Abstract
Mitochondria are found in all eukaryotic cells and contain their own genome (mtDNA). Unlike the nuclear genome which is derived from both the egg and sperm at fertilization, the mtDNA in the embryo are derived almost exclusively from the egg, i.e. is of maternal origin. Mutations in mtDNA contribute to a diverse range of still incurable human diseases and disorders. To establish preclinical models for new therapeutic approaches, we demonstrate here that the mitochondrial genome can be efficiently replaced in mature nonhuman primate oocytes by spindle-chromosomal complex transfer from one egg to an enucleated, mitochondrial-replete egg. The reconstructed oocytes with the mitochondrial replacement were capable of supporting normal fertilization, embryo development and produced healthy offspring. Genetic analysis confirmed that nuclear DNA in the three infants born so far originated from the spindle donors while mtDNA came from the cytoplast donors. No contribution of spindle donor mtDNA was detected in offspring. Spindle replacement is shown here as an efficient protocol replacing the full complement of mitochondria in newly generated embryonic stem cell lines. This approach may offer a reproductive option to prevent mtDNA disease transmission in affected families.
Mitochondria play important roles in cellular processes, e.g., production of cellular energy in the form of adenosine triphosphate (ATP) and programmed cell death (apoptosis). Each mitochondrion contains between 2 and 10 copies of mtDNA, and since cells have numerous mitochondria, a cell may harbor several thousand mtDNA copies. Mutations in mtDNA occur at a 10-fold or higher rate than in nuclear DNA possibly due to a high concentration of free oxygen radicals, lack of histones and limited mtDNA repair mechanisms. Diseases caused by mtDNA mutations were first described in 19881–3. Since then, over 150 mutations (including 100 deletions and approximately 50 point mutations) have been identified that are associated with serious human disorders, including myopathies, neurodegenerative diseases, diabetes, cancer and infertility4. Interest in mitochondrial DNA mutations has grown due to the increasing number of associated diseases and because they can affect patients throughout life. In addition, mtDNA mutations are increasingly implicated in a range of prevalent public health conditions, including Alzheimer's, Parkinson's and Huntington's diseases5–9.
Typically a cell contains only one type of mtDNA (homoplasmy). If an individual cell contains two or more types of mtDNA, i.e. as a mixture of normal and mutant mtDNA, the phenomenon is known as heteroplasmy. Heteroplasmy allows lethal mutations to persist and most importantly to pass to the next generation. MtDNA is maternally inherited through the egg's cytoplasm whereas sperm mitochondria constitute a minor fraction of the zygote's cohort and are rapidly eliminated after fertilization10. It is estimated that 1in 3,500–6000 people have either mtDNA disease or are at risk for development of mtDNA based disorders11–13. At present, there are no cures for mitochondrial disorders and available treatments only alleviate symptoms and slow disease progression. Preimplantation genetic diagnosis has previously been applied, in few cases, to identify and transfer embryos devoid of pathogenic mtDNA mutations and resulted in the birth of a healthy baby14. However, genetic counseling in most patients at risk of maternally inherited mtDNA mutations is challenging due to limitations in assessing the extent of mtDNA heteroplasmy and accurately predicting risks15. Therefore, there is a significant need to consider new therapeutic approaches that could prevent transmission of mtDNA mutations from mother to child.
Spindle-Chromosomal Complex Transfer in Mature Monkey Oocytes
The complete replacement of mutant mtDNA in patient's eggs with healthy mtDNA would be the most reliable method to avoid recurrence of mtDNA diseases but neither the feasibility nor safety of such a substitution has been evaluated. We hypothesized that mtDNA can be efficiently replaced by a novel approach, i.e., spindle-chromosomal complex transfer (ST) in mature eggs (metaphase II, or MII oocytes) without perturbing subsequent fertilization and developmental competence. To this end, the nuclear genetic material from a patient's egg containing mtDNA mutations could be removed, and transplanted into an enucleated egg containing normal mtDNA donated by a healthy female. A child born following fertilization with the husband's sperm would be free of risk from maternal mtDNA mutations as well as the authentic biological child of the patients (Suppl. Fig 1).
