Abstract
The flavin-binding BLUF domain of AppA represents a new class of blue light photoreceptors that are present in a number of bacterial and algal species. The dark state X-ray structure of this domain was determined at 2.3 Å resolution. The domain demonstrates a new function for the common ferredoxin-like fold; two long α-helices flank the flavin, which is bound with its isoalloxazine ring perpendicular to a five-stranded β-sheet. The hydrogen bond network and the overall protein topology of the BLUF domain (but not its sequence) bear some resemblance to LOV domains, a subset of PAS domains widely involved in signaling. Nearly all residues conserved in BLUF domains surround the flavin chromophore, many of which are involved in an intricate hydrogen bond network. Photoactivation may induce a rearrangement in this network via reorientation of the Gln63 side chain to form a new hydrogen bond to the flavin O4 position. This shift would also break a hydrogen bond to the Trp104 side chain, which may be critical in induction of global structural change in AppA.
Sensing blue light is important for a great variety of organisms. Plants, algae, and photosynthetic bacteria use blue light to control development and optimize photosynthesis. Organisms such as mice and Drosophila use blue light to mediate circadian rhythms (1), and the presence of blue light photoreceptors in nonphotosynthetic prokaryotes implies many other functions (2, 3). Two well-documented classes of photoreceptors utilize flavin for blue light perception, cryptochromes (4) and the LOV1 subset of PAS domains (5). The photoreceptor domain of AppA has been identified as the prototype of a new, third class of flavin-binding, blue light photoreceptors (3). These so-called BLUF (blue light sensing using FAD) domains have been identified by sequence homology in purple photosynthetic bacteria, in cyanobacteria, and in the unicellular eukaryote, Euglena gracilis.
Like PAS domains (6), BLUF domains are considered to be sensing domains that modify the activity of covalently attached effector domains (7) such as adenyl cyclase, GGDEF (8), and EAL domains (9). A subclass of small BLUF domain proteins contains an extension of 20–70 amino acids with no homology to any known protein, motifs, or effector function (3). The best studied proteins containing BLUF domains include AppA from the bacterium Rhodobacter sphaeroides (10–12), the α and β subunits of photoactivated adenylate cyclase (PAC) in Euglena gracilis (13), YcgF in Escherichia coli (10), and Slr1694 in Synechocystis sp. PCC6803 (14).
AppA functions as a negative repressor of photosynthetic gene expression via interaction with its noncovalently bound partner protein, PpsR (11). PpsR directly binds to DNA as a tetramer to promote expression of photosynthetic machinery. In its dark state, AppA catalyzes reduction of a disulfide bond in PpsR and converts PpsR from an active tetramer into an inactive dimer. Blue light absorption by AppA induces a conformation change that prevents it from interacting with PpsR (12). AppA contains two domains, an N-terminal BLUF domain (amino acids 1–110) that non-covalently binds the blue light absorbing chromophore, flavin adenine dinucleotide (FAD), and a catalytic cysteine-rich C-terminal domain (amino acids 111–450) responsible for reduction of the disulfide bond in PpsR (10).
The isolated N-terminal BLUF domain exhibits a photo-cycle identical to that observed with full-length AppA (12). Photoexcitation of AppA is proposed to involve a singlet excited state in the flavin, which leads to formation of a red-shifted intermediate state after 10 ns (15) that slowly decays back to the ground state after 30 min (11). This red shift may be caused by alterations in π–π stacking and hydrogen bonding between FAD and Tyr21 (12). The photochemical reaction of AppA is distinctly different from that observed in the flavin-binding domains of the other well-characterized blue light photoreceptors, phototropin and cryptochrome (16, 17). The phototropin photocycle involves the formation of a transient, covalent flavin–cysteinyl adduct (18–20) in its LOV2 domain whereas the photocycle of cryptochrome may involve formation of a flavin radical (21).
Visualizing the light-excited structure of the AppA BLUF domain should give much insight into the mechanism by which this newly defined class of photoreceptors transduces photon absorption into a biologically important change in structure. Although the AppA BLUF domain is photoactive in the crystal, data quality is at present severely compromised by photoexcitation-induced disorder. We are currently investigating different crystal forms to overcome this difficulty.
Here we describe the three-dimensional dark state structure of the BLUF domain of AppA from Rb. sphaeroides. We demonstrate that this domain represents a new class of flavin-binding proteins involved in light sensing, compare its structure to that of other related proteins, and discuss implications of the structure for signaling mechanisms.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
An AppA construct containing amino acids 1–156 was expressed and purified as described (12). A second construct containing amino acids 17–133 was expressed with an intein-chitin-binding domain (CBD) in a pTYB12 vector (New England Biolabs) using E. coli BL21(DE3) cells induced with 1 mM isopropyl β-D-thiogalactopyranoside. Protein was alternatively expressed with Se-Met under the same conditions. Initial purification was performed using affinity chromatography with chitin beads (New England Biolabs). Although the intein-CBD tag was subsequently removed, three additional amino acids remained on the N-terminus after cleavage. Protein was further purified using size exclusion chromatography on Superose 12 (Pharmacia) in 20 mM Tris-HCl, pH 8.0, and 0.1 M NaCl. The final protein was estimated to be >95% pure by both SDS–PAGE and MALDI.
