Abstract
A dual pressure linear ion trap mass spectrometer was modified to permit infrared multiphoton dissociation (IRMPD) in each of the two cells - the first a high pressure cell operated at nominally 5 × 10-3 Torr and the second a low pressure cell operated at nominally 3 × 10-4 Torr. When IRMPD was performed in the high pressure cell, most peptide ions did not undergo significant photodissociation; however, in the low pressure cell peptide cations were efficiently dissociated with less than 25 ms of IR irradiation regardless of charge state. IRMPD of peptide cations allowed the detection of low m/z product ions including the y1 fragments and immonium ions which are not typically observed by ion trap collision induced dissociation (CID). Photodissociation efficiencies of ~100% and MS/MS (tandem mass spectrometry) efficiencies of greater than 60% were observed for both multiply and singly protonated peptides. In general, higher sequence coverage of peptides was obtained using IRMPD over CID. Further, greater than 90% of the product ion current in the IRMPD mass spectra of doubly charged peptide ions was composed of singly charged product ions compared to the CID mass spectra in which the abundances of the multiply and singly charged product ions were equally divided. Highly charged primary product ions also underwent efficient photodissociation to yield singly charged secondary product ions, thus simplifying the IRMPD product ion mass spectra.
INTRODUCTION
As the field of proteomics continues to expand, there has become a growing need for more advanced mass spectrometric techniques for the characterization of biological molecules.1, 2 Tandem mass spectrometry methods have played a crucial role in the sequencing of proteolytic peptides and identification of proteins.3, 4 The most common means of fragmenting gas-phase peptide ions is collision induced dissociation (CID) in which selected ions gain internal energy through collisions with rare gas atoms.5 However, the amino acid sequence and charge state can greatly influence dissociation; CID is often limited by incomplete backbone fragmentation, preferential backbone cleavage at proline or acidic residues,6, 7 or dominant loss of labile groups which results in low peptide sequence coverage.8 These limitations of CID have sparked interest in the development of alternative and more universal ion activation and dissociation methods in recent years.9, 10 New electron-based activation methods such as electron capture dissociation (ECD)11 and electron transfer dissociation (ETD)12 have shown significant promise as each method maintains labile side-chain modifications during ion activation; however, both techniques are limited to the analysis of multiply charged peptides.13 More recently, electron ionization dissociation (EID) has been developed which utilizes fast electrons to dissociate singly charged peptide cations but the backbone fragment yields obtained were generally less than 5%.14
There has also been considerable interest in exploiting photons for ion activation, including infrared multiphoton dissociation (IRMPD),15-19 ultraviolet photodissociation (UVPD),20-24 and more recently the use of femtosecond pulsed lasers,25 and hybrid UVPD-CID methods.26 One of the primary advantages of performing IRMPD in quadrupole ion traps (QITs)16-19, 27-29 is the ability to fine-tune the energy deposition by varying the irradiation time or number of laser pulses, the laser power or flux, and wavelength. In addition, compared to CID, photodissociation is largely independent of the rf trapping voltage during ion activation which provides a broader m/z trapping range,18, 30 an advantage also recognized for ETD. Energy deposition by conventional CID is more efficient at higher rf trapping voltages (i.e., q-values) which prevents storage of product ions of m/z below the low-mass cut-off (LMCO), which is approximately the lower 28% of the m/z trapping range relative to the precursor ion. CID can be performed at lower q-values but with lower dissociation efficiencies. In addition, pulsed Q-dissociation (PQD)31 and high amplitude short time excitation (HASTE)32 have shown promise as alternative collision based fragmentation methods which do not suffer as greatly from the low-mass cut-off problem; however both of these techniques afford modest MS/MS efficiencies. IRMPD can be efficiently performed at low rf trapping voltages because the activation process is not defined by the rf trapping parameters, allowing observation of many informative low-mass fragments including immonium ions and y1 fragment ions of peptides. Photodissociation is also a non-resonant process in which the precursor ion and all product ions are irradiated and activated simultaneously; thus, dead-end dissociation pathways such as dehydration are largely circumvented as these primary product ions can undergo secondary dissociation to yield more diagnostic fragments. In addition, the photon absorption process does not affect the translational motion of ions and alleviates ion losses due to unstable trajectories that might be encountered in traditional CID.
Infrared multiphoton dissociation has not been widely implemented in quadrupole ion traps due to low fragmentation efficiencies of certain analytes including peptides and proteins. For optimal performance, quadrupole ion trap mass spectrometers are typically operated at a pressure of ~1 - 5 mTorr of He to effectively trap ions and reduce their kinetic energy via collisions with the buffer gas atoms.33 This high pressure, however, can cause ion deactivation through collisional cooling at a nearly equivalent or greater rate than that of ion activation by IR photon absorption, ultimately resulting in the observation of low dissociation efficiencies for IRMPD.10, 34 Several methods have been developed to improve the efficiency of IRMPD in quadrupole ion traps. The pressure in the QIT has been adjusted during the scan function through pulsed gas introduction35 or dynamic pressure control36 in which the bath gas is introduced during ion accumulation and then pumped away prior to IR irradiation. Payne et al. utilized thermally-assisted IRMPD30 and Hashimoto and coworkers collisionally-activated the precursor ions during IR irradiation37 to increase the internal energy of the ions to provide efficient photodissociation at standard QIT operating pressures. Newsome and Glish recently reported focusing the IR laser to the center of the ion trap as a simple means to improve IRMPD of peptides by increasing the overlap of the ion cloud with the laser beam and exposing the ions to a higher photon flux.19 Previous work in the Brodbelt group has focused on the development of chemical derivatization techniques that attach chromogenic labels to increase the IR absorption cross sections or reduce the critical energies for dissociation.38-40 Super-charged peptides can also be efficiently photodissociated in a linear quadrupole ion trap due to a reduction in the dissociation threshold and an increase in the number of mobile protons.41
Here we demonstrate that unmodified peptides can be efficiently and rapidly dissociated by IR irradiation in the low pressure cell of a dual pressure linear ion trap (LIT). The dual pressure LIT comprises a low pressure cell and high pressure cell to combine the advantages of both pressure regimes.42 High trapping, isolation, and CID efficiencies are obtained in the high pressure cell (~5×10-3 Torr), while the low pressure cell (~3×10-4 Torr) offers improved scanning speed and mass resolution. Little photodissociation of peptide ions occurs in the high pressure cell, while highly efficient IRMPD is observed in the low pressure cell yielding complete dissociation of the precursor ions with less than 25 ms of irradiation. Higher sequence coverage of peptides is obtained by performing IRMPD in the low pressure cell as compared to traditional CID in the high pressure cell.
