Abstract
Genes coding for putative CrtJ and AerR homologs were identified and characterized in the purple photosynthetic bacterium Rhodospirillum centenum (also known as Rhodocista centenaria), an organism that synthesizes photopigments even under highly aerated conditions. Mutational analysis indicated that in Rsp. centenum, gene crtJ codes for a repressor for photosynthesis gene expression as in Rhodobacter capsulatus, which exhibits a high level of oxygen repression of photosystem synthesis. In contrast to Rba. capsulatus, AerR in Rsp. centenum appears to be an aerobic activator; an aerR mutation resulted in significantly reduced levels of photopigment synthesis. Both aerR and crtJ mutants retained essentially normal levels of photosystem synthesis under anaerobic conditions, indicating that their activities are specific for aerobic photosystem synthesis. The readthrough transcript from crtE promoter, which is regulated by AerR and CrtJ, seems to be significant in maintaining the expression levels of the light harvesting I (puf) genes in Rsp. centenum. We suggest that AerR and CrtJ regulate aerobic photosystem synthesis primarily through controlling activity of the transcriptional readthrough.
Introduction
Photosystem synthesis in non-sulfur purple bacteria is regulated by oxygen tension and light intensity. Many studies on the regulatory system have been conducted using Rhodobacter capsulatus and Rhodobacter sphaeroides, in which photosystem synthesis is almost completely repressed under aerobic growth conditions.1,2 However, it is now known that there is a wide range of responses to oxygen among different species of purple bacteria. In contrast to Rhodobacter species, in Rhodospirillum centenum and Rhodovulum sulfidophilum photosystem synthesis is repressed only slightly when these organisms are grown aerobically.3,4 Previous studies with Rdv. sulfidophilum indicated that the mRNA of some photosynthesis genes are kept at high levels even under aerobic conditions, indicating that oxygen may differentially regulate the transcription of photosynthesis genes depending on the species of purple bacteria.5,6 However, the mechanism(s) of how the oxygen-insensitive photosystem synthesis is achieved is not fully understood.
The key regulators of photosystem synthesis under aerobic conditions are CrtJ and AerR in Rba. capsulatus and Rba. sphaeroides (called PpsR and PpaA, respectively, in Rba. sphaeroides).7–10 Biochemical and molecular genetic analyses have indicated that CrtJ binds under aerobic conditions to the conserved palindromes (TGT-N12-ACA) which are positioned in the promoters of bacteriochlorophyll (bch), carotenoid (crt), and light harvesting II (puc) genes.8,11–13 AerR also binds to the promoter regions, and co-operatively interacts with CrtJ.9 The homologous genes of crtJ (ppsR) and aerR (ppaA) have been identified in other purple bacteria, including Rubrivivax gelatinosus and Bradyrhizobium ORS278.14–16 This suggests that CrtJ and AerR are widely conserved in purple bacteria, although they exhibit different regulatory features in each bacterium: they can activate and/or repress the transcription of photosynthesis genes, and their activities are modulated or not by the redox conditions.17,18 Thus, characterization of crtJ and aerR in bacteria that show different features in photopigment synthesis seem to be useful to elucidate the regulatory mechanisms.
In this study, we identified the homologous genes of crtJ and aerR in Rsp. centenum. Mutational analyses indicated that CrtJ and AerR act as aerobic repressor and activator, respectively, in this bacterium. Our results also suggest that the readthrough transcript from crtEF genes into the puf operon is significant in maintaining the mRNA levels of the light harvesting I (pufB and pufA) genes.
Materials and methods
Bacterial strains and growth conditions
The wild-type strain of Rsp. centenum (ATCC 43720), the aerR-disrupted strain AERR1, and the crtJ-disrupted strain CRTJ1 were grown at 42 °C in CENS medium as described previously.19 For anaerobic growth, cultures were incubated in screw-cap tubes filled with CENS medium, and routinely illuminated by banks of 60 W incandescent Lumiline lamps. For aerobic growth, 15 mL cultures were incubated in 250 mL sidearm flasks with shaking (500 rpm). Cell growth was monitored by measuring turbidity with a Klett-Sumerson spectrophotometer. Escherichia coli strains DH5α and DH10B (Bethesda Research Laboratories) were used as host strains for routine cloning procedures, and the strain S17-120 was used as a mobilization strain. They were grown in Luria broth at 37 °C. Gentamycin, ampicillin, and spectinomycin were used for E. coli at 10, 150, and 50 μg mL−1, respectively. Gentamycin and spectinomycin were used for Rsp. centenum at concentration of 10 μg mL−1 each.
