Abstract
Recent discoveries in the science of ageing indicate that lifespan in model organisms such as yeast, nematodes, flies and mice is plastic and can be manipulated by genetic, nutritional or pharmacological intervention. A better understanding of the targets of such interventions, as well as the proximate causes of ageing-related degeneration and disease, is essential before we can evaluate if abrogation of human senescence is a realistic prospect.
The inevitability of ageing and death has preoccupied humanity for more than 5,000 years. In the epic named after him, Gilgamesh, the Sumerian king of Uruk, seeks to escape death, but ultimately concludes this is futile and turns to lasting works of culture to achieve immortality. Nowadays, disease—not death and the opposite, eternal life—preoccupies most biomedical scientists. Nevertheless, advances in understanding basic ageing mechanisms have made it difficult to ignore the issue of whether biomedical interventions to postpone ageing substantially are scientifically plausible. The overarching question is not whether mean human lifespan will increase modestly over the next decades. It almost certainly will, assuming continued success in reducing old-age morbidity and mortality1. Rather, the issue is whether postponing human ageing and natural death for many decades, possibly indefinitely, is feasible. Here, we discuss what we do and do not know about the mechanisms of ageing, and whether we have sufficient knowledge to foresee substantially retarding, halting or even reversing the degeneration and disease that limit human lifespan.
Lifespan is plastic
Ageing research has suffered more than disease-oriented research from unsubstantiated claims of potential cures. The field is also rife with opposing claims—that human life cannot be extended beyond a soft limit (120–125 years). For example, predictions in 1990 claimed that declines in death rates would not reach levels required for life expectancy at birth to exceed 85 years (ref. 2). However, Japanese females have already surpassed this limit (http://miranda.sourceoecd.org/vl=2231472/cl=20/nw=1/rpsv/health2007/g2-1-02.htm), and life expectancy in developed countries is now predicted to exceed 85 years by the year 2050 (ref. 1). A question remains as to whether science can free us from the bonds that seem to fix our lifespan.
With the discovery in the 1980s that mutations in single genes can significantly extend lifespan in the nematode Caenorhabditis elegans3,4, ageing began to be viewed as malleable by methods used to understand and manipulate development and disease. At present, hundreds of mutant genes can increase longevity in model organisms, including nematodes, yeast (Saccharomyces cerevisiae), fruitflies (Drosophila melanogaster) and mice (Mus musculus). Most act in evolutionarily conserved pathways that regulate growth, energy metabolism, nutrition sensing and/or reproduction5. Examples include genes encoding components of the insulin/insulin-like growth factor 1 (IGF-I) signalling (IIS) pathway, the target of rapamycin (TOR) pathway, and the mitochondrial electron transport chain (Fig. 1). In most cases, lifespan extension occurs when activity of the component is diminished. This abatement is thought to reduce somatic damage and/or increase somatic maintenance and repair5. Most pro-longevity mutations are discovered by mutagen or RNA interference screens, which mainly uncover inactivated or diminished gene functions, so further longevity mutations, resulting from enhanced gene function, may yet be discovered.
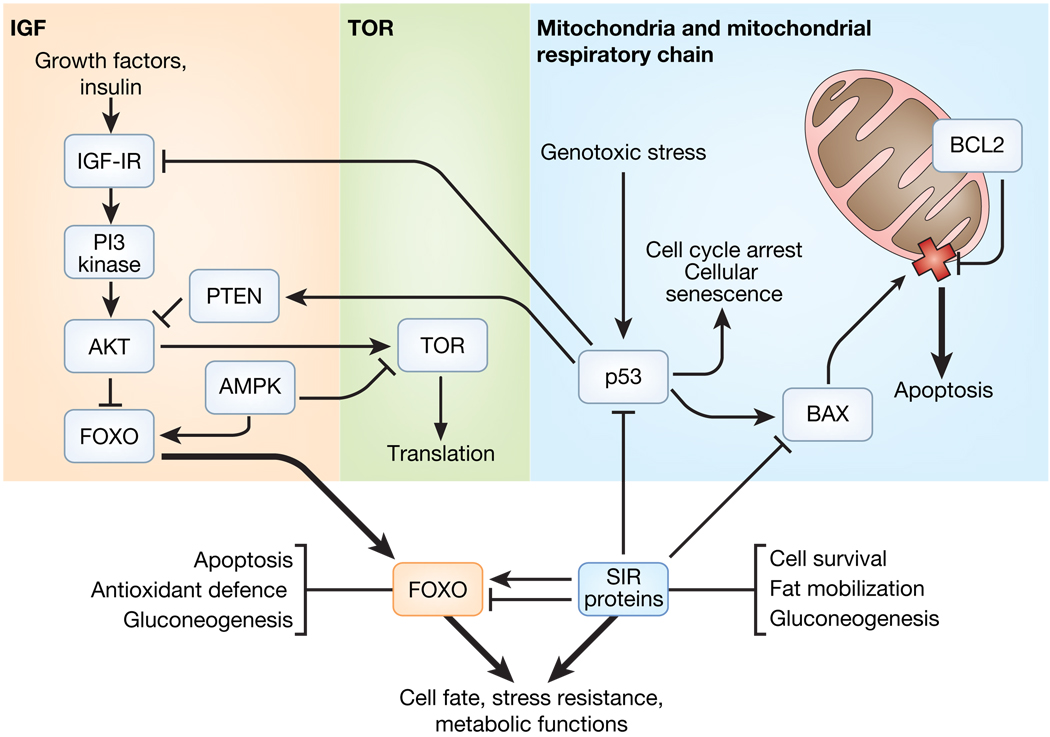
Figure 1. Potentially conserved pro-ageing pathways, their interconnections and possible targets for intervention.