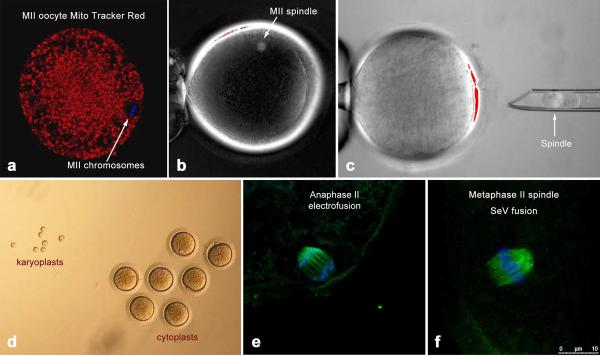
Initially, we investigated the distribution of active mitochondria in monkey oocytes by labeling with MitoTracker Red and monitoring with confocal laser scanning microscopy. In mature MII-stage oocytes, mitochondria were distributed relatively uniformly throughout the cytoplasm but spindles and metaphase chromosomes were devoid of mitochondria (Fig. 1a). These results suggested that the isolation and transfer of MII spindle-chromosomal complexes can be accomplished without significant mtDNA carryover from the nuclear donor oocyte. Until now, two major technical obstacles hampered the feasibility and success of this approach: (i) difficulties in visualization and isolation of intact MII chromosomes and (ii) the susceptibility of meiotic spindles and chromosomes to damage secondary to premature oocyte activation during manipulations. The visualization of DNA in mature oocytes is difficult because the nucleus is no longer evident after reinitiation of meiosis and breakdown of nuclear membrane. However, enucleating MII oocytes became routine after development of techniques for DNA staining with fluorophores (such as Hoechst 33342) and subsequent visualization under UV light, notably during cloning of embryos by somatic cell nuclear transfer (SCNT). However, unlike in cloning where the extracted chromosomes are discarded, the viability and integrity of the oocyte spindle associated DNA during ST must be maintained.
Figure 1. Spindle-chromosomal complex transfer and meiotic analysis of reconstructed monkey oocytes.
a, confocal microscopy of a rhesus monkey MII oocyte labelled with DAPI (blue) to depict chromosomes and MitoTracker Red to label active mitochondria. b, MII spindle visualization with Oosight™ Imaging System. c, karyoplast isolation. d, isolated karyoplasts and cytoplasts. e, confocal microscopy of progression to the anaphase II stage induced by electrofusion. f, intact metaphase II spindle after SeV fusion. Spindles are labelled with DAPI (blue) to depict chromosomes and with B-tubulin (green) to show microtubules
We recently accomplished successful SCNT in primates with adult skin cells as nuclear donors and went on to isolate functional embryonic stem (ES) cells16,17. The key for this success was implementation of several technical modifications to avoid damage to the cytoplast during spindle removal. This involved noninvasive visualization of the metaphase spindle during enucleation with polarized microscopy to avoid deleterious effects of Hoechst staining and UV exposure (Fig. 1b). We reasoned that the modified enucleation technique should be applicable to the isolation of intact MII spindle-chromosomal complexes and their subsequent transfer to spindle-free, donor cytoplasts. Indeed, we were able to isolate intact spindle-chromosomal complexes surrounded by a small amount of cytoplasm and cell membrane (karyoplast) with 100% efficiency (Fig. 1c, Suppl. video). We measured diameters of both karyoplasts and enucleated oocytes (cytoplasts) and calculated that the average volume of a karyoplast was 11.3 ± 1.2pL (mean ± SEM; n=7) while the average volume of a cytoplast was 752.1 ± 18.3pL (Fig. 1d). Thus, a karyoplast contained approximately 1.5% of the volume of a cytoplast, suggesting that a negligible amount of mitochondria/mtDNA surrounding the spindle would be carried over during ST.
