoxygenic Photosynthetic Bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 153–177. [Google Scholar]
- Beale SI. Tetrapyrrole biosynthesis in bacteria. In: Lederberg J, editor. Encyclopedia of Microbiology. 2. Vol. 4. San Diego, CA: Academic Press; 2000. pp. 558–570. [Google Scholar]
- Beer-Romero P, Gest H. Heliobacillus mobilis, a peritrichously flagellated anoxyphototroph containing bacteriochlorophyll g. FEMS Microbiol Lett. 1987;41:109–114. [Google Scholar]
- Bocs S, Cruveiller S, Vallenet D, Nuel G, Médigue C. AMIGENE: annotation of microbial genes. Nucleic Acids Res. 2003;31:3723–3726. doi: 10.1093/nar/gkg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Mol Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- de Boer AP, Reijnders WN, Kuenen JG, Stouthamer AH, van Spanning RJ. Isolation, sequencing and mutational analysis of a gene cluster involved in nitrite reduction in Paracoccus denitrificans. Antonie Van Leeuwenhoek. 1994;66:111–127. doi: 10.1007/BF00871635. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Frankenberg N, Schobert M, Moser J, Raux E, Graham R, Warren MJ, Jahn D. The biosynthesis of hemes, siroheme, vitamin B12 and linear tetrapyrroles in Pseudomonas. In: Ramos J-L, editor. Pseudomonas. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 111–146. [Google Scholar]
- Fujino E, Fujino T, Karita S, Sakka K, Ohmiya K. Cloning and sequencing of some genes responsible for porphyrin biosynthesis from the anaerobic bacterium Clostridium josui. J Bacteriol. 1995;177:5169–5175. doi: 10.1128/jb.177.17.5169-5175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gest H. Discovery of heliobacteria. Photosynth Res. 1994;41:17–21. doi: 10.1007/BF02184140. [DOI] [PubMed] [Google Scholar]
- Gest H, Favinger JL. Heliobacterium chlorum, an anoxygenic brownish-green photosynthetic bacterium containing a ’new’ form of bacteriochlorophyll. Arch Microbiol. 1983;136:11–16. [Google Scholar]
- Glockner AB, Zumft WG. Sequence analysis of an internal 9.72-kb segment from the 30-kb denitrification gene cluster of Pseudomonas stutzeri. Biochim Biophys Acta. 1996;1277:6–12. doi: 10.1016/s0005-2728(96)00108-9. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guo H, Xiong J. A specific and versatile genome walking technique. Gene. 2006;381:18–23. doi: 10.1016/j.gene.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Hamel PP, Dreyfuss BW, Xie Z, Gabilly ST, Merchant S. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J Biol Chem. 2003;278:2593–2603. doi: 10.1074/jbc.M208651200. [DOI] [PubMed] [Google Scholar]
- Hansson M, Rutberg L, Schröder I, Hederstedt L. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J Bacteriol. 1991;173:2590–2599. doi: 10.1128/jb.173.8.2590-2599.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci U S A. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Hederstedt L. Organization of genes for tetrapyrrole biosynthesis in Gram-positive bacteria. Microbiology. 1999;145:529–538. doi: 10.1099/13500872-145-3-529. [DOI] [PubMed] [Google Scholar]
- Kafala B, Sasarman A. Isolation of the Staphylococcus aureus hemCDBL gene cluster coding for early steps in heme biosynthesis. Gene. 1997;199:231–239. doi: 10.1016/s0378-1119(97)00372-7. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Arai H, Kodama T, Igarashi Y. Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa: sequencing and identification of a locus for heme d1 biosynthesis. J Bacteriol. 1997;179:235–242. doi: 10.1128/jb.179.1.235-242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble LK, Madigan MT. Nitrogen fixation and nitrogen metabolism in heliobacteria. Arch Microbiol. 1992;158:155–161. [Google Scholar]
- Koike H, Ishijima SA, Clowney L, Suzuki M. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc Natl Acad Sci U S A. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy N, Brown DP, Kirshner D, Sjolander K. PhyloFacts: an online structural phylogenomic encyclopedia for protein functional and structural classification. Genome Biol. 2006;7:R83. doi: 10.1186/gb-2006-7-9-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 2003;22:6214–6224. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Ormerod JG. Taxonomy, physiology and ecology of heliobacteria. In: Blankenship RE, Madigan MT, Bauer CE, editors. Anoxygenic Photosynthetic Bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 17–30. [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Nakamoto SS, Hamel P, Merchant S. Assembly of chloroplast cytochromes b and c. Biochimie. 2000;82:603–614. doi: 10.1016/s0300-9084(00)00605-2. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod JG, Kimble LK, Nesbakken T, Torgersen YA, Woese CR, Madigan MT. Heliophilum fasciatum gen. nov. sp. nov. and Heliobacterium gestii sp. nov.: endospore-forming heliobacteria from rice field soils. Arch Microbiol. 1996;165:226–234. doi: 10.1007/s002030050320. [DOI] [PubMed] [Google Scholar]