In this very simplified depiction, three main pathways, the IIS, TOR and mitochondrial pathway, are indicated. The pro-ageing activities of these pathways are conserved across species, with energy sensors, such as AMPK, as potentially important hubs in the complex networks that integrate them. However, it is important to note potential dissimilarities among species as well. Most, if not all, defects in the mitochondrial respiratory chain are lethal or cause disease in humans62, but can increase lifespan in nematodes or yeast. In mammals, mitochondria play an important part in signalling apoptosis, which can either drive or retard ageing, depending on the cell type. There is evidence that many longevity signals converge on members of the FOXO and sirtuin protein families, which can interact. Note that SIR proteins can both activate and repress FOXO. Moreover, the effects of FOXO and SIR2 in cells can be either beneficial (for example, increasing antioxidant defence) or detrimental (for example, apoptosis), and may or may not promote organismal survival. For example, in mammals, SIRT1 dampens apoptosis by repressing FOXO, but also by repressing BAX activation, thereby preventing its oligomerization into the mitochondria outer membrane (cross), which normally triggers permeabilization of the membrane and release of soluble apoptogenic factors, such as cytochrome c, into the cytosol. Apoptosis can be beneficial, for example, by eliminating damaged cells and preventing cancer, or can be detrimental, by eliminating irreplaceable cells, such as neurons.
Many pro-longevity mutations mimic dietary restriction—under-feeding without malnutrition—which was shown to extend lifespan in laboratory rodents almost a century ago6. Dietary restriction increases longevity in many species, including yeast, nematodes, spiders and dogs. Although dietary restriction phenotypes often overlap with those conferred by dampening pro-ageing pathways, in some cases dietary restriction synergizes with pro-longevity mutations, indicating that dietary restriction can act independently7.
The appreciation that lifespan is plastic and under negative influence by genes that favour growth or procreation fuels hopes of finding small molecules that target the pathways affected by dietary restriction or pro-longevity mutations8,9. Compounds have been identified that show promise in this regard (Box 1), although none has yet shown major effects on lifespan in healthy mice10. Most, however, are in clinical trials to treat ageing-related diseases, such as diabetes and cancer, and already had the attention of the pharmaceutical industry. Of note, the US Food and Drug Administration will not approve agents that merely impede ageing. Nevertheless, it is remarkable that barely two decades ago human lifespan extension was a fantasy, whereas now pharmacological strategies are being considered for exactly that purpose. For some, the discovery that mutations in evolutionarily conserved pathways can greatly extend lifespan in model organisms is a step towards curing ageing in humans. As we argue, this viewpoint may be premature.
Box 1 | Strategies to counter intrinsic ageing
Evidence is emerging that identified pro-ageing pathways can be manipulated to extend lifespan by pharmacological means, although the mechanism by which such interventions act is not always clear. Small polyphenolics such as resveratrol and fisiten increase SIR2 deacetylase activity in vitro, and increase lifespan in a Sir2-dependent manner in yeast, C. elegans and Drosophila9. However, resveratrol may also be an antioxidant and suppress oxidative damage. Resveratrol increases insulin sensitivity and the survival of mice fed a high caloric diet64. The antifungal antibiotic rapamycin inhibits the pro-ageing TOR pathway in yeast and human cells. It is not known if TOR inhibition extends lifespan in mammals, but mechanistic overlap with metformin, an antidiabetic drug, suggests that it might. Metformin increases insulin sensitivity by reducing blood glucose, and might increase lifespan by activating AMPK, thereby inhibiting the TOR pathway and IGF-1 signalling65. Glucose metabolism is a principal target of prolongevity interventions, on the basis of the evidence that inhibiting energy pathways might mimic dietary restriction or dampen IIS. An example is 2-deoxy-D-glucose (2DG), which produces beneficial effects reminiscent of dietary restriction8. It is not yet known if 2DG increases lifespan, but it is at present in phase 1 clinical trial to treat solid tumours, which depend on glycolysis for survival and are sensitive to 2DG.
Natural antioxidants, such as vitamins C and E and β-carotene, have been extensively tested for their potential to extend lifespan in mice, but with no significant effects66. In humans, high dietary antioxidant intake correlates with decreased disease risk, but clinical trials of vitamin E and β-carotene supplementation failed to improve disease outcome67. Synthetic ROS scavengers have been tested in C. elegans68,69 and mice70, with conflicting results.
Free radical spin traps, such as phenyl tert-butyl-nitrone (PBN), block or reverse damage associated with a variety of disease states in animal models71. Thus far, however, a PBN derivative has not shown promise for extending longevity in mice10.
Certain AGE crosslinks can be cleaved by chemical agents. At present, known crosslink breakers (for example, the thiazolium halide alagebrium) are only partially effective because they break only a small subset of crosslinked structures72. Nonetheless, alagebrium alleviated cardiovascular stiffening in monkeys73 and is at present in advanced testing in humans, primarily for cardiovascular and diabetes-associated renal disease.
Rapid advances in stem cell research makes cell replacement therapy a promising approach to regenerate intrinsically aged, functionally declining tissues. Because endogenous stem cells show functional declines with age, a logical option is to differentiate pluripotent cells—obtained from embryos or generated by reprogramming of somatic cells—into specialized cells ex vivo. Before this approach becomes a realistic option, we must understand much more about how adult stem cells contribute to tissue maintenance during ageing, how ageing alters the stem cell microenvironment or niche, and how well the capacity of stem cells to regenerate functional tissues is maintained during differentiation and expansion in culture74.
Diminishing returns of complexity and idiosyncratic models
There is ample evidence that much of basic biology is similar across species as divergent as yeast and humans11. This is also true for ageing. Studies of yeast, nematodes and flies undoubtedly illuminate our understanding of the evolutionary and mechanistic bases of human ageing. Still, the response of simple organisms to interventions might not be predictive when complexity increases, or when physiology deviates significantly from humans.