Modified mini-Tn5 mediated mutagenesis
Wild type Rsp. centenum was mutagenized using a modified mini-Tn5 with an ΩSpr cassette replacing the Kmr gene. Mutagenized culture was spread onto agar-solidified CENS media and grown aerobically in the dark. Pigment mutants were isolated based on colony color. The disrupted genes were cloned by isolating genomic DNA from each mutant strain and digesting the genomic DNA with various restriction enzymes in separate reactions. Digested DNA was ligated into pBluescript and transformed into E. coli strain DH10B. Strains containing the pBluescript plasmid with the appropriate insert were selected for by plating on LB medium containing both ampicillin and spectinomycin. The regions flanking the transposon were sequenced using primers to the spectinomycin gene. Sequence analysis was performed using the program, GCG.
Construction of aerR-disrupted strain, AERR1, from Rsp. centenum
Two 500-bp DNA fragments consisted of N-terminal and C-terminal regions of AerR were amplified by PCR using pfu DNA polymerase. Two sets of primers were used. One of the two is a forward primer AerRsacIfor (5′-GGGAGCTCTTCCGGGAAG-GCTAGACGTG-3′) and a reverse primer AerRecoVrev (5′-GG-GATATCGAATCAGGGACGCGAACCGG-3′). The other set is a forward primer AerRecoVfor (5′-GGGATATCCCGGCCA-GATCTGATCAAGAG-3′) and a reverse primer AerRxbaIrev (5′-GGTCTAGACGCGGACCCGGTTTGTCAAG-3′). SacI, EcoRV, and XbaI restriction sites were designed at additional polynucleotides of AerRsacIfor, AerRecoVrev and AerRecoVfor, and AerRxbaIrev primers, respectively (underlined). The first PCR fragment was digested with SacI and EcoRV. The second PCR fragment was digested with XbaI and EcoRV. After the digestion, these two fragments were mixed and ligated together into SacI-XbaI-cut pUC18, resulting in pUCAerR. The pUCAerR and the Gmr-suicide vector pZJD29A21 were digested with XbaI at their unique sites, and ligated together to construct a plasmid pAerRGm. The plasmid pZJD29A has a sacB gene encoding the levansucrase of Bacillus subtilis. The expression of sacB in the presence of sucrose is lethal for many Gram negative bacteria.22 The resulting plasmid, pAerRGm was transferred into Rsp. centenum cells by conjugation with the mobilizing strain S17-1, and cells undergoing single crossover events were selected by plating exconjugants on CENS plates containing Gm. The streak-purified Gmr cells were grown for over night in liquid medium without Gm, and then these cultures were plated onto CENS plates containing 5% sucrose. Several single colonies were picked up, and AerR deletion was confirmed by PCR (about half of these colonies were wild type as expected). The resulting aerR-deleted strain was named AERR1. The predicted amino acid sequence for AERR1 is 14 amino acids long (216 amino acids for the native protein).
Construction of crtJ-disrupted strain, CRTJ1, from Rsp. centenum
Two 500-bp DNA fragments consisted of N-terminal and C-terminal regions of CrtJ were amplified by PCR using pfu DNA polymerase. Two sets of primers were used. One of the two sets is a forward primer CrtJsacIfor (5′-GGGAGCTCGCAT-GCGCCAGCCTTAACGC-3′) and a reverse primer CrtJecoVrev (5′-GGGATATCGACTGCTGCGCTTCCACCAG-3′). The other set is a forward primer CrtJecoVfor (5′-GGGATATC-GCTCTATGCCACCACCCTGC-3′) and a reverse primer CrtJxbaIrev (5′-GGTCTAGACGGGCATCGCTCACCTGC-TG-3′). SacI, EcoRV, and XbaI restriction sites were designed at additional polynucleotides of AerRsacIfor, AerRecoVrev and AerRecoVfor, and AerRxbaIrev primers, respectively (underlined). The first fragment was digested with SacI and EcoRV, and the second fragment was digested with XbaI and EcoRV. After the digestion, these two fragments were mixed and ligated into SacI-XbaI-cut pUC18, resulting in pUCCrtJ. Further procedure is as same as that for aerR-disrupted strain construction (described above). The resulting crtJ-deleted strain, named CRTJ1, has the 201 amino acids in-frame deletion at crtJ locus (the native protein has a predicted length of 468 amino acids).