EXPERIMENTAL SECTION
Chemicals and Reagents
All materials and peptides were used without further purification. The peptides RPKPQQFFGLM-NH2 (substance P), FSWGAEGQR, YGGFLK, ASHLGLAR, and AGCKNFFWKTFTSC (somatostatin-14) were obtained from Bachem (King of Prussia, PA). DRVYIHPF (angiotensin II), ADSGEGDFLAEGGGVR (fibrinopeptide A), and RPPGFSPFR (bradykinin) were purchased from American Peptide Co. (Sunnyvale, CA). All solvents and other chemicals including melittin (honey bee venom) were from Sigma-Aldrich (St. Louis, MO).
Mass Spectrometry and Infrared Multiphoton Dissociation
All samples were prepared at ~10 μM in 49.5:49.5:1 H2O/MeOH/acetic acid (v/v/v) and infused at 3 μL/min using the standard electrospray ionization source. Unless indicated otherwise, all mass spectrometry experiments were performed on a modified LTQ Velos dual pressure linear ion trap (Thermo Fisher Scientific, San Jose, CA). A schematic of the instrument is shown in Supporting Information Figure 1; details of the instrument have been described previously.42 In brief, the octapole preceding the linear ion trap was reduced in length to accommodate an additional linear ion trap. The two traps are of a symmetrical design and have independent DC power supplies for the six total sections, but have a single common rf power supply. The pressure of each cell of the dual pressure LIT was regulated by the conductance limits of the front, center, and back lenses. The high pressure cell was maintained at 5 to 6 mTorr He and the low pressure cell at ~0.3 mTorr He. Precursor ions were isolated and cooled in the high pressure cell while mass analysis was performed in the low pressure cell; ion activation and dissociation could be performed in either cell. The ion source optics include a progressively spaced stacked ring ion guide for the improved ion transmission.43 Photodissociation was implemented on this dual LIT in a manner similar to that described previously.44 The back flange of the mass spectrometer was modified with a ZnSe window to transmit IR irradiation through a 1.8 mm stainless steel aperture on-axis with the dual pressure LIT. For all IRMPD experiments, the laser power was maintained at 100% (50 W); both the irradiation time (0 - 250 ms) and the q-value (0.05 - 0.25) of the precursor ion were varied. Collision-induced dissociation experiments were conducted solely in the high pressure cell; ions were activated for 30 ms at a q-value of 0.25. IRMPD was performed with a model 48-5 Synrad 50-W CO2 continuous wave laser (Mukilteo, WA). Other time-resolved IRMPD and MS3 experiments were performed on a standard LTQ XL mass spectrometer (i.e. single trap) modified for photodissociation operating at ~2.5 mTorr He.
Data Analysis
IRMPD was performed in each of the two cells. Dissociation efficiencies were calculated according to the following formula:
| (1) |
where P is the abundance of the surviving precursor ion after ion activation, and Fi is the abundance of each product ion. The efficiency of the tandem MS experiment (i.e., MS/MS efficiency) is defined below:
| (2) |
where P0 is the abundance of the precursor ion prior to any IR irradiation or collisional activation, and Fi is the abundance of each product ion.45 The sum of the product ion abundance (i.e., ΣFi) was determined by subtracting the abundance of any remaining precursor ion from the total ion abundance for the efficiency calculations. Ion abundances were determined by calculating the peak areas using Origin 7.0. Time-resolved IRMPD experiments were performed by incrementally increasing the irradiation time and the abundances of different ion types were plotted against irradiation time. Similarly, q-value resolved IRMPD was performed by sequentially raising the activation q-value and acquiring IRMPD mass spectra at each q-value. All time-resolved and q-value resolved IRMPD experiments were performed in triplicate. The precursor ion abundance was plotted relative to the initial precursor abundance without any ion activation or irradiation (P0), while the product ion abundances were plotted relative to the total ion current (sum of all fragment ions and precursor, P) in each IRMPD mass spectrum. The abundances and number of product ions in each charge state were also determined by summing the abundances (i.e., peak areas) or counting the number of product ions in each charge state. If a peak’s charge state could not be resolved, its area was not included in the abundance calculation nor was it counted. All spectra are an average of ten scans. Peptide fragment nomenclature includes superscripts “*” and “°” to designate ammonia loss and water loss, respectively. Immonium ions are labeled with the single letter amino acid residue with a subscript i.
RESULTS AND DISCUSSION
IRMPD in the Low Pressure Cell vs. High Pressure Cell
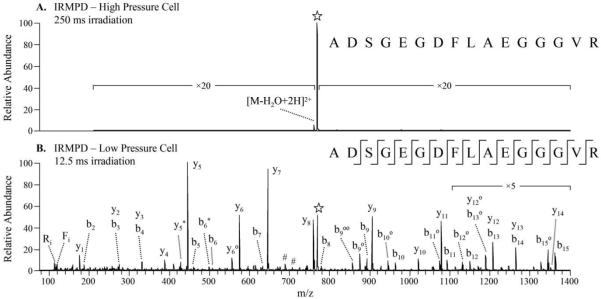
The primary drawback to performing IRMPD in quadrupole ion traps has been low dissociation efficiencies due to competition between ion activation through IR photon absorption and deactivation through collisions with the buffer gas. To evaluate the performance of IRMPD in the low pressure cell of the dual pressure LIT, a series of model peptides were analyzed. Product ion mass spectra of each peptide in various charge states were acquired by performing IRMPD in each of the two cells. IRMPD of doubly protonated ADSGEGDFLAEGGGVR (fibrinopeptide A) performed in the high pressure cell using 250 ms of irradiation (Figure 1A) resulted in less than 1.5% dissociation efficiency (precursor to product); in fact, only a single product ion was observed corresponding to dehydration of the precursor ion. The results in the high pressure cell mirror those obtained in a single linear ion trap operated at ~2.5 mTorr He (using the same laser and similar set-up) in which peptides were not observed to photodissociate even with long irradiation times of 250 ms44 due to the high rate of collisional cooling. In contrast, when IRMPD was performed in the low pressure cell, only 12.5 ms of irradiation was required to dissociate the precursor to less than 6% relative abundance (Figure 1B). All possible b-type ions and 14 out of 15 possible y-type ions were observed in the IRMPD tandem mass spectrum acquired in the low pressure cell. Low m/z products including immonium ions, which can be produced through secondary dissociation of larger b- and y-type ions, are observed. The non-resonant nature of IRMPD also resulted in a high degree of secondary neutral losses from b- and y-type fragment ions, including multiple losses of water (e.g., b9o and b9oo in Figure 1B).