The impact of complexity is illustrated by the IIS pathway. Invertebrates have a single receptor that binds ligands similar to insulin or IGF-1. Mutations that partially blunt signalling from this receptor extend lifespan in nematodes and flies5. However, mammals have distinct receptors for insulin and IGF-1, with different but overlapping functions. IGF-I primarily controls growth, whereas insulin regulates metabolism. In mammals, defective insulin signalling causes insulin resistance and diabetes. Defective IGF-I signalling causes protein breakdown and muscle degeneration. Indeed, IGF-I overexpression reduces ageing-associated cardiac dysfunction12 and improves muscle regeneration13. Nonetheless, reduced insulin signalling specifically in adipose tissue, or reduced IGF-I signalling throughout the animal, modestly extends lifespan in mice14,15. So, tissue-specific modulation of particular signalling pathways might retard ageing in humans.
The impact of complexity is also illustrated by interactions of two longevity-modulating protein families: forkhead (FOXO) transcription factors and silent information regulator (SIR) protein deacetylases (sirtuins). FOXO proteins (DAF-16 in nematodes) are required for the lifespan extension conferred by IIS mutations, and overexpression of SIR2 orthologues increases lifespan in yeast, nematodes and flies5. Mammals have multiple FOXO and SIR proteins. Some FOXO proteins initiate stress-induced cell death (apoptosis), which eliminates damaged or dysfunctional cells. FOXO proteins also upregulate antioxidant defence and DNA repair-facilitating genes16. FOXO is deacetylated by the human SIR2 orthologue SIRT1, which increases FOXO-dependent resistance to oxidative stress and cell-cycle arrest, but inhibits FOXO-dependent apoptosis17. SIRT1 also deacetylates the p53 tumour suppressor protein, attenuating its transcriptional activity and suppressing stress-induced apoptosis and cellular senescence (irreversible cell cycle arrest)18.
A priori, it is difficult to predict whether increased FOXO or sirtuin activity should increase or decrease mammalian longevity. Mammals rely on apoptosis and senescence to suppress cancer19, which is a major age-related disease and challenge to the longevity of mammals. This is not the case for yeast, flies and nematodes, which do not or rarely develop cancer because they are unicellular or as adults they contain largely post-mitotic somatic cells.
The magnitude of longevity extensions is worth noting. Genetic dampening of IIS routinely increases lifespan in nematodes twofold5, and one mutation increases lifespan tenfold20. In flies, however, single-gene mutations in the IIS or other pro-ageing pathways generally extend lifespan by only 25%–30%21. In mice, inactivating mutations in the Pou1f1, Prop1, or growth hormone receptor genes (which reduce IGF-I signals) increase lifespan by 40%22, whereas mutations that directly affect IIS extend lifespan by 20% or less14,15.
Thus, reduced IIS can substantially increase lifespan in nematodes, but much less so in the more complex fly and mouse. We know very little about mechanisms responsible for these species-specific differences. Within a species, genetic background, environment and sex differences matter. For example, lifespan extension in the IIS mutant fly chico depends on food concentration23. In transgenic flies overexpressing human superoxide dismutase in motor neurons, longevity benefits varied considerably among ten wild-derived genotypes, as well as by sex24.
Although it is possible that we have not yet defined optimal conditions for downregulating conserved ageing pathways in organisms more complex than nematodes, other characteristics set nematodes apart. First, many of the initial longevity mutations identified in nematodes affect an alternative developmental stage termed dauer, which suspends reproduction and alters metabolism. Hibernation, which temporarily suspends mammalian metabolism and reproduction, never lasts longer than the lifespan, in contrast to the dauer state in nematodes. Second, aerobic respiration is less critical for nematodes than for flies and mammals. This may explain why RNA interference screens for increased lifespan in nematodes identified multiple genes encoding mitochondrial respiratory chain subunits25. Downregulating these genes may increase lifespan by reducing mitochondrial function and its toxic by-products. In mammals, similar downregulation might be lethal or cause serious disease.
Translation to humans
Although disrupting conserved pro-ageing pathways identified in model organisms seems a realistic starting point for human lifespan extension, we first must determine whether these pathways modulate ageing in our own species. An initial approach is to identify associations between polymorphisms in or around conserved genes and human longevity. Extreme human longevity is genetically controlled, as indicated by the higher chance of siblings of centenarians to survive more than 100 years and moderate familial clustering of extreme longevity26. Thus far, however, linkage analyses are inconclusive, possibly because studies were underpowered or because of admixture in control populations27.
Attempts to associate candidate genes with extreme human longevity have mainly identified variants in lipoprotein metabolism genes as overrepresented in centenarians28. Furthermore, variants in FOXO1 and FOXO3 genes segregated with survival to age 85 and older29, and, in females, gene variants that reduce insulin/IGF-I signalling are associated with long survival30. Recently, heterozygous mutations in the IGF-IR, which markedly reduced IGF-IR activity, were found overrepresented in centenarians31.
Although these results are promising, more research is needed to confirm that humans and model organisms use similar longevity-modulating pathways. Even if these pathways are conserved in Homo sapiens, their natural variation evidently does not extend lifespan as much as laboratory-generated mutations in model organisms, notably nematodes. It is possible that organismal complexity will limit how much lifespan extension can be achieved by manipulating metabolic pathways; or there could be other layers of control or pathways that are yet to be discovered in complex animals. In predicting lifespan extension in humans, it is important to remember that these are crucial questions and their answers are unknown.