Crystallization
Initial crystallization trials of the AppA FAD-binding domain were performed on the 1–156 amino acid construct, a protein that dimerized in solution and maintained the same photocycle as the full-length protein (11, 12). Protein was concentrated to 7 mg/mL, and crystallization conditions were explored using a sparse matrix screen (Hampton Research); crystallization was performed in the dark in order to maintain the protein in its dark state. MALDI analysis of tetragonal crystals in the P41212 space group showed that the protein was partially hydrolyzed during crystallization to a fragment composed of amino acids 17–133. A new construct containing amino acids 17–133 was expressed, which also dimerized and maintained the same photocycle as full-length AppA (data not shown). This construct crystallized at 16 °C in 3% PEG 3350, 0.1 M Tris-HCl, pH 8.0, 1.5 mM N-dodecyl-N,N-dimethylglycine, and 24% ethylene glycol, yielding 300–500 μm crystals with the same morphology as the hydrolyzed fragment. Protein expressed with Se-Met was crystallized under identical conditions but with the addition of 1 mM EDTA and 5 mM β-mercaptoethanol to the mother liquor.
MALDI Mass Spectroscopy
Analysis of proteins and their fragments was done on Bruker Biflex III MALDI-TOF instrument at the mass spectroscopy facility of Indiana University. Crystals were dissolved in 25 mM NH4HCO3 prior to tryptic digestion (trypsin ratio of 1:20) at 37 °C for 3–4 h. The mixture was desalted using Zip-Tips (Millipore) and suspended in 60% acetonitrile/0.5% trifluoroacetic acid prior to MALDI. The molecular weight of fragments was calculated and matched to observed peaks using ExPASy proteomic tools (22).
TLC of Extracted Flavin
The flavin cofactor was extracted from both AppA crystals dissolved in distilled water and freshly prepared protein by heating the samples for 10 min in 70% ethanol at 60 °C. The dissolved cofactors from both samples were spotted on a thin-layer chromatographic silica plate and chromatographed with FAD, FMN, and riboflavin as standards in 1-butanol/acetic acid/water at a ratio of 5:2: 3. Flavins were visualized by excitation with far-UV light at 365 nm.
Data Collection
All X-ray diffraction data were collected using the monochromatic oscillation technique at the Advanced Photon Source BioCARS beamlines, Argonne National Laboratory. MAD data were collected from a single crystal on beamline 14BM-D using an ADSC Quantum 4 detector at three wavelengths: 0.97939, 0.97962, and 0.96113 Å. Native data were collected on beamline 14BM-C at a wavelength of 0.9 Å. Crystals were frozen in liquid nitrogen prior to data collection and all data collected at 105 K.
All sources of light were minimized in order to maintain the protein in its dark state during crystal freezing, sample mounting, and data collection. Although the crystal was exposed to some ambient light during data collection, the intensity was at a very low level (7–10 μmol m−2 s−1). This amount of light would only photoexcite a small fraction of molecules in dilute solution (11) and would therefore be insufficient to photoexcite a significant volume of the crystal, which was of high optical density.
Data Processing and Refinement
The HKL2000 program suite (23) was used for indexing, integration, scaling, and merging of monochromatic oscillation data. Initial phases were determined to 3.0 Å using SOLVE (24) and then improved via density modification with RESOLVE (25). A strong anomalous signal was present for 12 Se sites. The initial maps showed very clear density for the protein backbone and cofactor of all three monomers in the asymmetric unit. Initial phases were applied to a higher resolution, dark dataset collected on a native AppA crystal to 2.3 Å resolution. The NCS operators for the three monomers were determined from the heavy atom sites using the CCP4 program Professs, and density modification was performed using the CCP4 program DM.
The protein backbone was manually fit into the monomer with the best ordered electron density using XtalView (26). The amino acid sequence was identified and aligned using the aromatic side chains as reference and then copied to the other two monomers. Subsequent examination showed that all 12 heavy atom sites aligned exactly with the positions of methionine side chains. The models were initially refined without the cofactor as a check against model bias using the CCP4 program Refmac5 (27). Out of 121 residues expressed in a single monomer, the asymmetric unit included ordered electron density for 118 residues in the A chain, 116 in the B chain, and 115 in the C chain. The Lys98 side chain was disordered in all three monomers, but an additional 7 side chains were disordered in the C chain. Ordered electron density was also visualized for the FMN component of the FAD cofactor in each monomer, 6 molecules of N-dodecyl-N,N-dimethylglycine detergent, and 97 waters. The data have a high Wilson B-factor (62 Å2), which is presumably a consequence of static disorder caused by inconsistent detergent packing between molecules in the crystal lattice. Consequently, the average B-factors for the BLUF domain (17–109) are quite high: 60.2 for the A chain, 71.3 for the B chain, and 73.6 for the C chain.