Construction of crtE::lacZ and puf::lacZ fusions, and β-galactosidase assay
A DNA fragment consisted of 342-bp upstream and 57-bp downstream from crtE start codon was amplified from Rsp. centenum genomic DNA by PCR using pfu DNA polymerase. For the PCR reaction, forward and reverse primers (crtE-F: 5′-TCCA-GCCCCGGGAGCATG-3′ and crtE-R: 5′-AGACCGAATATC-GCATCGCC-3′, respectively) were designed. The amplified DNA fragment was cloned into the promoter testing vector, pCF1010,23 at the StuI site, resulting in crtE::lacZ transcriptional fusion named pRC200. A 1-kbp Rsp. centenum puf upstream region was amplified by PCR from the plasmid pRCPUF8 (kindly provided by Joann Williams, Arizona State University) with pfu DNA polymerase. The forward and reverse primers (M13-M4: 5′-GTTTTCCCAGTCACGAC-3′ and RcenPuf-R: 5′-GAGGTATTTTCAGGCATTGG-3′, respectively) were designed for the PCR reaction. The amplified DNA fragment was cloned into a promoter testing vector, pCF1010, at the StuI site, resulting in puf::lacZ transcriptional fusion named pRC100. These plasmids were mobilized into Rsp. centenum cells by congugation with the mobilizing strain E. coli S17-1. The β-galactosidase activity was determined as essentially described.24 Cell growth was monitored by measuring the optical density (OD) of the culture at 660 nm with a Klett-Summerson photometer. Final results were obtained as the amount of o-nitrophenyl-β-galactoside (ONPG) hydrolyzed per min per OD660.
Materials
Restriction endonucleases and T4 ligase were purchased from New England Biolabs. Synthetic oligonucleotide primers were purchased from MWG Biotech, Inc.
Accession numbers
The nucleotide sequence data reported in this study are available in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB369262.
Results and discussion
Identification of crtJ and aerR genes in Rsp. centenum
Rsp. centenum is one of a few photosynthetic bacteria that express photosynthetic pigments under aerobic conditions. To determine the regulatory elements that control expression of photosynthetic pigments in this organism, transposon mutagenesis with a modified mini-Tn5 was utilised to isolate mutants with altered expression patterns. Since Rsp. centenum is able to express its photosystem under aerobic conditions, we could directly screen for photosystem mutants based on colony color, under conditions where expression of the photosystem is not required for growth. Colonies with decreased photopigment expression (pink or yellow) were isolated, as well as colonies with increased photopigments (dark red). Out of 10 000 colonies screened, 20 mutants showed a yellow or pink phenotype and 2 mutants showed a dark-red phenotype. In this report, two of these mutants are discussed as follows.
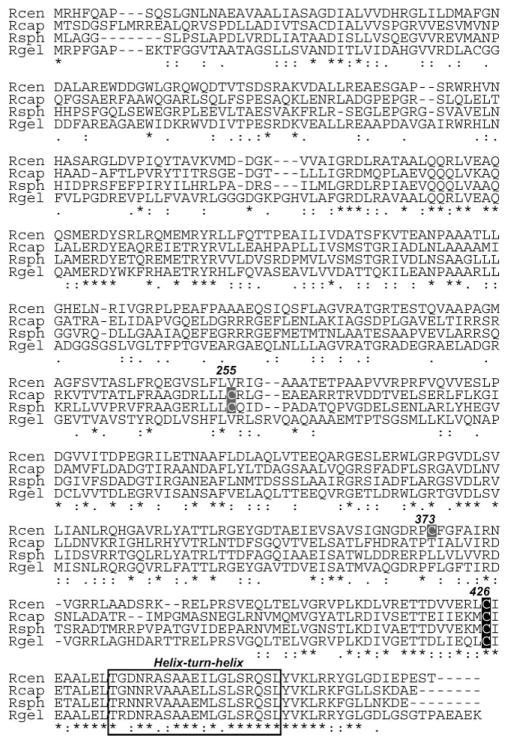
Strain JBC003 forms colonies with a dark-red color indicative of derepression of the photosystem. The disruption in this mutant was cloned and sequenced and mapped to an open reading frame of 1404 base pairs. A search of the BLAST database with the predicated amino acid sequence of this gene showed similarity to CrtJ and PpsR from Rba. capsulatus and Rba. sphaeroides, respectively (Fig. 1), which are known to be aerobic repressors for photosynthesis gene transcription.8,11–13 We also found that CrtJ/PpsR homolog of Rsp. centenum show much higher similarity to those of Thiocapsa roseopersicina and Rhodopseudomonas palustris (~55% identity) than to those of Rhodobacter species (~30% identity). This is in agreement with the phylogenetic distances between each bacterium based on the 16S rRNA sequences that showed that Rsp. centenum, Tca. roseoperisicina and Rps. palustris belong to α1-subgroup of proteobacteria; whereas, Rba. capsulatus and Rba. sphaeroides belong to α3-subgroup of proteobacteria.25 Tca. roseoperisicina and Rps. palustris contain two CrtJ/PpsR homologs,18,26 suggesting that Rsp. centenum possesses also two CrtJ/PpsR homologs, although, at present, no additional crtJ/ppsR gene has been found on the partially sequenced genome of Rsp. centenum (available at http://genomes.tgen.org/index.html) (data not shown).