Life extension in model organisms may be an artefact to some extent. None of the laboratory animals considered ‘wild type’ has the genetic diversity of true wild strains, nor is the laboratory a natural habitat. For example, dietary restriction does not substantially increase longevity in some wild mice. Thus, laboratory breeding might select for a robust dietary restriction response32. Two longitudinal dietary restriction studies in rhesus monkeys were initiated in the late 1980s. Interim results suggest that dietary restriction improves health (for example, less body fat, higher insulin sensitivity and favourable circulating lipids), but there is no evidence yet that dietary restriction increases lifespan to the extent that it does in laboratory rodents33,34. Moreover, in monkeys (and by extension, humans) some benefits of dietary restriction, such as low IGF-I levels, may decrease cancer risk, but also increase the risk of osteoporotic fractures35. Thus, it might be necessary to reduce IGF-I signalling during early adulthood to prevent cancer, but increase it at older ages to prevent non-cancerous diseases36.
Can we expect interventions that target the human IIS pathway, even with proper spatio-temporal regulation, to extend lifespan to the extent that they do in simple models? Among the pro-longevity effects of dampening IGF-I signalling is upregulated stress resistance. Notably, stress response is generally superior in cells from long-lived compared to short-lived species37. In short-lived species, there is evidently sufficient opportunity for enhancing protective mechanisms. However, in long-lived species, there may be fewer such opportunities 38.
Furthermore, human physiology obviously differs from that of yeast, nematodes or flies. Perhaps less obvious are the differences between humans and mice. We should keep in mind that many anti-cancer therapies are successful in mice but fail in humans. Moreover, side or off-target effects of drugs that affect complex physiological pathways are already a problem. For example, cholesteryl ester transfer protein inhibitors, developed to increase high-density lipoprotein cholesterol, did not decrease but increased the risk of heart disease39.
Before we can rationally evaluate the potential impact of interventions to increase human lifespan substantially, we will need to understand the primary causes of ageing, which leads to the important question of why and how we age.
Evolutionary logic of ageing
According to Dobzhansky40, “nothing in biology makes sense except in the light of evolution”; so it is in the biology of ageing. Most scientists now accept that ageing results from the greater weight placed by natural selection on early survival and reproduction than on vigour at later ages. This age-related decline in the force of natural selection, first articulated by Medawar41, is due to high mortality caused by extrinsic hazards in natural environments, resulting in a relative scarcity of older individuals. When these hazards make survival to old age rare, natural selection favours gene variants that promote early growth and reproduction. In less hazardous environments, survival increases and gene variants that promote somatic maintenance can propagate. Hence, species-specific lifespan is determined by a trade-off between somatic maintenance and early growth or reproduction (Fig. 2)42. For example, genes that ensure a powerful immune response to infection promote early life survival, but later contribute to inflammation, a major age-related phenotype and risk for developing many diseases43.
Figure 2. Balancing somatic maintenance with growth and reproduction may determine lifespan.
According to the ‘disposable soma theory’63, organisms must compromise between energy allocation to growth and reproduction or somatic maintenance and repair.
Obviously, huge differences in longevity can arise as a result of evolution—consider the longevity difference between nematodes (weeks) and mammals (years), or even between mice (~3 yr) and humans (~100 yr). Were these differences achieved evolutionarily by discarding pro-ageing pathways, or by creating new longevity assurance pathways? The notable conservation among known longevity-modulating pathways, and similarities between organisms such as mice and humans in genomic structure and organization, argue against this possibility. Of course, unique non-conserved pathways may yet be discovered. It is more probable that significant longevity was achieved by subtle changes in many genes over the course of evolution, not by single mutations with large effects, which often increase lifespan at a cost to reproduction or survival under stress45. If so, interventions that target a single mammalian gene or even a single pathway may not increase longevity to the extent achieved by natural selection. This should not discourage the search for pharmacological interventions, but rather underscores how the shallowness of our knowledge about comparative evolutionary mechanisms can severely hamper efforts in this area.
Despite a general consensus regarding the evolutionary basis of why we age, we still know little about the primary causes of ageing and its relation to disease, which is generally the cause of death.
The ageing phenotype and the relationship to disease
Emphasis on lifespan can distract from understanding ageing itself. In nematodes and flies, we know much about genes that determine lifespan, but little about how these animals die. This is due to the complexity of ageing phenotypes42 and our limited ability to define phenotype, in contrast to the relative ease of defining genotype.
Table 1 lists some of the best-known ageing phenotypes in humans, which are increasingly evident in laboratory models as they are scrutinized. There are remarkable similarities among species, but there are also marked differences. For example, amyloid plaques in the brain and atherosclerotic plaques in blood vessels are hallmarks of human ageing, but are virtually lacking in mice. Even examination of shared phenotypes can uncover differences. For example, kyphosis (spinal curvature) is caused by osteoporosis in humans, but can have other causes, such as growth plate abnormalities, in mice. Importantly, ageing phenotypes—from hair greying to cancer susceptibility—vary among individual humans, and among inbred mouse strains.
Table 1.
Conserved ageing phenotypes
| Phenotype | H. sapiens | M. musculus | D. melanogaster | C. elegans |
|---|---|---|---|---|
| Decreased cardiac function | Yes | Yes | Yes | NA |
| Apoptosis, senescence (somatic cells) | Yes | Yes | Yes | ? |
| Cancer, hyperplasia | Yes | Yes | No | No |
| Genome instability | Yes | Yes | Yes | Yes |
| Macromolecular aggregates | Yes | Yes | Yes | Yes |
| Reduced memory and learning | Yes | Yes | Yes | NA |
| Decline in GH, DHEA, testosterone, IGF | Yes | Yes | ? | ? |
| Increase in gonadotropins, insulin | Yes | Yes | ? | ? |
| Decreased thyroid function | Yes | Yes | NA | NA |
| Decrease in innate immunity | Yes | Yes | Yes | Yes |
| Increase in inflammation | Yes | Yes | No | No |
| Skin/cuticle morphology changes | Yes | Yes | ? | Yes |
| Decreased mitochondrial function | Yes | Yes | Yes | Yes |
| Sarcopenia | Yes | Yes | Yes | Yes |
| Osteoporosis | Yes | Yes | NA | NA |
| Abnormal sleep/rest patterns | Yes | Yes | Yes | ? |
| Decrease in vision | Yes | Yes | ? | NA |
| Demyelination | Yes | Yes | ? | No |
| Decreased fitness | Yes | Yes | Yes | Yes |
| Arteriosclerosis | Yes | Yes | NA | NA |
| Changes in fat* | Yes | Yes | ? | ? |
Although changes in fat content and distribution have been reported for long-lived invertebrate mutants, at present there are no data on fat-related changes during normal ageing in these organisms. GH, growth hormone; DHEA, dehydroandrosterone; NA, not applicable.