Next, we investigated the re-introduction of spindle-chromosomal complexes into enucleated oocytes derived from unrelated females. Karyoplasts were placed into the perivitelline space of cytoplasts, on the side opposite the 1st polar body and fusion with the cytoplast was accomplished by electroporation. Approximately 1h after fusion, reconstructed oocytes were fixed and analyzed by immunocytochemistry for spindle-chromosomal complex integrity. A spindle was present in all analyzed oocytes, but the majority of oocytes unexpectedly resumed the meiotic division and progressed to the anaphase of meiosis II or completed meiosis and formed a 2nd polar body (Fig. 1e).
We hypothesized that fusion by electroporation may have triggered premature activation and subsequent resumption of meiosis. To test this assumption, we employed an alternative karyoplast fusion technique using an extract from Sendai virus (SeV). Isolated karyoplasts were briefly exposed to SeV extract and then placed into the perivitelline space of cytoplasts (Suppl. video) where fusion was observed within 20–30 min. Analysis of reconstructed oocytes demonstrated that spindle-chromosomal complexes were maintained in the MII stage and had morphology similar to intact controls (Fig. 1f). Resumption of meiosis and separation of the 2nd polar body was observed in SeV group only after fertilization. Thus, these results indicate that electrofusion pulse induces premature activation and resumption of meiosis during spindle introduction. In contrast, this artifact was prevented by using SeV-assisted fusion.
Developmental Potential of Reconstructed Oocytes
We next determined the developmental competence of ST-generated oocytes produced by electrofusion or SeV, following fertilization by intracytoplasmic sperm injection (ISCI) and in vitro embryo culture. Karyoplast-cytoplast fusion rates were comparable between the two ST approaches. However, pronuclear formation in the electrofusion group was not observed, instead sperm injected oocytes cleaved prematurely by the morning after ICSI and arrested between the 8-cell and morula stages. In contrast, fertilization, cleavage and blastocyst rates in the SeV group were similar to those of intact control oocytes (Table 1). These results are consistent with the conclusion that spindle transfer in MII oocytes employing SeV does not compromise subsequent fertilization and in vitro embryonic developmental competence. In order to assess the quality of the blastocysts produced, we carried out cell counts by labeling with DAPI (all cells) and NANOG (the inner cells mass, ICM) (Suppl. Fig. 2). Five expanded or hatched blastocysts for each ST and corresponding control group were analyzed. The mean number of total cells and ICM cells in ST blastocysts were 152±34 and 21±10, respectively, and similar to controls (127±79 and 23±10, respectively; P>0.05).
Table 1.
In vitro development of embryos after spindle-chromosomal complex transfer (ST) into enucleated oocytes and ICSI as compared to controls
| Treatment | N | Fusion(%) | Fertilization (%) | 8-cell (%)a | Morula (%)a | Blastocyst (%)a |
|---|---|---|---|---|---|---|
| ST (SeV) | 87 | 78 (90) | 74 (95) | 69 (93) | 58 (78)b | 45 (61)b |
| ST (elect) | 15 | 11 (73) | 11 (100) | 8 (73) | 2 (18)c | 1 (9)c |
| Control | 72 | NA | 68 (94) | 57(84) | 51 (75)b | 41 (60) b |
The percentage of 8-cell, morula and blastocyst stage embryos was calculated based on the number of fertilized embryos
Different superscript indicates significant difference (P<0.05). Data were analysed using X2 test.
Different superscript indicates significant difference (P<0.05). Data were analysed using X2 test.
ISCI; intracytoplasmic sperm injection.
SeV and elect indicate fusion with Sendai virus extract or electrofusion, respectively.