Figure 1.
IRMPD of [ADSGEGDFLAEGGGVR + 2H]2+ of m/z 769.2 performed in the (A) high pressure cell (250 ms irradiation, q = 0.25, 50 W) and (B) low pressure cell (12.5 ms irradiation, q = 0.09, 50 W). The precursor ion is indicated with a star ([✰]) and internal ions are labeled with #.
The IRMPD efficiencies obtained for each of the model peptides when dissociated in the low pressure and high pressure cell are shown in Table 1. In the high pressure cell, only three of the peptide ions ([RPPGFSPFR + 3H]3+, [RPKPQQFFGLM-NH2 + 3H]3+, and [ADSGEGDFLAEGGGVR + 3H]3+) exhibited a photodissociation efficiency of greater than 50%, even using a long irradiation time of 250 ms and a more optimum q-value of 0.25 which should promote better overlap between the ion cloud and the laser beam (discussed in more detail in a later section). The greatest IRMPD efficiencies in the high pressure cell were obtained for three peptides in their highest charge states, an observation consistent with previous work on super-charged peptide cations,41 while a majority of the lower-charged peptide cations exhibited dissociation efficiencies of less than 5% in the high pressure cell. When performing IRMPD in the low pressure cell, only one of the peptides dissociated with an efficiency less than 70%. All of these IRMPD mass spectra were acquired using less than 25 ms irradiation and using low q-values, the latter factor allowing successful storage and detection of the very lowest m/z product ions. In fact, 23 of the 25 peptide precursor ions displayed dissociation efficiencies of greater than 90% at q-values of 0.10 or lower. In general, the more highly charged peptides require shorter irradiation times to yield dissociation efficiencies of ~90%; however, to a lesser extent the q-value of the precursor ion also influences the irradiation time necessary to produce high IRMPD efficiencies.
Table 1.
IRMPD efficiencies of model peptides dissociated in the low pressure and high pressure cells of the dual LIT
| Peptide | MW (Da) |
Charge State |
Low Pressure Cell | High Pressure Cell | ||||
|---|---|---|---|---|---|---|---|---|
| Irradiation Time (ms) |
q-value | Dissociation Efficiency (%)a |
Irradiation Time (ms) |
q-value | Dissociation Efficiency (%)a |
|||
| YGGFLK | 638.4 | 2+ | 15.0 | 0.10 | 98 | 250.0 | 0.25 | 2.6 |
| 1+ | 25.0 | 0.10 | 94 | 250.0 | 0.25 | 1.5 | ||
| 1+ | 35.0 | 0.07 | 6 | --- | --- | --- | ||
| 1+ | 50.0 | 0.07 | 94 | --- | --- | --- | ||
| ASHLGLAR | 823.5 | 2+ | 12.5 | 0.10 | 93 | 250.0 | 0.25 | 2.9 |
| 1+ | 40.0 | 0.06 | 70 | 250.0 | 0.25 | 3.0 | ||
| 1+ | 50.0 | 0.06 | 82 | --- | --- | --- | ||
| FSWGAEGQR | 1035.5 | 2+ | 12.5 | 0.09 | 98 | 250.0 | 0.25 | 6.3 |
| 1+ | 25.0 | 0.06 | 80 | 250.0 | 0.25 | 0.8 | ||
| DRVYIHPF (angiotensin II) |
1045.5 | 3+ | 7.5 | 0.10 | 97 | 250.0 | 0.25 | 26 |
| 2+ | 15.0 | 0.10 | 96 | 250.0 | 0.25 | 1.3 | ||
| 1+ | 25.0 | 0.10 | 92 | 250.0 | 0.25 | 4.3 | ||
| RPPGFSPFR (bradykinin) |
1059.6 | 3+ | 7.5 | 0.10 | 98 | 250.0 | 0.25 | 94 |
| 2+ | 17.5 | 0.09 | 92 | 250.0 | 0.25 | 0.9 | ||
| 1+ | 20.0 | 0.09 | 96 | 250.0 | 0.25 | 4.9 | ||
| RPKPQQFFGLM-NH2 (substance P) |
1346.7 | 3+ | 5.0 | 0.10 | 97 | 250.0 | 0.25 | 96 |
| 3+ | --- | --- | --- | 50.0 | 0.10 | 87 | ||
| 2+ | 17.5 | 0.07 | 93 | 250.0 | 0.25 | 2.6 | ||
| 1+ | 30.0 | 0.07 | 96 | 250.0 | 0.25 | 2.7 | ||
| ADSGEGDFLAEGGGVR (fibrinopeptide A) |
1535.7 | 3+ | 12.5 | 0.09 | 99 | 250.0 | 0.25 | 57 |
| 2+ | 25.0 | 0.06 | 99 | 250.0 | 0.25 | 1.3 | ||
| 2+ | 12.5 | 0.09 | 94 | --- | --- | --- | ||
| 1+ | 25.0 | 0.06 | 91 | 250.0 | 0.25 | 3.0 | ||
| AGCKNFFWKTFTSC (somatostatin-14) |
1636.7 | 3+ | 12.5 | 0.09 | 97 | 250.0 | 0.25 | 5.4 |
| 2+ | 13.5 | 0.09 | 90 | 250.0 | 0.25 | 0.9 | ||
| 2+ | 22.5 | 0.06 | 90 | --- | --- | --- | ||
| 1+ | 25.0 | 0.06 | 93 | 250.0 | 0.25 | 22 | ||
| GIGAVLKVLTTGLPALIS WIKRKRQQ-NH2 (melittin) |
2844.8 | 5+ | 9.0 | 0.09 | 96 | 250.0 | 0.25 | 17 |
| 4+ | 20.0 | 0.07 | 98 | 250.0 | 0.25 | 1.3 | ||
| 3+ | 15.0 | 0.10 | 96 | 250.0 | 0.25 | 3.0 | ||
| 2+ | 25.0 | 0.07 | 98 | 250.0 | 0.25 | 14 | ||
Dissociation efficiencies were calculated using eq. (1) for each individual IRMPD mass spectrum.