A prominent age-related phenotype in humans and mice, absent in nematodes and flies, is cancer. Cancer arises from renewable somatic tissues, which are largely lacking in invertebrates. Cancer is sometimes considered as the opposite of ageing because it entails more vigorous growth. Indeed, cellular senescence, the irreversible cessation of growth, was once considered a model for ageing in vivo, but is now known to be a stress and tumour suppressive response19. Senescent cells increase with age in mice, non-human primates and humans, but comprise only a fraction of cells in renewable tissues19. Cellular senescence may be another evolutionary trade-off, as it suppresses cancer at early ages, but may promote ageing by exhausting stem cells or altering their niches. Senescent cells secrete inflammatory cytokines and other molecules that alter tissue microenvironments, and could stimulate the growth of cells that harbour preneoplastic mutations46. On the other hand, increased cellular senescence (and decreased proliferative potential) could also explain the decrease in cancer incidence rate at very old age, that is, after age 80 (ref. 47). Hence, cellular senescence can act both as a carcinogen and as an anti-carcinogen.
Little is known about ageing phenotypes and the causes of death in C. elegans. The nervous system is remarkably preserved, but ageing nematodes show slower movement, lower pharyngeal pumping rates (due to muscle deterioration resembling human sarcopenia) and increased lipofuscin48. Notably, there is extensive variability in age-related degeneration among genetically identical animals and cells of the same type within an individual48. This finding emphasizes the potentially important role of stochastic events in ageing, a point to which we will return.
Also in Drosophila, little is known about ageing or the causes of death. Nonetheless, this organism is emerging as a powerful model for human age-related diseases, such as Parkinson’s disease49. Ageing flies also develop sarcopenia and accumulate lipofuscin, so these traits may be universal ageing phenotypes50. Flies also show signs of cognitive dysfunction (increased time to accomplish a task), sharing this phenotype with mice and humans (Table 1).
Should we distinguish between ageing and disease? The answer to this question, which is still debated, depends on the disease and how its mechanism relates to ‘intrinsic ageing’; that is, ageing-related changes that are not determined primarily by external factors or genetic predisposition. Early-onset diseases, such as sickle cell anaemia, caused by a heritable β-globin gene mutation, can result at young ages in vascular constriction and increased risk of infection, also common in older people. But because these phenotypes occur within the realm of natural selection, sickle cell anaemia is not an ageing-related disease as its causes have little to do with ageing. Such a mechanistic distinction is much more difficult for late-onset diseases. Many would distinguish potentially fatal vascular degeneration from benign greying of hair. However, both phenotypes could have the same cause: intrinsic ageing. On the other hand, different mechanisms might produce the same disease-related phenotype at old age. For example, intrinsic endothelial cell ageing might contribute to atherosclerosis, as do mutations or polymorphisms in genes encoding the low-density lipoprotein receptor or ApoB. Statins can lower cholesterol and suppress atherosclerosis in individuals with high-risk low-density lipoprotein receptor or ApoB alleles, but cannot prevent intrinsic endothelial cell ageing.
Diseases are the main causes of death in elderly humans. Arteriosclerosis, diabetes, dementia, osteoporosis, osteoarthritis and cancer are particularly prominent pathologies, and some account for much of mortality at old age. Among the old who escape these diseases, the cause of death is often unknown. However, because interactions among ageing phenotypes are complex, natural death may ultimately be traced to disease, even if occult. For example, subtle tissue atrophies, neuropathies or microvascular leakage may underlie the deaths of old people subjected to stress. It is not clear whether successful intervention in overt disease will ameliorate intrinsic ageing, and thus significantly extend human lifespan.
Ageing is influenced by genetic and environmental factors that may be unrelated to each other or to intrinsic ageing. Irrespective of possible intrinsic ageing mechanisms, alleles that promote ageing will penetrate the germ line as long as their adverse effects manifest late enough, creating a diversity of genetic risk factors. Even among inbred individuals, for example, monozygotic human twins, genetic diversity occurs in somatic cells by mutation and epimutation at very early ages51,52. Similarly, environmental or lifestyle factors (for example, sunlight or smoking) can accelerate intrinsic ageing in specific tissues. It is conceivable that individuals of extreme longevity (that is, 100 years and older) are primarily those who managed to escape these genetic and environmental risks. This possibility is supported by the decelerated mortality rate seen at old age in invertebrate and human populations, indicating the survival of increasingly less frail individuals53. A question remains as to whether these survivors that escape the genetic and environmental risks that normally eliminate individuals through disease succumb to intrinsic ageing. Is there really an intrinsic ageing mechanism(s) to which eventually every cell or tissue falls prey? If so, what is its basis?
Intrinsic ageing
Ageing entails numerous functional and structural changes, many, but not all of which, adversely affect survival. A universal process of ‘intrinsic ageing’ might explain common ageing phenotypes among animals. One characteristic shared by all species studied thus far is the accumulation of unrepaired somatic damage. Thus, lifelong accumulation of various types of damage, along with random errors in bioinformational processes, might underlie intrinsic ageing (Fig. 3). As discussed, attenuation of such damage could explain the longevity conferred by mutations that dampen normal metabolic processes. Moreover, defence systems that keep damage in check might differ in efficacy among species, thereby dictating their lifespan37.