Fig. 1.
Sequence alignment of the predicted CrtJ proteins of Rsp. centenum to homologous proteins in other purple bacteria. Abbreviations and their meaning: Rcen; Rsp. centenum, Rcap; Rba. capsulatus, Rsph; Rba. sphaeroides, Rgel; Rbv. gelatinosus. Accession numbers of the sequences are AB369262, P26167, ABA79455 and BAA94062, respectively.
It was shown that Rba. capsulatus CrtJ makes an intramolecular disulfide bond under aerobic growth conditions, which results in stimulation of DNA-binding activity for target promoters.12 Sequence alignment of the Rsp. centenum crtJ gene product with the CrtJ (PpsR) proteins from other species reveals the presence of several conserved regions including the highly conserved C-terminal helix-turn-helix DNA-binding domain (Fig. 1). Also of note is the conserved cysteine residue at position 426, which has been shown to be essential for formation of an intramolecular disulfide bond in Rba. capsulatus. Interestingly, the other cysteine residue implicated in disulfide bond formation with cysteine 426 in Rba. capsulatus CrtJ12 is not conserved in Rsp. centenum at the corresponding position (valine 255), suggesting a different oxygen-sensitivity of this protein in this bacterium. If the function of the Rsp. centenum CrtJ protein requires the formation of an intramolecular disulfide bond, it must occur at the only other cysteine residue in the predicted amino acid sequence at position 373 (Fig. 1).
Strain JBC315 forms pale colonies indicative of decreased expression of photosynthetic pigments. The disruption in this mutant was cloned and sequenced and mapped to an open reading frame of 648 base pairs. A search of the BLAST database with the predicated amino acid sequence of this gene showed similarity to AerR from Rba. capsulatus and PpaA from Rba. sphaeroides respectively. AerR/PpaA functions as a repressor in Rba. capsulatus and an activator in Rba. sphaeroides for transcription of photosynthetic genes.9,10 To investigate the roles of AerR as well as CrtJ in controlling aerobic photosystem synthesis in Rsp. centenum, we constructed aerR- and crtJ-deletion strains of this bacterium as follows.
Mutational analyses of AerR and CrtJ in Rsp. centenum
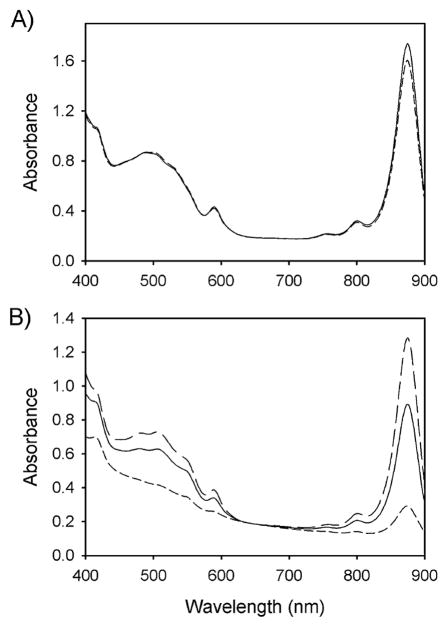
The aerR and crtJ deletion strains were constructed by site-specific recombination (designed as AERR1 and CRTJ1, respectively). To avoid the complicating effects of expression of downstream or upstream genes, no antibiotic-resistance cassette was employed for the construction (see Materials and methods). Phenotypic differences between aerobically grown colonies of these mutants and wild type were striking. The AERR1 strain exhibits poorly pigmented (pale pink) colonies whereas CRTJ1 strain makes colonies that are more pigmented (dark red) than observed with wild type cells (data not shown). Fig. 2A and B show the spectral analyses of Rsp. centenum wild-type (solid lines), AERR1 (short-dashed lines), and CRTJ1 (long-dashed line) strains grown under anaerobic light and aerobic dark conditions, respectively. CRTJ1 cells grown under aerobic conditions synthesized higher levels of photopigments than the wild type strain whereas aerobically grown AERR1 cells synthesized significant reduced amounts of photopigments (Fig. 2B). Spectral analysis of phosynthetically grown cells indicated that CRTJ1 had the same amounts of photopigments as the wild type strain (data not shown). Similarly, AERR1 showed just slightly reduced levels of photopigment synthesis (Fig. 2A). These results indicated that CrtJ represses and AerR activates photosystem synthesis only under aerobic conditions.
Fig. 2.
Absorption spectra of Rsp. centenum wild type (solid lines), AERR1 (short dashed lines) and CRTJ1 (long dashed line) grown anaerobically (panel A) or aerobically (panel B). AERR1 is the aerR-disrupted strain, and CRTJ1 is the crtJ-disrupted strain. The spectrum for CRTJ1 cells grown anaerobically is not shown, but identical to that of the wild type cells (panel A, solid line).