- Palmedo G, Seither P, Korner H, Matthews JC, Burkhalter RS, Timkovich R, Zumft WG. Resolution of the nirD locus for heme d1 synthesis of cytochrome cd1 (respiratory nitrite reductase) from Pseudomonas stutzeri. Eur J Biochem. 1995;232:737–746. [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- Ren J, Sainsbury S, Combs SE, Capper RG, Jordan PW, Berrow NS, Stammers DK, Saunders NJ, Owens RJ. The structure and transcriptional analysis of a global regulator from Neisseria meningitidis. J Biol Chem. 2007;282:14655–14664. doi: 10.1074/jbc.M701082200. [DOI] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modelling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasson O, Vaaknin A, Fleischer H, Portugaly E, Bilu Y, Linial N, Linial M. ProtoNet: hierarchical classification of the protein space. Nucleic Acids Res. 2003;31:348–352. doi: 10.1093/nar/gkg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiex T, Gouzy J, Moisan A, de Oliveira Y. FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res. 2003;31:3738–3741. doi: 10.1093/nar/gkg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP–DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Shmatkov AM, Melikyan AM, Chernousko FL, Borodovsky M. Finding prokaryotic genes by the “frame-by-frame” algorithm: targeting gene starts and overlapping genes. Bioinformatics. 1999;15:874–886. doi: 10.1093/bioinformatics/15.11.874. [DOI] [PubMed] [Google Scholar]
- Simossis VA, Kleinjung J, Heringa J. Homology-extended sequence alignment. Nucleic Acids Res. 2005;33:816–824. doi: 10.1093/nar/gki233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe ME, Leech HK, Daniels DS, Warren MJ, Getzoff ED. CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat Struct Biol. 2003;10:1064–1073. doi: 10.1038/nsb1007. [DOI] [PubMed] [Google Scholar]
- Suharti &, de Vries S. Membrane-bound denitrification in the Gram-positive bacterium Bacillus azotoformans. Biochem Soc Trans. 2005;33:130–133. doi: 10.1042/BST0330130. [DOI] [PubMed] [Google Scholar]
- Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Mi H, Lewis S. Ontology annotation: mapping genomic regions to biological function. Curr Opin Chem Biol. 2007;11:4–11. doi: 10.1016/j.cbpa.2006.11.039. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timkovich R. The family of d-type hemes: tetrapyrroles with unusual substitutents. In: Kadish KM, Smith KM, Guilard R, editors. The Porphyrin Handbook. San Diego, CA: Academic Press; 2003. pp. 123–156. [Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Trawick JD, Yamamoto R, Zamudio C. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 2004;32:3689–3702. doi: 10.1093/nar/gkh694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren MJ, Bolt EL, Roessner CA, Scott AI, Spencer JB, Woodcock SC. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem J. 1994;302:837–844. doi: 10.1042/bj3020837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Woodcock SC, Raux E, Levillayer F, Thermes C, Rambach A, Warren MJ. Effect of mutations in the transmethylase and dehydrogenase/chelatase domains of sirohaem synthase (CysG) on sirohaem and cobalamin biosynthesis. Biochem J. 1998;330:121–129. doi: 10.1042/bj3300121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Inoue K, Bauer CE. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci U S A. 1998;95:14851–14856. doi: 10.1073/pnas.95.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap-Bondoc F, Bondoc LL, Timovich R, Baker DC, Hebbler A. C-methylation occurs during the biosynthesis of heme d1. J Biol Chem. 1990;265:13498–13500. [PubMed] [Google Scholar]
- Youn HS, Liang Q, Cha JK, Cai M, Timkovich R. Compound 800, a natural product isolated from genetically engineered Pseudomonas: proposed structure, reactivity, and putative relation to heme d1. Biochemistry. 2004;43:10730–10738. doi: 10.1021/bi0491954. [DOI] [PubMed] [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]