Flavin Analysis
Since the flavin is oxidized in native AppA, X-ray-induced photoreduction of the flavin during data collection could perturb the dark state structure (28). To address this issue, the MAD data were examined for X-ray dose-dependent changes in electron density since these data were extremely redundant and collected from a single crystal. Data were binned into six complete datasets according to the dose absorbed by the crystal (29). Difference electron density maps between the first and subsequent datasets were examined for evidence of structural changes in the flavin-binding pocket that might arise from photoreduction. Although there were some signs of X-ray radiation damage on the Se-Met and acidic side chains, no significant structural changes were observed in the flavin-binding pocket or on the ribityl side chain and phosphate moiety (data not shown). If there is indeed X-ray-induced photoreduction of the flavin, either it results in a subtle structural change that is not apparent at 2.7 Å resolution or it is occurring rapidly during acquisition of the first binned dataset, which is accompanied by absorption of 8 × 105 Gy.
RESULTS AND DISCUSSION
The dark state AppA BLUF domain was visualized using a native dataset collected to 2.3 Å resolution on crystals of the 17–133 amino acid construct. The initial phase set was calculated from a three-wavelength MAD experiment collected to 2.7 Å on protein expressed with Se-Met (Table 1). Composite omit maps were generated as a check against model bias during refinement. Initial refinement produced an R-factor of 35% and Rfree of 39%. The flavin cofactor and waters were added to the model during the later stages of refinement, which along with further model adjustment reduced the R-factor to 27% and Rfree to 30%. The final R-factor was achieved by the application of TLS refinement (30) using the CCP4 program Refmac5, with each monomer broken into two units (residues 17–109 and 110–130). Coordinates have been deposited in the Protein Data Bank under accession code 1YRX.
Table 1.
Crystallographic Data and Refinement Statistics
| native | Se, peak | Se, inflection | Se, remote | |
|---|---|---|---|---|
| space group | P41212 | P41212 | P41212 | P41212 |
| wavelength (Å) | 0.9 | 0.97939 | 0.97962 | 0.96113 |
| resolution (Å) | 2.30 | 2.70 | 2.70 | 2.70 |
| a = b, c (Å) | 98.7, 127.8 | 98.7, 127.8 | 98.7, 127.8 | 98.7, 127.8 |
| total observations | 377634 | 306126 | 301893 | 445995 |
| unique reflections | 27558 | 31708 | 31649 | 31678 |
| redundancy | 13.7 | 9.7 | 9.5 | 14.1 |
| resolution (Å) (last shell) | 50–2.30 (2.38–2.30) | 50–2.70 (2.80–2.70) | 50–2.70 (2.38–2.70) | 50–2.70 (2.38–2.70) |
| completeness %a | 95.5 (98.4) | 95.5 (97.4) | 95.3 (97.2) | 95.5 (97.6) |
| I/σ(I)a | 24.0 (3.6) | 32.6 (3.5) | 41.3 (4.8) | 41.2 (4.9) |
| Rmergea,b | 6.0 (43.1) | 4.7 (49.3) | 3.9 (35.4) | 3.5 (32.5) |
| Rcrystc | 24.1 | |||
| Rfreed | 27.3 | |||
| no. of ordered waters | 97 | |||
| no. of atoms | 2965 | |||
| RMSD bond lengths (Å) | 0.014 | |||
| RMSD bond angles (deg) | 1.4 | |||
| Ramachandran distribution | ||||
| most favored (%) | 96.8 | |||
| allowed (%) | 99.4 | |||
| PDB accession code | 1YRX | |||
Values in parentheses refer to the last shell.
Rmerge = ΣhklΣi|Ii − 〈I〉|/ΣhklΣiIi for all data.
Rcryst = Σhkl||Fo − Fc||/Σhkl|Fo| includes all data.
Rfree uses 5% of data for the test set.
BLUF Domain Fold
The AppA BLUF domain is an α + β sandwich with two long α-helices located on one side of a primarily antiparallel, five-stranded β-sheet with a slight right-handed twist (Figure 1A). The BLUF domain was identified to have a ferredoxin-like fold using the Structural Classification of Proteins database, SCOP (31). The fold is extremely common and currently contains 48 characterized superfamilies. Ferredoxin-like folds serve a diverse array of function in a variety of proteins and are found as soluble monomers, homodimers, homotrimers, and component domains of larger proteins. The AppA BLUF domain is the first example of the ferredoxin-like fold functioning as a sensor domain via a flavin cofactor. Proteins belonging to the superfamily of FAD-linked oxidases have two domains with a ferredoxin-like fold similar to that in the AppA BLUF domain. However, the flavin cofactor in FAD-linked oxidases is not associated with either of these domains (32). The AppA BLUF domain displays greatest structural homology with bovine acylphosphatase (PDB entry 2ACY) despite having only 14% sequence identity (33), with a 2.0 Å RMSD between 90 corresponding Cα atoms and a Z-score of 10.9 according to SCOP. Acylphosphatases do not bind a cofactor but utilize the ferredoxin-like fold as a conserved phosphate binding motif (34), in which similar proteins bind inorganic ions in the same active site despite little or no sequence homology (35). The AppA BLUF domain has no functional similarity with acylphosphatases; AppA lacks the conserved phosphate binding loop and replaces the tightly packed hydrophobic core in the acylphosphatases with its noncovalently bound flavin chromophore.