We next tested the effect of aerR and crtJ mutations on gene expression by measuring β-galactosidase activities in strains that harbored the crtE::lacZ or puf::lacZ reporter plasmids. As indicated in Fig. 3, disruption of aerR results in lower (0.7 fold) crtE expression than that of wild type when grown aerobically. In contrast, CRTJ1 showed high levels (1.7 fold) of activity. There is no significant difference in the crtE expression between these mutant strains and wild type when grown photosynthetically. These results indicated that AerR and CrtJ act as activator and repressor, respectively, for the crtE transcription under aerobic conditions. The aerR mutation, but not crtJ mutation, affected the puf operon expression. The AERR1 cells grown aerobically demonstrated 0.4-fold levels of puf expression, but anaerobically grown cells showed 1.7-fold expression levels. These results indicated that AerR is a bifunctional regulator for puf operon expression which activates transcription under aerobic conditions, but represses under anaerobic conditions.
Fig. 3.
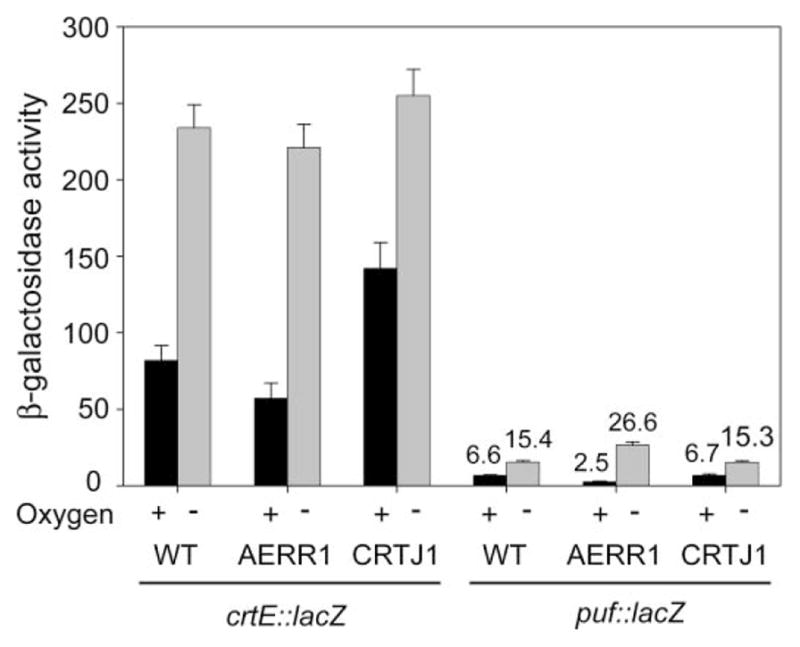
β-galactosidase activity measurements of the crtE::lacZ and puf::lacZ fusions in wild type, ΔaerR (AERR1) and ΔcrtJ (CRTJ1) strains. Cultures were grown either aerobically (black bars) or anaerobically (gray bars) in CENS medium and then assayed for β-galactosidase activity. Units of β-galactosidase activity represent μmol of ONPG (o-nitrophenyl-β-D-galactopyranoside) hydrolyzed/min/OD660. The data represent the mean of at least three independent experiments (error bars indicate ±S.D.).
Identification of CrtJ binding site in the Rsp. centenum crt promoters
As shown in Fig. 4, a CrtJ-recognition site (TGT-N12-ACA) with a −35 σ70-recognition motif (TTGACA) which was well matched with the consensus sequence shown in the Rhodobacter species, were detected in the promoter regions of Rsp. centenum crtE and crtD genes. This observation suggests that Rsp. centenum CrtJ binds to the same consensus motifs shown in Rba. capsulatus for its repressor activity. The CrtJ-repressed promoters often contain two CrtJ-recognition sequences in tandem as observed in Rba capsulatus bchC promoter (Fig. 4). The two palindromic sites were critical for cooperative binding of CrtJ, because CrtJ works as tetramer (dimmer of dimmer).27 However, Rsp. centenum crtE and crtD promoters contain only one CrtJ-recognition site as shown in Rba. capsulatus crtE, crtI and bchF promoters (Fig. 4). In Rba. capsulatus, CrtJ still binds to the promoters in a cooperative manner with a distantly separated palindrome located in a different promoter through forming a looped DNA structure.28 Given that Rsp. centenum crtE and crtD genes adjoin on the genome (Fig. 5) and crtE promoter activity is influenced by the crtJ mutation (Fig. 3), Rsp. centenum CrtJ may bind to the crtE promoter region cooperatively with the crtD promoter.
Fig. 4.