Figure 3. The causes of intrinsic ageing.
Although ultimately stochastic in nature, the proximal causes of ageing involve both programmed and random mechanisms.
Prominent causes of somatic damage include reactive oxygen species (ROS) and reducing sugars. ROS (by-products of respiration and other metabolic processes54) can damage and crosslink DNA, proteins and lipids. Reducing sugars react with carbohydrates and free amino groups, resulting in difficult-to-degrade advanced glycation end products (AGEs)55. AGEs accumulate in long-lived structural proteins, such as collagen and elastin. They increase the stiffness of blood vessels, joints and the bladder, and impair function in the kidney, heart, retina and other organs.
Interventions that remove damage might successfully counter the adverse effects of ROS and AGEs, thereby postponing ageing indefinitely 56 (see Box 1). However, macromolecular damage comes in many forms and all key lesions may not have been identified. Moreover, we do not know their relative contributions to intrinsic ageing, or how various components of the damage spectrum interact.
It might seem more efficacious to eliminate damaging molecules, rather than damage itself. However, some damaging molecules are crucial for normal cellular function. Two examples are glucose, which clearly cannot be eliminated, and ROS, which are also signalling molecules57. Trade-offs between beneficial and deleterious effects of damaging molecules will complicate strategies aimed at extending longevity by neutralizing them.
A similar trade-off might be cellular processes that defend us against cancer. Tissue regeneration elevates cancer risk by increasing the chance of acquiring DNA mutations or epimutations, which occur frequently in every organism as a consequence of errors during the repair or replication of a damaged template. Tumour suppressor mechanisms either eliminate cells that acquired extensive damage (apoptosis) or permanently prevent their proliferation (senescence). These responses, however, may gradually cause tissue atrophy, and therefore loss of organ function and regenerative capacity (Fig. 3)58. In principle, stem cell transplantation could counter the adverse effects of these damage responses.
When damage levels are not high enough to elicit an apoptotic or senescence response, a potentially more serious situation ensues that is difficult to counter: the gradual accumulation of random changes in DNA or protein, turning tissues into cellular mosaics. Such stochastic changes can reset gene regulatory loops, and randomly alter gene expression patterns on a cell by cell basis59,60. These changes, in aggregate, might compromise tissue function, without eliciting immediate cellular responses (Fig. 3). Stochastic gene regulatory drift would be difficult to correct and could even occur in stem cells in vivo or ex vivo during expansion for transplantation therapy.
Furthermore, developmental pathways that are essential for early life fitness or reproduction might be deleterious in adult tissues—for example, pathways that drive ductal morphogenesis in the developing or pregnant mammary gland might gradually promote ductal hyperplasia in the adult gland, predisposing it to cancer.
Future prospects
There is abundant evidence for mortality decline and increased lifespan throughout the developed world1. Before 1970, this probably reflected improvements in the food supply and sanitation, and achievements in medicine, notably vaccination and antibiotics. After 1970, the mortality decline probably reflects preventive medicine, lifestyle changes, routine use of anti-hypertensive and other drugs, and so on. It is impossible to predict, at this stage, whether increasingly sophisticated interventions will negate most or all ageing phenotypes. However, there are reasons for caution.
First, pharmacological intervention on the basis of pathways identified in model organisms may be an illusion because gains in longevity achieved in these organisms seem to decline with organismal complexity or depend on idiosyncratic physiology. Furthermore, lifespan in some organisms may be less plastic than in others. In addition, cancer poses a challenge to longevity that is distinct from age-related degeneration, and may be suppressed by mechanisms that are also pro-ageing. Third, there are still enormous gaps in our knowledge about how metabolic pathways operate and interact; serious side effects may constrain the effectiveness of pharmacological interventions.
Repair of macromolecular damage may prove more promising, especially in unison with improved anti-cancer therapies. However, it is not clear that all toxic lesions have been identified, or whether practical strategies exist to eliminate them. For example, it would be impossible to counter (epi)genomic drift pharmacologically, and transplanted organs and cells are also subject to loss of (epi)genomic integrity. Moreover, we do not know if macromolecular damage is the sole cause of ageing. Even in simple organisms, it is clear that longevity-modulating pathways entail exquisitely balanced interactions, regulated by numerous genetic elements61. And the large number of genomic transactions makes errors—many that are irreversible—inevitable. Indeed, there is evidence that ageing entails a gradual drift towards more random patterns of gene expression59, which might cause organ/tissue failure that cannot be undone by pharmacological or biological intervention.
In theory, interventions could be designed to alter the orchestrated networks of cell–cell interaction to increase lifespan. This is essentially what evolution has done to produce long-lived species. The question is, can we mimic the evolutionary process to the extent that senescence becomes essentially negligible? At this stage, the answer must be that we do not know. Although there is no scientific reason for not striving to cure ageing—similar to what we profess to do for cancer and other diseases—our current understanding makes it impossible to assert that indefinite postponement is feasible. Rather, we need to use the current momentum to intensify research aimed at resolving major outstanding questions that hinder a more complete understanding of basic ageing mechanisms and their relationship to disease (Box 2). Only this will allow us to generate sophisticated, integrated strategies to increase human health and lifespan.
Box 2 | Future research
Comparative phenotyping
In view of the general agreement that any ageing intervention should primarily target the quality of late life, it is imperative to learn more about ageing phenotypes and how the human phenotypes compare to those in model animals. This goal hinges on good descriptive research, traditionally less appreciated by the scientific community than hypothesis testing, but in this case it is critically important. This type of research should include a search for biomarkers, not as long-sought but illusory predictors of individual mortality risk, but to better define and characterize the degenerative processes underlying mortality. Furthermore, progress in this area is unthinkable without the development and use of advanced, interactive database systems75.