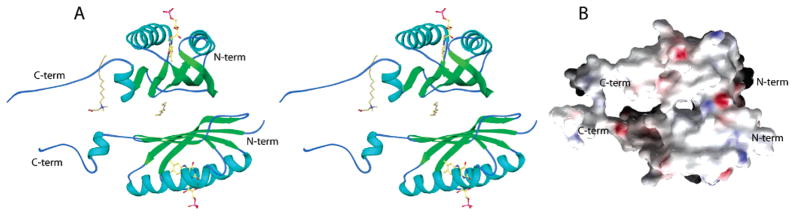
Figure 1.
Protein fold and relative orientation of the A and B chain monomers of AppA forming the noncrystallographic dimer. The C-terminal helix and tail are not part of the BLUF domain. (A) Stereo ribbon diagram including models for the flavin cofactor and two detergent molecules. (B) The dimer in a slightly different orientation with the molecular surface color-coded according to electrostatic potential, red denoting negative and blue positive. The apparent hole between monomers is the location of the partially ordered detergent molecule shown in panel A.
Residues 17–109 in AppA form the compact ferredoxin-like fold, which comprises the flavin-binding BLUF domain. Although amino acids 110–133 are present in this construct, they are only loosely associated with the ferredoxin-like fold and are likely to form part of the C-terminal effector domain in full-length AppA (Figure 1). Residues 112–117 form a solvent-exposed amphipathic helix that does not pack tightly against the rest of the protein. Exposed hydrophobic side chains from this helix are in contact with ordered detergent molecules, explaining the unusual necessity for detergent in the mother liquor. Residues 120–129 have no ordered secondary structure but make extensive hydrogen bonds with the same sequence on another molecule.
Protein Dimerization
As in solution, AppA is found as a dimer in the crystal (Figure 1). There are three molecules in the asymmetric unit; two monomers (the A and B chains) form a noncrystallographic dimer and the third (the C chain) forms half of a crystallographic dimer. The A and B chains are related by a noncrystallographic 2-fold symmetry axis, while two copies of the C chain are related by a 2-fold crystallographic axis. Given this unusual arrangement of dimers, we investigated the possibility of pseudosymmetry by solving the structure in P41 with three sets of dimers per asymmetric unit. However, expanding the model and refining it with reduced symmetry did not improve Rfree nor produce clearer electron density. We conclude that the P41212 space group is correct. The C chain is disordered relative to the A and B chains, with no apparent electron density for eight side chains and very poor electron density for the loop from residues 97–100 that connects the β4 and β5 stands. Applications of NCS constraints between the C chain and the other two chains increased the value of Rfree and were not used.
Two monomers pack together via the exposed side of the β-sheet in the same manner as members of the dimeric α βbarrel superfamily of the ferredoxin-like fold (36). Since eight hydrophobic side chains are exposed on the surface of the β-sheet (Figure 2), the interface is largely hydrophobic; 17% of the surface area is buried in the interface between the A and B chains (2043 Å2), and 14% of the surface area is buried between the two copies of the C chain (1631 Å2). The moderate size of this contact region is on the borderline for consideration of the AppA BLUF domain as an obligate dimer (37), but the interaction between monomers is presumably more extensive in the full-length protein. Although this large hydrophobic patch could potentially interact with the C-terminal domain in native AppA, we assumed that it was buried in the dimer interface since both native AppA and the isolated BLUF domain are dimeric in solution. The C-terminal domain is likely to interact with another hydrophobic patch composed of the Leu31, Leu34, Val38, Met106, and Leu108 side chains. All of these hydrophobic residues are shielded from solvent by either the C3′ helix (not part of the BLUF domain) or ordered detergent.
Figure 2.
Secondary structure of the AppA BLUF domain. Residues that are absolutely conserved in BLUF domains are highlighted in red, and those that are partly conserved with one alternate residue are highlighted in green (3). Arrows designate amino acids in the flavin-binding pocket and double lines those in the dimer interface. The C-terminal extra-domain region (Figure 1) is highlighted in gray.
Ordered detergent molecules are embedded in the hydrophobic interface of both the noncrystallographic and crystallographic dimers. Bound detergent creates the apparent hole in the dimer interface between the two monomers (Figure 1B). Since detergent disturbs the interface, it is not possible to identify the fully native orientation between monomers. Phe101 and Leu91 are both conserved residues buried in the hydrophobic interface (Figure 2). Although neither of these residues aligns with conserved residues in the other monomer, they may serve an important role in defining the dimer orientation in native AppA.
Flavin-Binding Pocket
The flavin cofactor is bound noncovalently in the core of the AppA BLUF domain (Figure 1A). Although there is very clear electron density for the flavin and ribityl side chain, there is no apparent density for the adenine moiety. Despite the fact that FAD is the cofactor in native AppA (11), the shorter BLUF fragment of AppA contains a mixture of flavins when expressed in E. coli (38). Cofactor extracted from freshly purified AppA 17–133 was composed of ~25% FAD, 70% FMN, and 5% riboflavin, while that extracted from crystals was ~20% FAD, 50% FMN, and 30% riboflavin (data not shown). Thus, the native FAD cofactor has been substantially replaced by FMN during expression, and the FMN was subsequently hydrolyzed to riboflavin during crystallization.