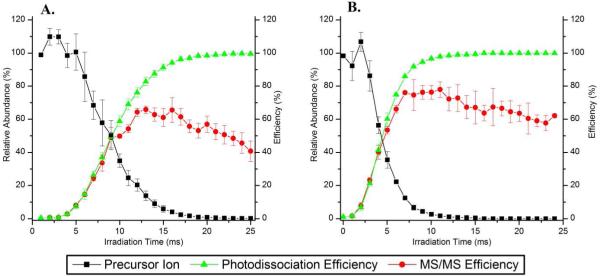
The time-resolved IRMPD plots in Figure 2 (for DRVYIHPF) and Supporting Information Figure S-2 (for ADSGEGDFLAEGGGVR) also show the effect of irradiation time on the photodissociation and MS/MS efficiencies. As shown in Figure 2A, [DRVYIHPF + 2H]2+ required less than 20 ms of irradiation to yield ~100% photodissociation efficiency and the corresponding triply charged precursor only required ~12 ms of irradiation (Figure 2B). The more highly charged peptides dissociated more readily and this is attributed to higher Coulombic repulsion and greater proton mobility. For ADSGEGDFLAEGGGVR (fibrinopeptide A), complete dissociation of the singly charged precursor ion required less than 25 ms of irradiation, and the doubly charged analog required ~20 ms of irradiation at a q-value of 0.10 (Supporting Information Figures S-2B and S-2D, respectively). At lower q-values, longer irradiation times were required to dissociate fibrinopeptide A, indicating a q-value dependence on the photodissociation efficiencies (Supporting Information Figures S-2A and S-2C). For both of these peptides - fibrinopeptide A and angiotensin II - maximum MS/MS efficiencies between 48% and 78% were obtained using a q-value of 0.10. The maximum MS/MS efficiencies were obtained when the dissociation efficiencies were between 75% and 96%. With longer irradiation times, the MS/MS efficiencies began to steadily decrease which was a result of a decrease in the total product ion abundance. This latter decrease is attributed to the production of low m/z fragment ions through secondary and higher order dissociation which fall below the LMCO in these spectra (~m/z 50 - 100). The MS/MS efficiencies obtained in the dual pressure LIT compare favorably to those measured in a standard three-dimensional quadrupole ion trap for singly charged peptide ions using a focused IR laser in which a maximum MS/MS efficiency of 43% was obtained.19
Figure 2.
Time-resolved IRMPD plots of DRVYIHPF (angiotensin II) activated at a q-value of 0.10. Precursor ion abundances (relative to initial precursor abundance without any irradiation, squares ■), photodissociation efficiencies (triangles ▲) and MS/MS efficiencies (circles ●) of (A) [M+2H]2+ and (B) [M+3H]3+.
Comparison of IRMPD to CID
IRMPD performed in the low pressure cell was compared to conventional CID results obtained in the high pressure cell for the series of model peptides. Shown in Supporting Information Figure S-3A is the IRMPD mass spectrum of [RPPGFSPFR + 3H]3+, and the complementary CID mass spectrum is shown in Supporting Information Figure S-3B. The CID mass spectrum is dominated by neutral water loss from the precursor ion, whereas the ion current is distributed among a variety of diagnostic product ions in the IRMPD mass spectrum. Immonium ions and several internal fragment ions are observed in the IRMPD mass spectrum that are not detected in the CID mass spectrum. Also, many y-type ions in conjunction with water losses are detected in the IRMPD spectrum which, presumably arise from secondary dissociation of the initial y-type ions upon absorption of additional IR photons. Experimental evidence suggests that a majority of the product ions observed in the IRMPD mass spectrum are actually produced upon secondary dissociation of [RPPGFSPFR - H2O + 3H]3+ ions. The [RPPGFSPFR - H2O + 3H]3+ ions were isolated and subjected to a discrete second stage of activation via IRMPD or CID, and the resulting MS3 product ion spectra are shown in the Supporting Information Figure S-4. CID of [RPPGFSPFR - H2O + 3H]3+ yielded three yo-type ions (i.e., y-type ions with a water loss) as well as the phenylalanine immonium ion (Fi) and the b2 ion (Figures S-4A - S-4C). In fact, most of the product ions observed in the MS3 product ion spectra match those detected in the IRMPD mass spectrum shown in Figure S-3A. These results indicate that triply charged bradykinin (RPPGFSPFR) primarily undergoes dehydration, and then this dehydrated product subsequently dissociates to yield informative sequence ions. Wideband CID of [RPPGFSPFR + 3H]3+ was also performed, and it provided higher sequence coverage than CID alone and similar coverage to MS3 and IRMPD experiments (Supporting Information Figure S-4D). However, as this technique employs a broadband excitation waveform to collisionally activate the m/z region between the precursor ion and neutral loss (e.g. H2O, NH3) product ions, no ions were observed between m/z 330 and 360 including any diagnostic sequences ions such as y62+ of m/z 355.7 which is detected in the IRMPD mass spectrum (Figure S-3A). For peptide ions which predominantly dissociate through a single pathway (e.g., dehydration), non-resonant ion activation techniques such as photodissociation provide a means to increase the number of diagnostic product ions.
The percentages of b- and y-type fragments, as defined by the number of b- and y-type fragment ions detected divided by the total number of possible b- and y-type fragments, observed in the IRMPD and CID mass spectra of the model peptides are summarized in Figure 3. (If a fragment was observed in two or more charge states, it was only counted once to more accurately give an account of the sequence coverage.) For almost every peptide investigated, a higher percentage of b- and y-type ions was observed by IRMPD. IRMPD showed significant increases in the number of b- and y-type fragments observed for all charge states of RPPGFSPFR (bradykinin) and RPKPQQFFGLM-NH2 (substance P), as well as for singly charged DRVYIHPF (angiotensin II) and ASHLGLAR. For [RPPGFSPFR + 3H]3+, the percentage of b- and y-type ions more than doubled for IRMPD in comparison to CID, with fourteen b- and y-type ions observed upon IRMPD but only six b- and y-type ions upon CID. In the two instances that CID outperformed IRMPD (i.e. singly and doubly charged FSWGAEGQR), it was due to formation of a single additional b- or y-type fragment.