Comparison of possible promoter sequences found in Rsp. centenum crtE and crtD genes and those reported in Rba. capsulatus photosynthesis genes. The number in parentheses indicates the number of nucleotides between the start codon, ATG, and the right end of the sequence. The proposed CrtJ-binding sites (TGT-N12-ACA) are shown in bold. Nucleotide sequences of the consensus E. coli σ70 recognition site are shown in the bottom.
Fig. 5.
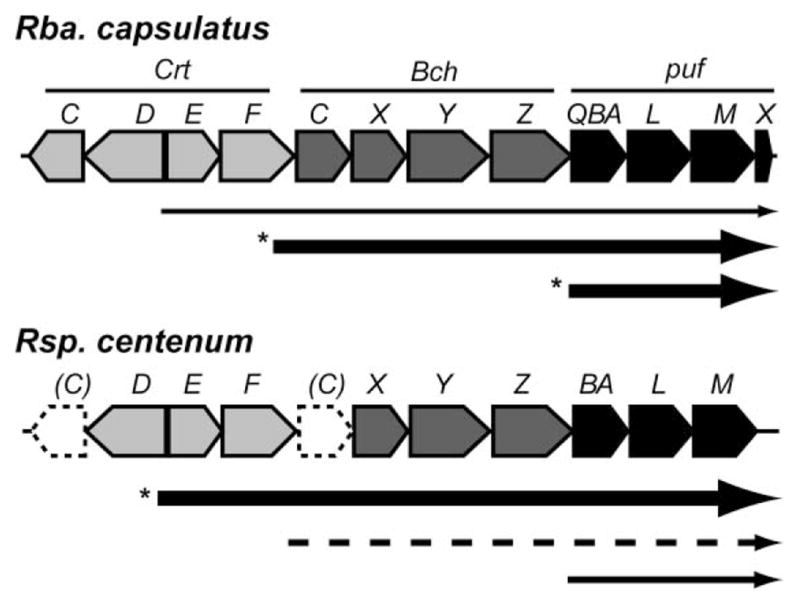
Comparison of readthrough transcription for puf operon between Rba. capsulatus and Rsp. centenum. Thickness of the arrows indicates the levels of transcription. The dotted lines donate possible gene locations and transcripts that have not been experimentally proven. Asterisks denote the major transcripts for puf operon.
No CrtJ binding motif was found within 1-kbp upstream of Rsp. centenum puf operon (data not shown). These observations are consistent with the lacZ reporter experiments, described above, in the sense that crtJ mutation affects the expression of crtE, but not of the puf operon. A previous study indicated that Rba. capsulatus AerR binds to the promoter regions of some photosynthesis genes including puf.9 Both puf and crtE expression are regulated by AerR in Rsp. centenum (Fig. 3), suggesting the same binding pattern in Rsp. centenum. However, any AerR recognition sequences have not been clear yet even in Rba. capsulatus.9
Readthrough transcript for the Rsp. centenum puf operon
In Rba. capsulatus, the puf operon is co-transcribed with the upstream crtEF and bchCXYZ genes, although a large amount of puf mRNA are also transcribed by its own promoter24,29 (Fig. 5). Specifically, based on the reporter gene fusion analysis, about 83% and 50% puf transcription come from bchC promoter under high- and low-oxygen conditions, respectively.30 The similar polycistronic transcription for puf was also reported in another purple bacterium Rbv. gelatinosus.31 A previous genetic study suggested the same gene arrangement in the Rsp. centenum puf upstream region (crtEF-bchCXYZ-puf)32 (Fig. 5). Sequence analysis indicated that the bchZ gene locates in the upstream of the Rsp. centenum puf operon, supporting the data (data not shown). The superoperonal structure (crtEF-bchCXYZ-puf) has been observed in all purple bacteria studied to date including Rba. spheroides, Rps. palustris and Bradyrhizobium ORS278,15,18,31,33 suggesting the importance of the gene arrangement for their regulated expressions. Interestingly, expression of Rsp. centenum crtE gene is at much higher levels than that of the puf specific promoter (15-fold higher than that of puf operon) (Fig. 3). In Rba. capsulatus, crtE expression is significantly lower (30-fold) than that of the puf specific promoter.24 These results suggest that most of puf transcription under aerobic conditions in Rsp. centenum is derived from the readthrough as observed in Rba. capsulatus (Fig. 5), although detailed mRNA analyses have not been performed. Given that the crtE expression is weakly repressed by oxygen in Rsp. centenum (about 3-fold) (Fig. 3), puf mRNA could be maintained at high levels even under aerobic conditions in this bacterium.