NA; not applicable (no fusion step)
To further define the developmental potential, we isolated ES cell lines from ST blastocysts. Two stable ES cell lines were established from 8 ST embryos (designated as STES-1 and -2) with derivation efficiency (25%) similar to controls (Suppl. Table 1). Both cell lines exhibited typical primate pluripotency markers and were able to differentiate into neuronal cell types and spontaneously contracting cardiomyocytes (Suppl. Fig. 3). To determine if the ST procedure induced lasting chromosomal abnormalities, we conducted a cytogenetic analysis of STES cell lines by G-banding. The analysis revealed that these cell lines contained normal rhesus macaque karyotypes (one male-42, XY and one female-42, XX) with no detectable chromosomal anomalies (Suppl. Fig. 4). Finally, we tested the in vivo developmental potential of ST embryos by transfer into the reproductive tract of recipient females. We transferred 15 ST embryos into oviducts of nine recipients; six females received either 1 or 2 blastocysts each whereas three recipients received 2 cleavage stage (4–8-cell) embryos each (Table 2).Three females became pregnant, one carrying twins and two with singletons, all derived from the transfer of ST embryos at the blastocyst stage (Table 2). Remarkably, the pregnancy (33%) and implantation rates (27%) with ST embryos were even higher than those previously reported for non-manipulated, ICSI-produced embryos (Suppl. Table 2)18. On April 24, 2009, the first pregnant monkey delivered a set of healthy twins by cesarean section and were named Mito and Tracker (Fig. 2). To our knowledge, these infants represent the world's first animals born after MII spindle-chromosomal complex transfer. The second pregnant monkey gave birth to a healthy infant on May 8, 2009 while the third is ongoing. All three ST infants are healthy and their birth weights and gestational lengths were within normal ranges for rhesus macaques (Suppl. Table 3)18. Overall, these results demonstrate that MII spindle-chromosomal complexes can be efficiently isolated and transplanted into enucleated oocytes. Reconstructed oocytes produced with this technology were suitable for fertilization and developed to blastocysts at rates similar to controls. Moreover, the isolation of normal ES cells and the birth of healthy offspring support further evaluation of this approach for mitigating the consequences of mtDNA defects.
Table 2.
Embryo transfers and pregnancies with ST embryos
| Spindle donor | Cytoplast donor | Embryo stage (age)a | No of embryos transferred | Recipient stageb | Pregnancy |
|---|---|---|---|---|---|
| female 1 | female 2 | ExB (D7) | 2 | D4 | Twin |
| female 2 | female 1 | ExB (D8) | 2 | D2 | no |
| female 4 | female 3 | 4-cell (D2) | 2 | D2 | no |
| female 3 | female 4 | EB( D7) | 1 | D4 | Single |
| female 13 | female 14 | 8-cell (D3) | 2 | D2 | no |
| female 14 | female 13 | 8-cell (D3) | 2 | D2 | no |
| female 13 | female 14 | ExB (D8) | 2 | D4 | no |
| female 15 | female 16 | ExB (D8) | 1 | D4 | no |
| female 8 | female 7 | ExB(D7) | 1 | D4 | Single |
Day of ICSI is calculated as Day 0 (DO).
The next day after estrogen surge is considered as ovulation day or Day 0. Embryo transfers were scheduled to synchronize the stage of early embryo development with uterine development leading to the implantation interval.
EB: early blastocyst, ExB: expanded blastocyst
Figure 2. Mito and Tracker, world's first primates produced by spindle-chromosomal complex transfer (ST) into enucleated oocytes followed by fertilization and embryo transfer.
Twin pregnancy was established by transfer of 2 ST-derived blastocysts into a recipient. Both infants are healthy and their growth and development is within a normal range for rhesus macaques. The photo was taken at 6 days of age.