The ageing–disease relationship
To evaluate the effects of ageing interventions, it is important to understand how the pathogenesis of age-related diseases (for example, cancer, diabetes, cardiovascular and neurodegenerative disorders), which account for most of late life mortality, relates to basic molecular processes of ageing. Although we need to abandon the concept that ageing is a genetically programmed process and diseases of ageing essentially separate entities, it is conceivable that many disease risk factors are independent of ageing per se, which may have only one or a few, rather than a multiplicity, of causes. It is important to begin deciphering how disease processes intersect with basic ageing cause(s).
A rational basis for interventions
Before developing interventions for humans, it is critically important to have a strong rationale for both the intervention and target—this rationale should derive from many independent lines of evidence, including mechanistic, animal model and human-based studies. In selecting targets, strong consideration should be given to potential side effects. This mission will necessarily require multidisciplinary teams of clinicians and basic scientists working in cooperation with the pharmaceutical industry and regulatory agencies.
Unravel the causes of ageing
All interventions will fail to increase longevity substantially if the ultimate cause(s) of ageing does not lend itself to available treatments. For example, if ageing involves a gradual drift away from regulatory constraints, with organs and tissues developing into cellular mosaics, only in situ tissue replacement may offer a long-term solution. Even if ultimately ageing has the singular cause of a gradual, lifelong accumulation of molecular and cellular damage, the large diversity in types of damage, their complex interactions and ultimate consequences require an integrative, systems-biology approach42.
Acknowledgements
We thank N. Barzilai, A. de Grey, G. Lithgow and M. Gough for comments on the manuscript and P. Kapahi, R. Shmookler Reis, L. Balducci and Y. Suh for discussions. The authors’ work is supported by the US National Institutes of Health and Ellison Medical Foundation.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Kinsella KG. Future longevity-demographic concerns and consequences. J. Am. Geriatr. Soc. 2005;53:S299–S303. doi: 10.1111/j.1532-5415.2005.53494.x. [DOI] [PubMed] [Google Scholar]
- 2.Olshansky SJ, Carnes BA, Cassel C. In search of Methuselah: estimating the upper limits to human longevity. Science. 1990;250:634–640. doi: 10.1126/science.2237414. [DOI] [PubMed] [Google Scholar]
- 3.Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech. Ageing Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- 4.Friedman DB, Johnson TE. Amutation inthe age-1 gene inCaenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Masoro EJ. Overview of caloric restriction and ageing. Mech. Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Gems D, Pletcher S, Partridge L. Interpreting interactions between treatments that slow aging. Aging Cell. 2002;1:1–9. doi: 10.1046/j.1474-9728.2002.00003.x. [DOI] [PubMed] [Google Scholar]
- 8.Ingram DK, et al. Calorie restriction mimetics: an emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol. Med. 2007;13:64–71. doi: 10.1016/j.molmed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Miller RA, et al. An aging interventions testing program: study design and interim report. Aging Cell. 2007;6:565–575. doi: 10.1111/j.1474-9726.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- 11.Botstein D, Chervitz SA, Cherry JM. Yeast as a model organism. Science. 1997;277:1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santini MP, et al. Enhancing repair of the mammalian heart. Circ. Res. 2007;100:1732–1740. doi: 10.1161/CIRCRESAHA.107.148791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelosi L, et al. Local expression of IGF-1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J. 2007;21:1393–1402. doi: 10.1096/fj.06-7690com. [DOI] [PubMed] [Google Scholar]
- 14.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 15.Selman C, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 16.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 17.Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Smith J. Human Sir2 and the ‘silencing’ of p53 activity. Trends Cell Biol. 2002;12:404–406. doi: 10.1016/s0962-8924(02)02342-5. [DOI] [PubMed] [Google Scholar]
- 19.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 20.Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2007;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 21.Broughton SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl Acad. Sci. USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartke A. Minireview: role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology. 2005;146:3718–3723. doi: 10.1210/en.2005-0411. [DOI] [PubMed] [Google Scholar]
- 23.Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- 24.Spencer CC, Howell CE, Wright AR, Promislow DE. Testing an ‘aging gene’ in long-lived Drosophila strains: increased longevity depends on sex and genetic background. Aging Cell. 2003;2:123–130. doi: 10.1046/j.1474-9728.2003.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 26.Perls T, et al. Survival of parents and siblings of supercentenarians. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:1028–1034. doi: 10.1093/gerona/62.9.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beekman M, et al. Chromosome 4q25, microsomal transfer protein gene, and human longevity: novel data and a meta-analysis of association studies. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:355–362. doi: 10.1093/gerona/61.4.355. [DOI] [PubMed] [Google Scholar]
- 28.Barzilai N, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. J. Am. Med. Assoc. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 29.Kuningas M, et al. Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur. J. Hum. Genet. 2007;15:294–301. doi: 10.1038/sj.ejhg.5201766. [DOI] [PubMed] [Google Scholar]
- 30.Capri M, et al. The genetics of human longevity. Ann. NY Acad. Sci. 2006;1067:252–263. doi: 10.1196/annals.1354.033. [DOI] [PubMed] [Google Scholar]