However, there are some conflicting observations. Specifically, the levels of light-harvesting complexes in AERR1 were about 20% of those in wild type under aerobic conditions (Fig. 2B); however, promoter activity of crtE in AERR1 was about 75% of that in wild type under the identical conditions (Fig. 3). These results suggest that there are still unknown mechanisms for controlling puf mRNA levels by AerR in Rsp. centenum. One possibility is that bchC promoter activity in Rsp. centenum is significant to influence puf transcription, as observed in Rba. capsulatus,30 and that the activity is also regulated by AerR.
Conclusion
In this study, we show that aerobic photosystem synthesis in Rsp. centenum is regulated by CrtJ and AerR regulatory factors. AerR is necessary for the aerobic induction of photosynthesis gene expression in Rsp. centenum. CrtJ, on the other hand, is an aerobic repressor for photosynthesis genes, as in Rhodobacter species. The typical CrtJ-binding motifs of Rhodobacter species are observed at Rsp. centenum crtE and crtD promoter regions (Fig. 4), indicating that they may recognise the same motif. However, one of the two conserved cysteine residues in the Rhodobacter CrtJ which make an intramolecular disulfide bond is not conserved in Rsp. centenum (Fig. 1), suggesting that CrtJ sensitivity to oxygen is somehow different between these two species.
The puf specific promoter activity in Rsp. centenum is much lower than that of the crtE promoter, which makes a superoperonal structure with the puf operon. As a result, the readthrough transcript may be high enough to influence puf mRNA levels, especially under aerobic conditions. The importance of readthrough for aerobic induction of photosynthesis genes in another purple bacterium, Rdv. sulfidophilum was also suggested previously.6 Perhaps, CrtJ and AerR are the main regulators for the aerobic synthesis of the photosynthetic apparatus through regulating superoperonal transcription in purple bacteria that aerobically synthesize photopigments. Finally, AerR works as an activator in Rsp. centenum, although it was reported to be a repressor in Rba. capsulatus.9 Further studies of Rsp. centenum AerR should be useful in clarifying the exact mechanism of how AerR regulates photosystem synthesis with respect to the phenotypic difference observed in this bacterium.
Acknowledgments
We thank Howard Gest for critical reading of the manuscript. We also thank Joann Williams for the gift of the plasmid pRCPUF8. This work was supported by Research Foundation For Opto-Science and Technology, Grant-in-aid from MEXT of Japan (to SM), and National institute of Health grant (to CEB).
Footnotes
This paper was published as part of the themed issue of contributions from the 7th International Conference on Tetrapyrrole Photoreceptors in Photosynthetic Organisms held in Kyoto, December 2007.
Notes and references
- 1.Bauer CE, Elsen S, Swem LR, Swem DL, Masuda S. Redox and light regulation of gene expression in photosynthetic prokaryotes. Philos Trans R Soc London, Ser B. 2003;358:147–153. doi: 10.1098/rstb.2002.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeilstra-Ryalls JH, Kaplan S. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell Mol Life Sci. 2004;61:417–436. doi: 10.1007/s00018-003-3242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi M, Shioi Y, Gad’on N, Golecki JR, Drews G. Spectroscopical studies on the light-harvesting pigment-protein complex II from dark-aerobic and light-anaerobic grown cells of Rhodobacter sulfidophilus. Biochim Biophys Acta. 1991;1058:235–241. [Google Scholar]
- 4.Yildiz FH, Gest H, Bauer CE. Attenuated effect of oxygen on photopigment synthesis in Rhodospirillum centenum. J Bacteriol. 1991;173:5502–5506. doi: 10.1128/jb.173.17.5502-5506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagemann GE, Katsiou EK, Forkl H, Steindorf ACJ, Tadros MH. Gene cloning and regulation of gene expression of the puc operon from Rhodovulum sulfidophilum. Biochim Biophys Acta. 1997;1351:341–358. doi: 10.1016/s0167-4781(96)00228-x. [DOI] [PubMed] [Google Scholar]
- 6.Masuda S, Nagashima KVP, Shimada K, Matsuura K. Transcriptional control of expression of genes for photosynthetic reaction center and light-harvesting proteins in the purple bacterium Rhodovulum sulfidophilum. J Bacteriol. 2000;182:2778–2786. doi: 10.1128/jb.182.10.2778-2786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penfold RJ, Pemberton JM. Sequencing, chromosomal inactivation, and functional expression of ppsR a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponnampalam SN, Buggy JJ, Bauer CE. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light-harvesting-II expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:2990–2997. doi: 10.1128/jb.177.11.2990-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong C, Elsen S, Swen LR, Bauer CE. AerR, a second aerobic repressor of photosynthesis gene expression in Rhodobacter capsulatus. J Bacteriol. 2002;184:2805–2814. doi: 10.1128/JB.184.10.2805-2814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomelsky L, Sram J, Moskvin OV, Horne IM, Dodd HN, Pemberton JM, McEwan AG, Kaplan S, Gomelsky M. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology. 2003;149:377–388. doi: 10.1099/mic.0.25972-0. [DOI] [PubMed] [Google Scholar]