Analysis of nuclear and mtDNA in ST offspring
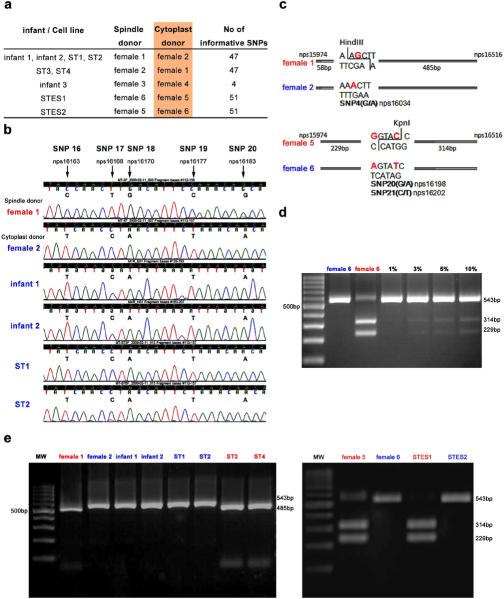
Finally, we investigated the degree of heteroplasmy extant in infants and cell lines as a result of mtDNA carryover during spindle transfer. We isolated DNA samples from ST infants and STES cell lines to analyze genomic DNA by microsatellite parentage analysis and mtDNA by direct sequencing of the mitochondrial D-loop hypervariable region 1(rhDHV1) as previously described16. We also collected and analyzed DNA from four differentiated cultures derived from the outgrowth of plated ST blastocysts (designated as ST1, 2, 3 and 4). Detailed analysis of nuclear DNA employing 41 microsatellite markers confirmed that all three infants and all cell lines inherited their nuclear genome from the spindle donor animals (Fig. 3a and Suppl. Table 4). Spindle and cytoplast donor animals contained multiple single nucleotide polymorphisms (SNPs) within the rhDHV1 region (nucleotides positions 15974–16516 of the Macaca mulata NCBI reference sequence NC_005943) that were informative and permitted to distinguish the mtDNA origin of infants and cell lines (Fig. 3a). Initially, we performed direct sequencing of rhDHV1 and the results confirmed that in all samples the mtDNA originated from the cytoplast donors (Fig. 3b). No contribution of spindle donor mtDNA was detected in any ST offspring using this approach. As indicated above, a small amount of cytoplasm is usually carried over with the spindle during the ST procedure. Therefore, we conducted three independent quantitative mtDNA assays. The first involved cloning of mtDNA PCR products containing rhDHV1region and the direct sequencing of multiple individual clones. We randomly selected and sequenced 20–25 clones for each sample and the results indicated that mtDNA in all analyzed clones was from cytoplast donors (Suppl. Table 5). Thus, if heteroplasmy existed, any contribution of spindle mtDNA in ST infants and cell lines is below detectable levels.
Figure 3. MtDNA analysis in ST offspring.
a, Origin of ST offspring and informative mtDNA SNPs. b, mtDNA chromatogram demonstrating SNPs. Infants 1 and 2 and ST1 and 2 cell lines were produced by transfer of spindles from female 1 to cytoplasts from female 2. C, Restriction enzyme recognition sites within SNP area. Female 1 mtDNA can be digested by HindIII while G to A nucleotide change precludes restriction of female 2 mtDNA. KpnI digests mtDNA from the female 5 but not from the female 6. d, mtDNA from females 5 and 6 were mixed at various proportions and were detectable at the level of 3%. e, RFLP analysis of ST offspring demonstrating undetectable contributions of spindle donor mtDNA.
The second approach employed mtDNA Restriction Fragment Length Polymorphism (RFLP)19. SNPs in the mitochondrial genome can result in the loss of a restriction site, thus digestion of PCR fragments with endonucleases can be employed to reveal the presence of a population of mtDNA originating from the spindle donors. We identified unique restriction enzyme recognition sites within SNP areas for all ST samples except for the third infant. Specifically, mtDNA of the female 1 (a spindle donor for infants 1 and 2 plus ST1 and ST2 cell lines) possessed a unique HindIII digestion site that includes the G/A SNP site (Fig. 3c). In contrast, female 2 (a cytoplast donor) carried mtDNA with an A allele at this SNP precluding recognition and digestion by HindIII (Fig. 3c). To confirm the sensitivity of this approach, we performed RFLP analysis of serial mixtures (1 to 100%) of mtDNA from females 5 and 6. The results indicate that RLFP can detect as little as 3% of heteroplasmy (Fig. 3d). MtDNA heteroplasmy was undetectable in any experimental sample by RFLP (Fig. 3e).