- 31.Suh Y, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl Acad. Sci. USA. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper JM, Leathers CW, Austad SN. Does caloric restriction extend life in wild mice? Aging Cell. 2006;5:441–449. doi: 10.1111/j.1474-9726.2006.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattison JA, Lane MA, Roth GS, Ingram DK. Calorie restriction in rhesus monkeys. Exp. Gerontol. 2003;38:35–46. doi: 10.1016/s0531-5565(02)00146-8. [DOI] [PubMed] [Google Scholar]
- 34.Weindruch R. Will dietary restriction work in primates? Biogerontology. 2006;7:169–171. doi: 10.1007/s10522-006-9007-0. [DOI] [PubMed] [Google Scholar]
- 35.Dirks AJ, Leeuwenburgh C. Caloric restriction in humans: potential pitfalls and health concerns. Mech. Ageing Dev. 2006;127:1–7. doi: 10.1016/j.mad.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Anzo M, Cohen P. Control of aging and longevity by IGF-I signaling. Exp. Gerontol. 2005;40:867–872. doi: 10.1016/j.exger.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Kapahi P, Boulton ME, Kirkwood TB. Positive correlation between mammalian lifespan and cellular resistance to stress. Free Radic. Biol. Med. 1999;26:495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- 38.Kirkwood TB, Rosenberger RF, Galas DJ, editors. Accuracy of Molecular Processes. Chapman & Hall; 1986. [Google Scholar]
- 39.Suckling K. The continuing complexities of high-density lipoprotein metabolism in drug discovery and development. Expert Opin. Ther. Targets. 2007;11:1133–1136. doi: 10.1517/14728222.11.9.1133. [DOI] [PubMed] [Google Scholar]
- 40.Dobzhansky T. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 1973;35:125–129. [Google Scholar]
- 41.Medawar PB. An Unsolved Problem in Biology. H. K. Lewis; 1952. [Google Scholar]
- 42.Kirkwood TB. A systematic look at an old problem. Nature. 2008;451:644–647. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- 43.Finch CE, Crimmins EM. Inflammatory exposure and historical changes in human life-spans. Science. 2004;305:1736–1739. doi: 10.1126/science.1092556. [DOI] [PubMed] [Google Scholar]
- 44.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Jenkins NL, McColl G, Lithgow GJ. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc. Biol. Sci. 2004;271:2523–2526. doi: 10.1098/rspb.2004.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl Acad. Sci. USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harding C, Pompei F, Lee EE, Wilson R. Cancer suppression at old age. Cancer Res. 2008;68:4465–4478. doi: 10.1158/0008-5472.CAN-07-1670. [DOI] [PubMed] [Google Scholar]
- 48.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 49.Wang C, et al. Drosophila overexpressing parkin R275W mutant exhibits dopaminergic neuron degeneration and mitochondrial abnormalities. J. Neurosci. 2007;27:8563–8570. doi: 10.1523/JNEUROSCI.0218-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grotewiel MS, Martin I, Bhandari P, Cook-Wiens E. Functional senescence in Drosophila melanogaster. Ageing Res. Rev. 2005;4:372–397. doi: 10.1016/j.arr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaupel JW, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–860. doi: 10.1126/science.280.5365.855. [DOI] [PubMed] [Google Scholar]
- 54.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic. Biol. Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich P, Cerami A. Protein glycation, diabetes, and aging. Recent Prog. Horm. Res. 2001;56:1–21. doi: 10.1210/rp.56.1.1. [DOI] [PubMed] [Google Scholar]
- 56.de Grey AD, et al. Time to talk SENS: critiquing the immutability of human aging. Ann. NY Acad. Sci. 2002;959:452–462. doi: 10.1111/j.1749-6632.2002.tb02115.x. [DOI] [PubMed] [Google Scholar]
- 57.Finkel T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 58.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nature Rev. Mol. Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 59.Bahar R, et al. Increased cell-to-cell variation in gene expression in aging mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 60.Vijg J. Aging of the Genome. Oxford Univ. Press; 2007. [Google Scholar]
- 61.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 63.Kirkwood TB. Evolution of ageing. Nature. 1977;270:301–304. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- 64.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarty MF. Chronic activation of AMP-activated kinase as a strategy for slowing aging. Med. Hypotheses. 2004;63:334–339. doi: 10.1016/j.mehy.2004.01.043. [DOI] [PubMed] [Google Scholar]
- 66.Meydani M, et al. The effect of long-term dietary supplementation with antioxidants. Ann. NY Acad. Sci. 1998;854:352–360. doi: 10.1111/j.1749-6632.1998.tb09915.x. [DOI] [PubMed] [Google Scholar]
- 67.Frei B. Efficacy of dietary antioxidants to prevent oxidative damage and inhibit chronic disease. J. Nutr. 2004;134:3196S–3198S. doi: 10.1093/jn/134.11.3196S. [DOI] [PubMed] [Google Scholar]
- 68.Melov S, et al. Extension of life-span with superoxide dismutase/catalase mimetics. Science. 2000;289:1567–1569. doi: 10.1126/science.289.5484.1567. [DOI] [PubMed] [Google Scholar]
- 69.Keaney M, Gems D. No increase in lifespan in Caenorhabditis elegans upon treatment with the superoxide dismutase mimetic EUK-8. Free Radic. Biol. Med. 2003;34:277–282. doi: 10.1016/s0891-5849(02)01290-x. [DOI] [PubMed] [Google Scholar]
- 70.Morten KJ, Ackrell BA, Melov S. Mitochondrial reactive oxygen species in mice lacking superoxide dismutase 2: attenuation via antioxidant treatment. J. Biol. Chem. 2006;281:3354–3359. doi: 10.1074/jbc.M509261200. [DOI] [PubMed] [Google Scholar]
- 71.Floyd RA. Nitrones as therapeutics in age-related diseases. Aging Cell. 2006;5:51–57. doi: 10.1111/j.1474-9726.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 72.Furber JD. Extracellular glycation crosslinks: prospects for removal. Rejuvenation Res. 2006;9:274–278. doi: 10.1089/rej.2006.9.274. [DOI] [PubMed] [Google Scholar]
- 73.Vaitkevicius PV, et al. A cross-link breaker has sustained effects on arterial and ventricular properties in older rhesus monkeys. Proc. Natl Acad. Sci. USA. 2001;98:1171–1175. doi: 10.1073/pnas.98.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 75.Calder RB, et al. MPHASYS: a mouse phenotype analysis system. BMC Bioinformatics. 2007;8:183. doi: 10.1186/1471-2105-8-183. [DOI] [PMC free article] [PubMed] [Google Scholar]