- 11.Ponnampalam SN, Bauer CE. DNA binding characteristics of CrtJ. J Biol Chem. 1997;272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- 12.Masuda S, Dong C, Swem D, Setterdahl AT, Knaff DB, Bauer CE. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc Natl Acad Sci USA. 2002;99:7078–7083. doi: 10.1073/pnas.102013099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi N, Harada J, Nagashima S, Matsuura K, Shimada K, Nagashima KVP. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J Mol Evol. 2001;52:333–341. doi: 10.1007/s002390010163. [DOI] [PubMed] [Google Scholar]
- 15.Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Verméglio A. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature. 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 16.Steunou A-S, Astier C, Ouchane S. Regulation of photosynthesis genes in Rubrivivax gelatinosus: transcription factor PpsR is involved in both negative and positive control. J Bacteriol. 2004;186:3133–3142. doi: 10.1128/JB.186.10.3133-3142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsen S, Jaubert M, Pignol D, Giraud E. PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol Microbiol. 2005;57:17–26. doi: 10.1111/j.1365-2958.2005.04655.x. [DOI] [PubMed] [Google Scholar]
- 18.Kovács ÁT, Rákhely G, Kovács KL. The PpsR regulator family. Res Microbiol. 2005;156:619–625. doi: 10.1016/j.resmic.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Stadtwald-Demchick R, Turner FR, Gest H. Physiological properties of the thermotolerant photosynthetic bacterium Rhodospirillum centenum. FEMS Microbiol Lett. 1990;67:139–144. [Google Scholar]
- 20.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering; transposon mutagenesis in Gram negative bacteria. Biotechnology. 1983;1:37–45. [Google Scholar]
- 21.Masuda S, Bauer CE. Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus. J Bacteriol. 2004;186:235–239. doi: 10.1128/JB.186.1.235-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gay P, LeCoq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee LK, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. J Biol Chem. 1995;270:20453–20458. [PubMed] [Google Scholar]
- 24.Young DA, Bauer CE, Williams JC, Marrs BL. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989;218:1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- 25.Imhoff JF. In: Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria in Anoxygenic Photosynthetic Bacteria. Blankenship RE, Madigan MT, Bauer CE, editors. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1995. pp. 1–15. [Google Scholar]
- 26.Giraud E, Verméglio A. Bacteriophytochromes in anoxygenic photosynthetic bacteria. Photosynth Res. 2008;97:141–153. doi: 10.1007/s11120-008-9323-0. [DOI] [PubMed] [Google Scholar]
- 27.Ponnampalam SN, Elsen S, Bauer CE. Aerobic repression of the Rhodobacter capsulatus bchC promoter involves cooperative interactions between CrtJ bound to neighboring palindromes. J Biol Chem. 1998;273:30757–30761. doi: 10.1074/jbc.273.46.30757. [DOI] [PubMed] [Google Scholar]
- 28.Elsen S, Ponnampalam SN, Bauer CE. CrtJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J Biol Chem. 1998;273:30762–30769. doi: 10.1074/jbc.273.46.30762. [DOI] [PubMed] [Google Scholar]
- 29.Wellington CL, Beatty JT. Overlapping mRNA transcripts of photosynthesis gene operons in Rhodobacter capsulatus. J Bacteriol. 1991;173:1432–1443. doi: 10.1128/jb.173.4.1432-1443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellington CL, Taggart AKP, Beatty JT. Functional significance of overlapping transcrips of crtEF, bchCA, and puf photosynthesis gene operons in Rhodobacter capsulatus. J Bacteriol. 1991;173:2954–2961. doi: 10.1128/jb.173.9.2954-2961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liotenberg S, Steunou A-S, Picaud M, Reiss-Husson F, Astier C, Ouchane S. Organization and expression of photosynthesis genes and operons in anoxygenic photosynthetic proteobacteria. Environ Microbiol. 2008;10:2267–2276. doi: 10.1111/j.1462-2920.2008.01649.x. [DOI] [PubMed] [Google Scholar]
- 32.Yildiz FH, Gest H, Bauer CE. Conservation of the photosynthetic gene cluster in Rhodospirillum centenum. Mol Microbiol. 1992;6:2683–2691. doi: 10.1111/j.1365-2958.1992.tb01445.x. [DOI] [PubMed] [Google Scholar]
- 33.Naylor GW, Addlesee HA, Gibson LCD, Hunter CN. The photosynthesis gene cluster of Rhodobacter sphaeroides. Photosynth Res. 1999;62:121–139. [Google Scholar]