Figure 3.
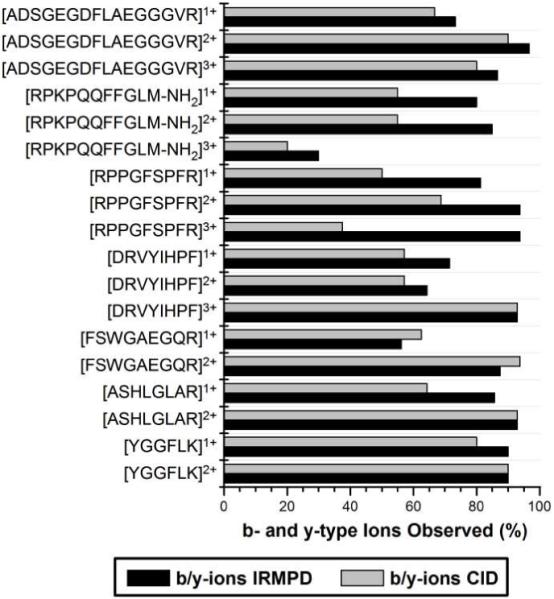
Percent of b- and y-type ions of model peptides observed by IRMPD (performed in the low pressure cell, black bars) and CID (performed in the high pressure cell, gray bars). IRMPD mass spectra were acquired using q-values between 0.07 and 0.10 (depending on precursor m/z) to reduce the low-mass cut-off to less than m/z 100. A value of 100% would indicate that all b- and y-ions were detected (including b1) as all amide bonds were considered to be potential sites of cleavage.
Increases in the CID activation time did not yield increased sequence coverage or significant changes in the relative abundances or types of product ions observed. For doubly charged fibrinopeptide A (ADSGEGDFLAEGGGVR), almost identical CID mass spectra were obtained when applying the collisional activation waveforms for between 25 - 100 ms; at shorter activation times, little dissociation was observed (data not shown). Similarly, variation in the normalized collision energy (20 - 100%) did not produce greater secondary dissociation or alter the product ion distribution for ADSGEGDFLAEGGGVR or DRVYIHPF peptide cations once the dissociation threshold (typically less than 20% NCE) was reached (data not shown).
Charge State Distribution of Product Ions
One of the more striking differences observed between the IRMPD and CID mass spectra of multiply protonated peptides is that almost all of the product ions in the IRMPD mass spectra are singly charged as compared to the CID mass spectra in which product ions of multiple charge states are typically observed. The IRMPD and CID mass spectra of [ADSGEGDFLAEGGGVR + 3H]3+ illustrates this observation in Figures 4A and 4B, respectively. In the CID mass spectrum, product ions are seen in the 2+ and 1+ charge states, with some sequence ions observed in both charge states (e.g., b9, b10, b11, and y10). However, in the IRMPD mass spectrum not a single product ion is observed to be multiply charged - all product ions are singly charged. One example of this lack of multiply charged peptide fragment ions is particularly evident in the range between m/z 440 and 610 in which more than ten abundant product ions are observed in the CID mass spectrum in the 1+ and 2+ charge states whereas only four fragment ions are detected in the IRMPD mass spectrum, all singly charged.
Figure 4.
(A) IRMPD (12.5 ms irradiation, q = 0.09, 50 W) and (B) CID (30 ms, q = 0.25, 25% normalized collision energy) mass spectra of [ADSGEGDFLAEGGGVR + 3H]3+ of m/z 512.9. The precursor ion is indicated with a star ([✰]) and internal ions are labeled with #.
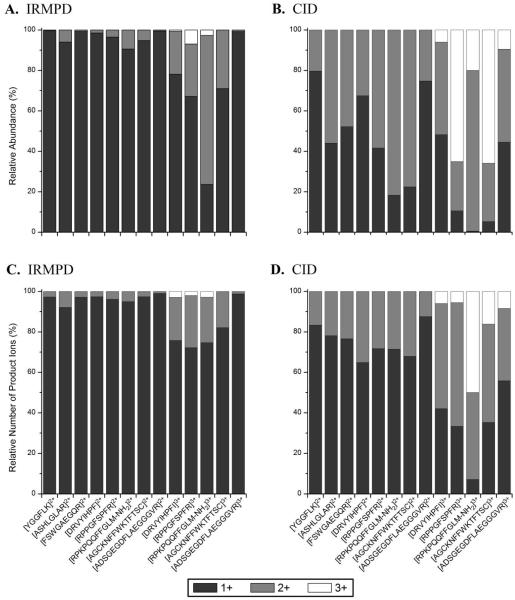
The distributions and numbers of singly, doubly, and triply charged product ions were determined for the IRMPD and CID mass spectra of an array of peptide ions. The distributions (abundances expressed as percentages) of product ions in each charge state are shown in Figures 5A and 5B for fragment ions produced by IRMPD and CID, respectively. Moreover, the number of products ions in each charge state (relative to the total number of fragment ions observed in each spectrum) are shown in Figures 5C and 5D (IRMPD and CID data, respectively). For the doubly charged precursor ions, the singly charged product ions comprise 96.6 ± 3.3% of the product ion current (defined as the total ion current minus the abundance of any surviving precursor) in the IRMPD mass spectra, whereas in the CID mass spectra the average abundance of singly charged fragment ions was 50 ± 23%. Doubly charged and singly charged product ions accounted for 30 ± 27% and 67 ± 30% of the product ion current, respectively, in the IRMPD mass spectra of the triply charged peptide ions; triply charged product ions accounted for an insignificant abundance. In contrast, the product ion current in the CID mass spectra, on average, was composed of 22 ± 23% of singly charged fragment ions, 45 ± 22% of doubly charged fragment ions, and 33 ± 30% of triply charged fragment ions. The lack of multiply charged product ions in the IRMPD mass spectra was further substantiated when the number of product ions in each charge state was counted and expressed as a percentage of the total number of product ions. Greater than 96% of the product ions were singly charged in the IRMPD mass spectra of doubly charged peptides, and more than 80% were singly charged in the IRMPD mass spectra of the triply charged peptides. In the CID mass spectra of doubly charged peptides, 75% of the product ions were singly charged, and for the triply charged peptides, the distribution of product ions was 35%, 48%, and 17% for singly, doubly, and triply charged product ions. Overall, CID yields product ions in a greater variety of charge states, whereas more than 80% of the fragment ions are singly charged in the IRMPD mass spectra with these ions accounting for over 85% of the product ion current. The lack of multiply charged product ions in the IRMPD mass spectra could be useful for potential de novo sequencing or database searches in which only singly charged fragment ions would be considered.