Despite the presence of ~20% FAD in the crystal, there is no ordered electron density for the adenine moiety in any of the three monomers. The N-terminus is located near the bound cofactor, and residues 1–16 may interact with the adenine and stabilize its position in full-length AppA. Density corresponding to the FMN phosphate moiety is more prominent in the C chain, indicating that it may have either lower B-factors or a higher occupancy due to a reduced rate of hydrolysis from FMN to riboflavin at this position in the crystal lattice. The ribityl side chain is stabilized by a hydrogen bond between Asp82 and the O2* hydroxyl, a hydrogen bond between Arg84 and the O3* hydroxyl, and a salt bridge between His78 and the phosphate moiety. Asp82 and Arg84 are both highly conserved residues, and His78 is a conserved polar residue that is positively charged in >50% of BLUF domains.
Not surprisingly, most of the highly conserved residues found in all BLUF domains surround the flavin cofactor (Figure 2). The flavin dimethylbenzene ring is primarily surrounded by hydrophobic side chains, while the pyrimidine ring is involved in an intricate hydrogen bond network with adjacent residues. Ile37, Phe61, His85, and the aliphatic portion of the Arg84 side chain surround the hydrophobic side of the flavin. Arg84 and His85 are highly conserved residues, Phe61 is present in BLUF homologues as either phenylalanine or isoleucine, and Ile37 is a conserved hydrophobic residue (3). The side chains of His44, Asn45, and Gln63 make four hydrogen bonds to the flavin. Asn45 and Gln63 are both highly conserved, and His44 is a conserved polar residue (3).
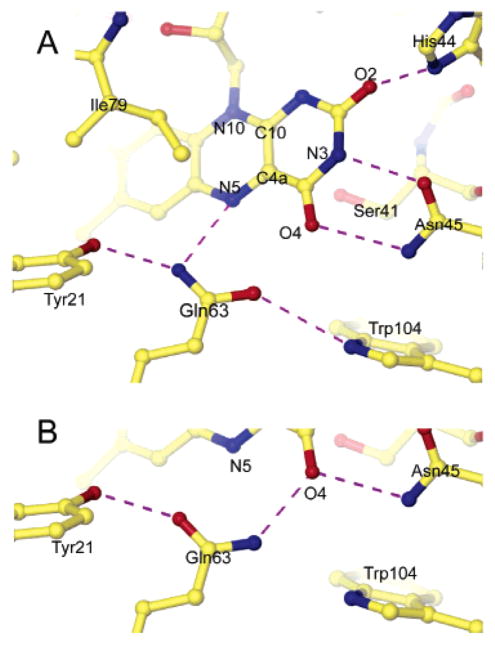
Gln63 appears to play a central role in AppA since its side chain has the potential to make hydrogen bonds to both the flavin and two other highly conserved residues, Tyr21 and Trp104. The Gln63 side chain can make two different sets of hydrogen bonds depending on its orientation (Figure 3), but this orientation cannot be directly determined from the electron density at this resolution nor by context from its position in the protein. Although both are sterically reasonable, Figure 3A shows the most likely orientation of Gln63 in the dark state. This orientation is presumably more stable because of an additional hydrogen bond to Gln63. Spectroscopic evidence also predicts the formation of a new hydrogen bond to the O4 position of the flavin upon photoactivation, which is consistent with the hydrogen bond rearrangement depicted in Figure 3B (39, 40). In the presumed dark orientation (Figure 3A), the Gln63 ε2N donates hydrogen bonds to the flavin N5 and Tyr21 ηO, while the Gln63 ε1O accepts a hydrogen bond from the ε1N of the highly conserved Trp104 (Figure 3A). In the putative light-excited orientation of the Gln63 side chain (Figure 3B), the hydrogen bonds to the flavin N5 and Trp104 are broken. A new hydrogen bond to the flavin O4 is made in lieu of that to N5, while Gln63 serves as a hydrogen bond acceptor for the Tyr21 ηO rather than a donor. This orientation would also be preferred if X-rays photoreduce FAD to FADH− since the protonated flavin N5 could now donate an additional hydrogen bond to the Gln63 ε1O.
Figure 3.
Hydrogen bond network to the flavin. Hydrogen bonds are shown as dashed lines. (A) Hydrogen bond network with the presumed dark state orientation of Gln63. (B) Alternate hydrogen bond network with flipped orientation of Gln63.
The highly conserved Asn45 side chain makes two hydrogen bonds to the flavin pyrimidine ring in both orientations of Gln63. The Asn45 δ2N donates a hydrogen bond to the flavin O4 carbonyl oxygen, while the Asn45 δ1O accepts a hydrogen bond from the flavin N3. Ser41 is a partly conserved residue that is always found as either serine or alanine. This residue probably does not play an important functional role in AppA, but its position under the plane of the isoalloxazine ring allows it to make a weak hydrogen bond to the center of the flavin pyrimidine ring.