Lastly, we employed a quantitative real-time PCR analysis based on existing SNP differences between spindle and cytoplast donor animals. Two different fluorescent probes were designed for identification of mtDNA from females 5 and 6 based on SNPs and specificity was validated (Suppl. Fig. 5a). To build a standard curve and establish sensitivity, we conducted real-time PCR analysis of mtDNA mixtures from the two females at various ratios. The technique detected heteroplasmy at the minimal level of 3% (Suppl. Fig. 5b). Next, we analyzed STES1 and 2 cell lines derived from these females and results confirmed that contribution of the spindle donor mtDNA is undetectable (Suppl. Fig. 5b). Similarly, we performed a real-time PCR analysis of mtDNA from infants 1 and 2 as well as ST1–4 cultures; the results indicated that all ST offspring is nearly homoplasmic.
These results provide strong support for the notion that mtDNA can be efficiently replaced in mature primate oocytes by ST resulting in offspring with undetectable or negligible amounts of spindle-derived mtDNA.
Karyoplast fusion using SeV extract was instrumental in our study but clinical applications of the viral extract may be problematic. Therefore, we analyzed SeV extract, ST infants and STES cells for presence of the SeV genetic material. We employed reverse transcription (RT)-PCR assay using commonly used primers for the F protein-coding sequence20. We did not detect any presence of viral genome in analyzed samples (Suppl. Fig. 6) indicating that the extract is inactivated and purified from the genomic RNA.
Discussion
Assisted reproductive technologies that could eliminate the transmission of mitochondrial diseases in affected families include cytoplasmic, germinal vesicle and pronuclear transfer15,21,22. Cytoplasmic transfer, the augmentation of patient eggs with a small amount of donor cytoplasm, was designed to improve viability and developmental competence of compromised, “ooplasmic deficient” oocytes. The introduction of large amounts of donor cytoplasm by electrofusion were unsuccessful23, however, the co-injection of a small volumes (1–5%) of donor cytoplasm with sperm as an extension of ICSI has been successful with acceptable IVF outcomes23–26. While heteroplasmy was detected in small amounts in six of 13 embryos studied and in two of four fetal cord blood samples27, this procedure is not suitable for women carrying mtDNA mutations since it would require transferring significantly larger, practically impossible, cytoplasmic volumes (30–50% of the final volume) to ensure adequate dilution of mutant mtDNA. A relatively high number of chromosomal abnormalities and birth defects in infants resulting from the initial application of this technique28 led to its banning in the United States by the Food and Drug Administration because of safety concerns.
Another possibility is transferring nuclear DNA from a mother with mtDNA disease to a cytoplast (or spindle-free oocyte) containing normal mtDNA as obtained from a healthy egg donor. Experiments in the mouse suggested that it is technically feasible to transfer DNA between immature oocytes where nuclear DNA is enclosed in a clearly visible germinal vesicle (GV)29. However, if this approach were applied to human oocytes, efficacy would be limited by the poor developmental competence of oocytes produced after the in-vitro maturation of GV-intact oocytes. Moreover, GV oocytes have polarized cyto-architecture with mitochondria concentrated in the perinuclear space30 and GV transplantation would inevitably result in the introduction of significant amounts of patient mtDNA into the donor cytoplasm. Pronuclear transfer is essentially the same procedure, except that the nuclear material, both the male and female pronucleus, is removed after fertilization. This technique in the mouse, has resulted in the birth of live offspring, however, reconstructed embryos contained karyoplast mtDNA (on average 19%) and progeny were all heteroplasmic31–33.