Figure 5.
Ion abundances of singly (black bars), doubly (gray bars), and triply charged fragment ions (white bars) relative to the total ion abundance (upper panels A and B) and number of fragment ions in each charge relative to the total number of product ions (lower panels C and D) observed in IRMPD (left panels A and C) and CID (right panels B and D) mass spectra.
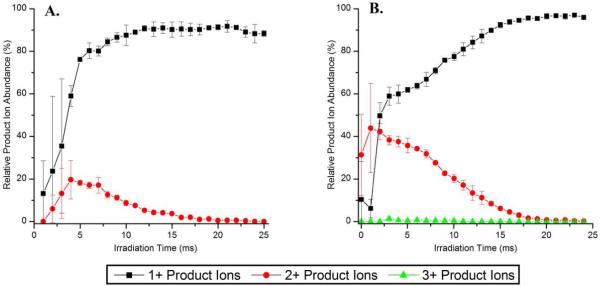
The relative product ion abundances of 1+, 2+, and 3+ charged product ions of [DRVYIHPF + 2H]2+ and [DRVYIHPF + 3H]3+ are plotted as a function of irradiation time in Figure 6. As the irradiation time increases from 0 to 4 ms, the abundances of both the 1+ and 2+ product ions of doubly protonated DRVYIHPF (angiotensin II) increase almost linearly; however, for irradiation times longer than 5 ms, the relative abundance of the 2+ charged product ions decreases to less than 1% of the total product ion current at irradiation times exceeding 20 ms (Figure 6A). For [DRVYIHPF + 3H]3+, at short irradiation times, doubly charged product ions are actually observed at a higher abundance than singly charged fragments (Figure 6B). At irradiation times longer than 3 ms, however, the abundance of 2+ product ions declines as these species are apparently converted into singly charged product ions via secondary dissociation. The only triply charged product ion observed in the IRMPD mass spectra of [DRVYIHPF+ 3H]3+ is the dehydrated precursor ion. By monitoring the abundances of individual product ions in specific charge states as produced upon IRMPD, it is observed that product ions in higher charge states consistently dissociate more readily than those same product ions in lower charge states (e.g. b112+ versus b11+ from ADSGEGDFLAEGGGVR, see Supporting Information Figures S5 and S6). The difference in PD efficiencies of doubly vs. singly charged product ions is attributed to a number of factors. In addition to experiencing greater Coulombic repulsion which could reduce the threshold for dissociation, most multiply charged product ions have a greater number of mobile protons which can facilitate their subsequent fragmentation.
Figure 6.
Time-resolved IRMPD plots of DRVYIHPF (angiotensin II) activated at a q-value of 0.10. Relative product ion abundances of singly (squares ■), doubly (circles ●), and triply charged product ions (triangles ▲) of (A) [M+2H]2+ and (B) [M+3H]3+.
Effect of Q-Value on IRMPD Efficiencies
The photodissociation efficiencies of peptides is dependent on the activation q-value. While it has been widely assumed that photodissociation is completely independent of ion trap operating parameters, recent work by Remes and Glish showed that the q-value (i.e., rf trapping voltage) greatly influences ion position within the ion trap.45 At higher q-values, which create greater potential well depths, the ions spend more time in the center of the trap where there is better overlap between the ion cloud and the photon beam. While a better overlap and consequently greater photodissociation efficiencies are observed at high q-values, these increases in performance are obtained at the expense of trapping low m/z fragment ions (i.e., at higher q-values the low mass cutoff is increased and a narrower range of ions can be effectively trapped and detected). To investigate the effect of the rf trapping level during photodissociation in the dual pressure LIT, IRMPD mass spectra were acquired at q-values between 0.10 and 0.24. The resulting plots for [DRVYIHPF + 2H]2+ and [DRVYIHPF + 3H]3+ are shown in Figures 7A and 7B, respectively. For these experiments, the MS/MS efficiencies could not be directly calculated as changes in the IRMPD q-value can result in changes in the trapping efficiency of the LIT. However, eq. (2) can be used to calculate MS/MS × trapping efficiencies in which P0 was determined from the IRMPD mass spectrum acquired at the lowest q-value; these values are plotted as a function of q-value in Figure 8. For [DRVYIHPF+ 2H]2+, as the q-value is increased from 0.05 to 0.10, there is a drastic decrease in the relative abundance of the precursor ions and a corresponding increase in photodissociation efficiencies from less than 5% to over 75% (Figure 7A). At q-values of greater than 0.12, PD efficiencies of ~100% are obtained. IRMPD experiments were not conducted at q-values of greater than 0.24 in these experiments as no increase in photodissociation efficiencies was observed and the MS/MS efficiencies were observed to decrease at greater q-values. In addition, at q-values above 0.25 effects of an increased low mass cutoff would be expected. The MS/MS × trapping efficiencies increase to greater than 95% at q-values between 0.08 and 0.11 for doubly protonated angiotensin II (Figure 8A). The total abundance of product ions falls at q-values greater than 0.09 as indicated by the decrease in the MS/MS × trapping efficiency. This decrease is attributed to secondary dissociation of product ions resulting in formation of smaller ions of m/z below that of the low mass cutoff. However, even at q-values as high as 0.24, the MS/MS × trapping efficiency is greater than 50%. Similar results are obtained for triply protonated DRVYIHPF (angiotensin II) as shown in Figure 7B. At q-values greater than 0.12, the photodissociation efficiencies are ~100%, and the precursor ion is completely dissociated. The MS/MS × trapping efficiencies varied somewhat more erratically due to larger variations in the total ion current of each mass spectrum, but the same general trend is observed in which the total abundance of the product ions increases with q-value and then gradually declines as lower m/z fragment ions are likely produced (but not detected). (The q-value was also varied for CID experiments conducted in a single linear ion trap mass spectrometer for [DRVYIHPF+ 2H]2+ and [DRVYIHPF+ 3H]3+, and at low q-values (< 0.15) almost no signal was observed. This is attributed to the low potential well-depth in which the precursor ion resides. Upon collisional activation, the precursor ions are lost due to unstable trajectories. Efficient dissociation (>75%) was observed only at q-values greater than 0.20; data not shown.)