His44 is a conserved polar residue that donates a hydrogen bond from the δ1N to the flavin O2 carbonyl oxygen, a hydrogen bond that could still be formed by most of the alternate residues at this weakly conserved position. The position of the His44 side chain places a positive charge adjacent to the N1–C2=O2 locus of the flavin. A positive charge in this position is found in nearly all flavoenzymes (41), where it is important in modulating the reduction potential of the flavin and stabilizes the anionic form of reduced flavin (42). Interestingly, a positive charge is also found at this position in >50% of BLUF domains, with the other BLUF domains have positively charged residues near this location in the structure that could potentially interact with the flavin. Fast transient absorption spectroscopy studies (manuscript in preparation) on the AppA BLUF domain and its photoinactive Y21F mutant suggest that a fast electron transfer from Tyr21 to the flavin takes place upon photo-excitation, followed by charge recombination on a picosecond time scale. The positive charge near the N1–C2=O2 locus may therefore be important for stabilization of a transient negative charge on the flavin during this process.
Flavin-Binding Photoreceptors
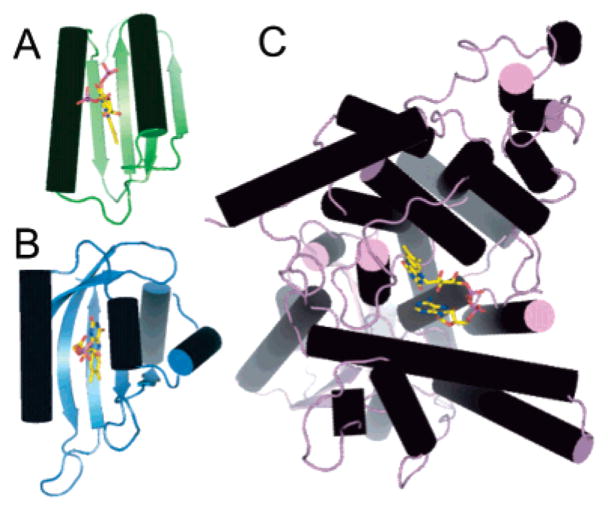
Cryptochrome, phototropin, and AppA are all blue light photoreceptors that utilize flavin as a chromophore (16, 17). Cryptochromes sense blue light with an N-terminal photolyase homology (PHR) domain. PHR domains are structurally similar to DNA photolyase but utilize light absorption by the FAD cofactor for blue light signaling rather than DNA repair (43). The AppA BLUF domain has no structural similarity to photolyase or PHR domains in either the overall fold or FAD binding pocket (Figure 4). The cryptochrome CRY-1 PHR domain from Arabidopsis thaliana (PDB entry 1U3C) binds the FAD cofactor in its purely helical α-domain (44). Its flavin makes several hydrophobic and hydrogen bond interactions with the protein backbone; there are no such interactions in AppA. There is no positive charge directly adjacent to the flavin N1–C2=O2 locus, but a salt bridge above the plane of the flavin pyrimidine ring may have a significant effect on redox potential.
Figure 4.
Overall protein fold of flavin-binding blue light photoreceptors: (A) AppA BLUF domain in green, (B) phy3 LOV2 in blue (PDB entry 1G28), and (C) cryptochrome 1 Phr domain in purple (PDB entry 1U3C).
Although the ferredoxin-like fold of AppA has no structural homology with the LOV domain subclass of the PAS family, there are notable similarities in the overall topology (Figure 4) and flavin-binding interactions (Figure 5) of these two flavin-containing photoreceptors. Both domains bind the flavin between two helices with the cofactor perpendicular to a five-stranded antiparallel β-sheet. The interactions between the protein and flavin are also remarkably similar. The flavin in both the AppA BLUF domain and phy3 phototropin LOV2 domain (PDB entry 1G28) from the fern Adiantum (19) is held via hydrophobic and hydrogen bond interactions with the surrounding amino acid side chains. Hydrophobic side chains are in very similar locations relative to the flavin: Ile37, Phe61, Arg84, and His85 in AppA correspond to Phe1010, Leu1012, Phe1025, and Thr934 in phy3 LOV2. The hydrogen bond network to the flavin in AppA is also similar to that in phy3 LOV2 but is more extensive (Figure 5). Asn45 in AppA replaces the two hydrogen bonds from Asn998 δ1O and Asn1008 δ2N in phy3 LOV2, the His44 δ1N replaces the hydrogen bond from the Asn998 δ2N, and the Gln63 ε2N replaces the hydrogen bond from the Gln1029 δ2N. Although there are no residues in phy3 LOV2 with the ability to readily form different sets of hydrogen bonds to the flavin via reorientation of a side chain, Gln1029 in phy3 LOV2 may play a similar role (Figure 5). Gln1029 is hydrogen bonded to the flavin O4 position in the ground state but switches its hydrogen bond to the N5 position when the flavin moves during photoactivation (45). This motion breaks a hydrogen bond from Gln1029 to Ser930, a residue that although not highly conserved is usually a hydrophilic residue capable of making a hydrogen bond to Gln1029 in the ground state.
Figure 5.
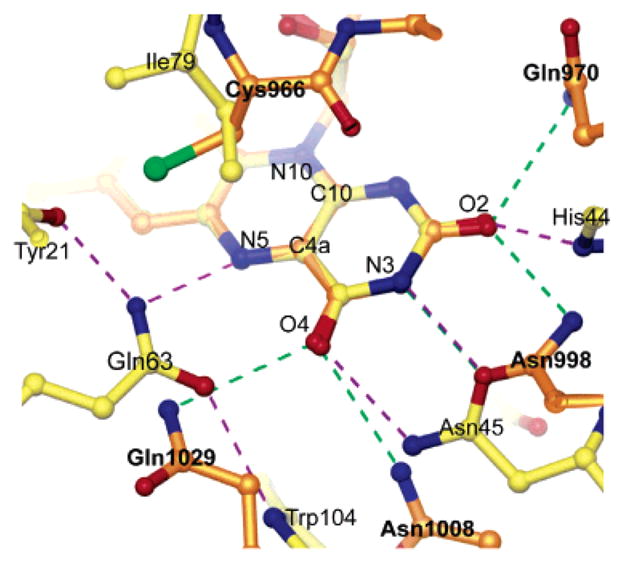
Overlay of hydrogen bond network to the flavin in AppA and phy3 LOV2 (PDB entry 1G28). The structures were aligned by superimposing the isoalloxazine rings. Hydrogen bonds are shown as dashed lines. AppA, yellow atoms with purple hydrogen bonds; LOV2, orange atoms with green hydrogen bonds.
The most obvious structural differences between the AppA BLUF domain and phy3 LOV2 are likely to be of functional significance. The long α1 helix in the AppA BLUF domain is replaced by the functionally important 310 α′A helix in phy3 LOV2. Photoactivation of phy3 LOV2 leads to the creation of a transient covalent adduct between Cys966 and the flavin C4a atom (45). The catalytic active site on the flavin is located above the plane of the central isoalloxazine ring, in the region between the lines connecting N5 to N10 and C4a to C10 (41). Cys966 is located directly above this region of the flavin in phy3 LOV2, a region that is occupied by the highly conserved but chemically inert Ile79 in AppA (Figure 5). Photoactivation in AppA is believed to be accompanied by changes to the hydrogen bond network with the flavin; it is therefore not surprising that the extended hydrogen bond network in AppA is more extensive than that in phy3 LOV2. There are no charged interactions with the flavin pyrimidine ring in phy3 LOV2 that would modulate the redox potential as in AppA.
Photoactivation in AppA
Even though a detailed understanding of the mechanism of photoactivation in BLUF domains remains to be determined, some insight can be provided from the dark state AppA BLUF structure coupled with solution spectroscopy. Difference NMR spectra between the photoexcited and dark states of AppA are consistent with changes to either the aromatic stacking or hydrogen bonding with the flavin isoalloxazine ring. Results from mutational analysis to Tyr21 led to the conclusion that this residue was involved in aromatic stacking to the flavin (12). Although Tyr21 is connected to the flavin via a hydrogen bond network, Tyr21 is too distant to pack against the flavin without major structural rearrangement. Since there are no obvious candidates to participate in aromatic stacking with the flavin, photoactivation must proceed via a rearrangement of the hydrogen bond network to the flavin. Difference FTIR spectra between the photoexcited and dark states of both AppA and the BLUF containing Slr1694 protein in Synochocystis imply that light absorption by the flavin strengthens a hydrogen bond to the flavin O4 carbonyl oxygen (39, 40).
In view of the position of Gln63 and its ability to readily create different sets of hydrogen bonds via reorientation of its side chain, this residue is very likely a critical player in the photoactivation process. Light-induced transient reduction of the flavin N5 would induce a reorientation of the Gln63 side chain to create the new hydrogen bond to the flavin O4 carbonyl oxygen as predicted by difference FTIR (39, 40), in addition to altering hydrogen bonding to the Tyr21 side chain implicated in AppA photoactivation via difference NMR (12). Reorientation of Gln63 causes the side chain to exchange its hydrogen bond to the flavin N5 atom for one to the flavin O4 carbonyl oxygen, while breaking its hydrogen bond to the highly conserved Trp104 side chain. Trp104 is part of the β5 strand at the C-terminus of the AppA BLUF domain. The residue lies beneath the plane of the chromophore, with its indole side chain in hydrophobic contact with Ser41, Gln42, Leu54, and Met106. The Trp104 side chain is close to the Val38 and Met106 side chains, residues that are involved in the hydrophobic patch presumably in contact with the attached C-terminal effector domain in full-length AppA. Breaking the anchor tying Trp104 to the flavin will allow increased motion in the β5 strand, a structural change that may be important in creating the global motion preventing light-activated AppA from interacting with PpsR.
Acknowledgments
We acknowledge the staff at BioCARS for help with data collection. Thanks also to Dr. Samantha Perez Miller for help with crystal freezing, Dr. Lingling Chen for the use of crystallographic equipment and useful suggestions, and Dr. Bogdan Dragnea for many fruitful discussions.