Figure 7.
Photodissociation efficiencies (triangles ▲), MS/MS × trapping efficiencies (circles ●), and relative precursor ion abundances (squares ■) as a function of q-value of (A) [DRVYIHPF + 2H]2+ using 15.0 ms of IR irradiation and (B) [DRVYIHPF + 3H]3+ using 7.5 ms of irradiation. Precursor ion abundances are relative to the initial precursor abundance at q = 0.05 without any irradiation.
The effect of the q-value on photodissociation is also shown in the time-resolved IRMPD plots of singly and doubly protonated ADSGEGDFLAEGGGVR (fibrinopeptide A) at two different q-values (Supporting Information Figure S-2). Approximately 19 ms of irradiation was required to reduce [ADSGEGDFLAEGGGVR + H]+ to 50% relative abundance when activated at a q-value of 0.06 (Figure S-2A), but only 12 ms of IR irradiation was required at a q-value of 0.10 (Figure S-2B). For [ADSGEGDFLAEGGGVR + 2H]2+, 16 ms of irradiation was necessary to reduce the precursor to 50% abundance at q = 0.06 and only 9 ms was required at q = 0.10 (Figures S-2C and S-2D, respectively). These results clearly suggest that at higher q-values there is a greater overlap between the ion cloud and the laser beam, which results in higher photodissociation efficiencies. Thus, upon further comparison of the IRMPD mass spectra in Figures 1A and 1B, the difference is even more striking as almost no dissociation was observed when IRMPD was performed in the high pressure cell even when the q-value was set to 0.25, whereas high sequence coverage and high PD efficiency were obtained in the low pressure cell at a q-value of 0.09 using only 12.5 ms of irradiation.
Further Application and Implementation of IRMPD
While these IRMPD studies were conducted on a small subset of model peptides, the results suggest that IRMPD on the dual pressure LIT would be well suited as a complementary technique to CID and ETD for the analysis of large scale proteomic samples (e.g. enzymatic digests of cell lysates). In general, shorter irradiation times (~5 - 10 ms) were required to completely dissociate triply charged peptide ions with masses of less than 2000 Da. Longer irradiation times and slightly higher q-values were necessary to dissociate doubly charged peptides (~10 - 15 ms) and singly charged peptides (~20 - 25 ms). When peptide ions were irradiated for significantly longer intervals, the product ion mass spectra became cluttered with internal ions and low mass ions which could not be identified. Further experiments need to be performed in order to determine appropriate irradiation conditions based on peptide precursor m/z to avoid extensive secondary dissociation for more widespread and commercial implementation of IRMPD, particularly for data-dependent LC-IRMPD-MS experiments. However, a parallel study by us has shown that intact proteins readily dissociate upon IR irradiation in the low pressure cell indicating the IRMPD is a potentially useful method for top-down protein identification in ion trap mass spectrometers.46
CONCLUSIONS
Infrared multiphoton dissociation has been successfully implemented in a dual pressure LIT mass spectrometer to afford rapid and efficient dissociation of peptides in the low pressure cell. At lower trap pressures the rate of activation through IR photon absorption is greater than the rate of collisional cooling and thus peptides are readily dissociated. Dissociation efficiencies approaching 100% and MS/MS efficiencies of greater than 60% were obtained using irradiation times of less than 25 ms at a q-value of 0.10. The dissociation efficiencies were observed to be dependent on the q-value; at higher q-values the ions spend more time near the center of the trap and the ion cloud has a greater overlap with the photon beam, resulting in more rapid dissociation. Compared to CID, IRMPD allows the observation of low-mass fragment ions such as immonium ions and provides slightly higher sequence coverage of the peptides. The higher sequence coverage was attributed to secondary and higher order dissociation inherent to non-resonant activation methods such as photodissociation. IRMPD predominantly yielded singly charged fragment ions which aids in de-cluttering the product ion spectra and simplifying spectral interpretation. Highly charged product ions were observed to photodissociate more readily than the singly charged species which was attributed to a combination of factors including higher Coulombic repulsion, as well as the influence of a greater number of mobile protons.
Supplementary Material
ACKNOWLEDGMENT
Funding from the NSF (CHE-0718320), NIH (R01GM0846) and the Welch Foundation (F1155) is gratefully acknowledged.