Footnotes
Abbreviations: BLUF, blue light sensing using FAD; PAS, Per-ARNT-Sim; LOV, light, oxygen, voltage.
This work was supported by National Institutes of Health Grants GM36452 (K.M.) and GM53940 (C.B.) and a Grant-in-Aid for Young Scientists (B) 16770046 from MEXT of Japan (S.M.). BioCARS is supported by NIH Grant RR07707 (K.M.) and the Advanced Photon Source by the Department of Energy.
The atomic coordinates for the BLUF domain have been deposited in the Protein Data Bank (PDB ID 1YRX).
References
- 1.Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- 2.Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 3.Gomelsky M, Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 4.Lin C. Plant blue-light receptors. Trends Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- 5.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han Y, Braatsch S, Osterloh L, Klug G. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci USA. 2004;101:12306–12311. doi: 10.1073/pnas.0403547101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei JM, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 9.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;204:213–214. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 10.Gomelsky M, Kaplan S. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J Biol Chem. 1998;273:35319–35325. doi: 10.1074/jbc.273.52.35319. [DOI] [PubMed] [Google Scholar]
- 11.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 12.Kraft BJ, Masuda S, Kikuchi J, Dragnea V, Tollin G, Zaleski JM, Bauer CE. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: a novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 2003;42:6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- 13.Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 14.Masuda S, Hasegawa K, Ishii A, Ono T. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of Synechocystis sp. PCC6803. Biochemistry. 2004;43:5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- 15.Gauden M, Yeremenko S, Laan W, van Stokkum IH, Ihalainen J, van Grondelle R, Hellingwerf KJ, Kennis JT. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 2005;44:3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- 16.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 17.Christie JM, Briggs WR. Blue light sensing in higher plants. J Biol Chem. 2001;276:11457–11460. doi: 10.1074/jbc.R100004200. [DOI] [PubMed] [Google Scholar]
- 18.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 19.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 21.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science. 1995;269:968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 22.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McRee DE. XtalView Xfit–A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Kort R, Komori H, Adachi S, Miki K, Eker A. DNA apophotolyase from Anacystis nidulans: 1.8 angstrom structure, 8-HDF reconstitution and X-ray-induced FAD reduction. Acta Crystallogr D. 2004;60:1205–1213. doi: 10.1107/S0907444904009321. [DOI] [PubMed] [Google Scholar]
- 29.Murray JW, Garman EF, Ravelli RBG. X-ray absorption by macromolecular crystals: the effects of wavelength and crystal composition on absorbed dose. J Appl Crystallogr. 2004;37:513–522. [Google Scholar]
- 30.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 31.Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraaije MW, van den Heuvel RHH, van Berkel WJH, Mattevi A. Structural analysis of flavinylation in vanillyl-alcohol oxidase. J Biol Chem. 2000;275:38654–38658. doi: 10.1074/jbc.M004753200. [DOI] [PubMed] [Google Scholar]
- 33.Thunnissen MM, Taddei N, Liguri G, Ramponi G, Nordlund P. Crystal structure of common type acylphosphatase from bovine testis. Structure. 1997;5:69–79. doi: 10.1016/s0969-2126(97)00167-6. [DOI] [PubMed] [Google Scholar]
- 34.Swindells MB, Orengo CA, Jones DT, Pearl LH, Thornton JM. Recurrence of a binding motif. Nature. 1993;362:299–299. doi: 10.1038/362299a0. [DOI] [PubMed] [Google Scholar]
- 35.Rosenzweig AC, O’Halloran TV. Structure and chemistry of the copper chaperone proteins. Curr Opin Chem Biol. 2000;4:140–147. doi: 10.1016/s1367-5931(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 36.Lytle BL, Peterson FC, Kjer KL, Frederick RO, Zhao Q, Thao S, Bingman C, Johnson KA, Phillips GN, Volkman BF. Structure of the hypothetical protein At3g17210 from Arabidopsis thaliana. J Biomol NMR. 2004;28:397–400. doi: 10.1023/B:JNMR.0000015385.54050.19. [DOI] [PubMed] [Google Scholar]
- 37.Nooren IMA, Thornton JM. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laan W, van der Horst MA, van Stokkum IH, Hellingwerf KJ. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine di-nucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: A key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem Photobiol. 2003;78:290–297. doi: 10.1562/0031-8655(2003)078<0290:icotpp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa K, Masuda S, Ono TA. Structural intermediate in the photocycle of a BLUF (sensor of blue light using FAD) protein Slr1694 in a cyanobacterium Synechocystis sp. PCC6803. Biochemistry. 2004;43:14979–14986. doi: 10.1021/bi048671n. [DOI] [PubMed] [Google Scholar]
- 40.Masuda S, Hasegawa K, Ono TA. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44:1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- 41.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 42.Ghisla S, Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 43.Deisenhofer J. DNA photolyases and cryptochromes. Mutat Res. 2000;460:143–149. doi: 10.1016/s0921-8777(00)00023-9. [DOI] [PubMed] [Google Scholar]
- 44.Brautigam CA, Smith BS, Ma ZQ, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]