REFERENCES
- (1).Winston RL, Fitzgerald MC. Mass Spectrom. Rev. 1997;16:165–179. doi: 10.1002/(SICI)1098-2787(1997)16:4<165::AID-MAS1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- (2).Godovac-Zimmermann J, Brown LR. Mass Spectrom. Rev. 2001;20:1–57. doi: 10.1002/1098-2787(2001)20:1<1::AID-MAS1001>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- (3).Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- (4).Biemann K. Methods Enzymol. 1990;193:455–479. doi: 10.1016/0076-6879(90)93433-l. [DOI] [PubMed] [Google Scholar]
- (5).Wells JM, McLuckey SA. Methods Enzymol. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- (6).Vaisar T, Urban J. J. Mass Spectrom. 1996;31:1185–1187. doi: 10.1002/(SICI)1096-9888(199610)31:10<1185::AID-JMS396>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- (7).Tsaprailis G, Somogyi A, Nikolaev EN, Wysocki VH. Int. J. Mass Spectrom. 2000;195/196:467–479. [Google Scholar]
- (8).Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O’Hair RA, Speed TP, Simpson RJ. Anal. Chem. 2003;75:6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- (9).Sleno L, Volmer DA. J. Mass Spectrom. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- (10).McLuckey SA, Goeringer DE. J. Mass Spectrom. 1997;32:461–474. [Google Scholar]
- (11).Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- (12).Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fung YME, Adams CM, Zubarev RA. J. Am. Chem. Soc. 2009;131:9977–9985. doi: 10.1021/ja8087407. [DOI] [PubMed] [Google Scholar]
- (15).Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- (16).Stephenson JL, Jr., Booth MM, Shalosky JA, Eyler JR, Yost RA. J. Am. Soc. Mass Spectrom. 1994;5:886–893. doi: 10.1016/1044-0305(94)87013-6. [DOI] [PubMed] [Google Scholar]
- (17).Brodbelt JS, Wilson JJ. Mass Spectrom. Rev. 2009;28:390–424. doi: 10.1002/mas.20216. [DOI] [PubMed] [Google Scholar]
- (18).Colorado A, Shen JX, Brodbelt J. Anal. Chem. 1996;68:4033–4043. doi: 10.1021/ac9600565. [DOI] [PubMed] [Google Scholar]
- (19).Newsome GA, Glish GL. J. Am. Soc. Mass Spectrom. 2009;20:1127–1131. doi: 10.1016/j.jasms.2009.02.003. [DOI] [PubMed] [Google Scholar]
- (20).Reilly JP. Mass Spectrom. Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Thompson MS, Cui W, Reilly JP. Angew. Chem., Int. Ed. 2004;43:4791–4794. doi: 10.1002/anie.200460788. [DOI] [PubMed] [Google Scholar]
- (22).Moon JH, Yoon SH, Kim MS. Rapid Commun. Mass Spectrom. 2005;19:3248–3252. doi: 10.1002/rcm.2184. [DOI] [PubMed] [Google Scholar]
- (23).Joly L, Antoine R, Broyer M, Dugourd P, Lemoine J. J. Mass Spectrom. 2007;42:818–824. doi: 10.1002/jms.1222. [DOI] [PubMed] [Google Scholar]
- (24).Wilson JJ, Brodbelt JS. Anal. Chem. 2007;79:7883–7892. doi: 10.1021/ac071241t. [DOI] [PubMed] [Google Scholar]
- (25).Kalcic CL, Gunaratne TC, Jones AD, Dantus M, Reid GE. J. Am. Chem. Soc. 2009;131:940–942. doi: 10.1021/ja8089119. [DOI] [PubMed] [Google Scholar]
- (26).Ly T, Julian RR. J. Am. Chem. Soc. 2008;130:351–358. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- (27).Gabryelski W, Li L. Rapid Commun. Mass Spectrom. 2002;16:1805–1811. doi: 10.1002/rcm.795. [DOI] [PubMed] [Google Scholar]
- (28).Goolsby BJ, Brodbelt JS. Anal. Chem. 2001;73:1270–1276. doi: 10.1021/ac001161o. [DOI] [PubMed] [Google Scholar]
- (29).Keller KM, Brodbelt JS. Anal. Biochem. 2004;326:200–210. doi: 10.1016/j.ab.2003.12.010. [DOI] [PubMed] [Google Scholar]
- (30).Payne AH, Glish GL. Anal. Chem. 2001;73:3542–3548. doi: 10.1021/ac010245+. [DOI] [PubMed] [Google Scholar]
- (31).Schwartz JC, Syka JEP, Quarmby ST. Proceedings of the 53th ASMS Conference on Mass Spectrometry and Allied Topics; San Antonio, TX. June 2005. [Google Scholar]
- (32).Cunningham C, Glish GL, Burinsky DJ. J. Am. Soc. Mass Spectrom. 2006;17:81–84. doi: 10.1016/j.jasms.2005.09.007. [DOI] [PubMed] [Google Scholar]
- (33).Stafford GC, Jr., Kelley PE, Syka JEP, Reynolds WE, Todd JFJ. Int. J. Mass Spectrom. Ion Processes. 1984;60:85–98. [Google Scholar]
- (34).Black DM, Payne AH, Glish GL. J. Am. Soc. Mass Spectrom. 2006;17:932–938. doi: 10.1016/j.jasms.2006.01.001. [DOI] [PubMed] [Google Scholar]
- (35).Boue SM, Stephenson JL, Jr., Yost RA. Rapid Commun. Mass Spectrom. 2000;14:1391–1397. doi: 10.1002/1097-0231(20000815)14:15<1391::AID-RCM36>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- (36).Hashimoto Y, Hasegawa H, Waki I. Rapid Commun. Mass Spectrom. 2004;18:2255–2259. doi: 10.1002/rcm.1619. [DOI] [PubMed] [Google Scholar]
- (37).Hashimoto Y, Hasegawa H, Yoshinari K, Waki I. Anal. Chem. 2003;75:420–425. doi: 10.1021/ac025866x. [DOI] [PubMed] [Google Scholar]
- (38).Pikulski M, Wilson JJ, Aguilar A, Brodbelt JS. Anal. Chem. 2006;78:8512–8517. doi: 10.1021/ac061472k. [DOI] [PubMed] [Google Scholar]
- (39).Wilson JJ, Brodbelt JS. Anal. Chem. 2006;78:6855–6862. doi: 10.1021/ac060760d. [DOI] [PubMed] [Google Scholar]
- (40).Vasicek LA, Wilson JJ, Brodbelt JS. J. Am. Soc. Mass Spectrom. 2009;20:377–384. doi: 10.1016/j.jasms.2008.10.016. [DOI] [PubMed] [Google Scholar]
- (41).Madsen JA, Brodbelt JS. J. Am. Soc. Mass Spectrom. 2009;20:349–358. doi: 10.1016/j.jasms.2008.10.018. [DOI] [PubMed] [Google Scholar]
- (42).Schwartz JC, Syka JEP, Remes PM. Proceedings of the 56th ASMS Conference on Mass Spectrometry and Allied Topics; Denver, CO. June 2008. [Google Scholar]
- (43).Wouters ER, Splendore M, Mullen C, Schwartz JC, Senko MW, Dunyach J. Proceedings of the 57th ASMS Conference on Mass Spectrometry and Allied Topics; Philadelphia, PA. June 2009. [Google Scholar]
- (44).Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Anal. Chem. 2008;80:4807–4819. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- (45).Remes PM, Glish GL. J. Phys. Chem. A. 2009;113:3447–3454. doi: 10.1021/jp808955w. [DOI] [PubMed] [Google Scholar]
- (46).Madsen JA, Gardner MW, Smith SI, Ledvina AR, Coon JJ, Schwartz JC, Stafford GC, Jr., Brodbelt JS. submitted. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.