Thus, these nuclear transfer approaches are associated with the transmission of a significant amount of mtDNA from the nuclear donor (patient) resulting in heteroplasmy rendering them inappropriate for the patient with mtDNA associated disease.
Chromosome transfer in mature oocytes was thought unattainable due to the unique biological characteristics of MII arrested oocytes until we introduced several procedural innovations that overcame these biological barriers12. Here we also provide evidence that the ST approach is efficient, safe and does not generate any detectable heteroplasmy. This could be due to the fact that spindle-chromosomal complexes were isolated into karyoplasts containing very small amounts of cytoplasm. Moreover, in primates, karyoplasts are mostly occupied by a spindle-chromosomal apparatus that is free of mitochondria. Another advantage is that ST does not require the in vitro oocyte maturation and can be performed with minimum procedural upgrades in a standard clinical IVF setting.
Assuming that our protocols are applicable to human oocytes, ST with cryopreserved oocytes would undoubtedly benefit future clinical applications. Current technology requires that the oocyte donor and recipient undergo synchronous ovarian stimulation protocols; however the increasing availability of oocyte cryopreservation protocols will abrogate this requirement.
As we show here, the ST strategy will likely contribute the least amount of the spindle-associated mtDNA compared to the other approaches discussed above. The sensitivity of our assays did not allow detection of mtDNA heteroplasmy levels below the 3% threshold. However, due to random genetic drift and segregation of mtDNA, some tissues and organs of ST offspring may contain higher degree of heteroplasmy. Another concern is the possibility of recurrence of mtDNA diseases in the next generation, in children of heteroplasmic females produced by ST, due to the phenomenon known as “genetic bottleneck”34,35. Future clinical applications of mtDNA replacement therapy will largely dependent upon the safety and efficiency of this technology. Thus, appropriate long-term preclinical studies addressing these important concerns in a clinically relevant nonhuman primate model are warranted before this technology can be applied to humans.
In summary, mtDNA mutations contribute to a diverse range of human diseases, encompassing both tissue-specific and multiple system disorders, with the onset of disease appearing throughout life. We demonstrate here that the transmission of mtDNA mutations from mother to child can be abrogated by the efficient transfer of mother's spindle-associated chromosomes into donor cytoplasts. In the rhesus monkey, spindle-associated chromosomal transfer did not show adverse effects on fertilization or on subsequent embryo/fetal development to term. Hence, we propose that this procedure may represent a new reliable therapeutic approach to prevent the transmission of mtDNA mutations in affected families.
Methods Summary
Mature MII oocytes were recovered from females undergoing controlled ovarian stimulations. Oocytes were transferred to a manipulation chamber containing medium with 5 μg ml cytochalasin B and MII spindle-chromosomal complexes were aspirated and isolated using Oosight imaging system (www.cri-inc.com) as described before16. Isolated karyoplasts were briefly exposed to SeV extract (www.cosmobio.co.jp) and placed into perivitelline space on the side opposite to the 1st polar body (Suppl. video). During manipulations, oocyte's zona pellucida was penetrated by laser assisted zona drilling (www.hamiltonthorne.com). After fusion, reconstructed oocytes were fertilized by ISCI and cultured as described before16. Detailed methods are described in Supplementary information at www.nature.com/nature.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the Division of Animal Resources, Surgical Team, Endocrine Technology Core, Imaging & Morphology Core and Molecular & Cellular Biology Core at the Oregon National Primate Research Center for providing expertise and services that contributed to this project. We are grateful to Dr. Warren Sanger and Marilu Nelson for karyotyping services, Dr. Cecilia Penedo for microsatellite analysis and Dr. Scott Wong for providing Sendai virus. We are indebted to Drs. Jon Hennebold, Richard Stouffer and Don Wolf for consulting, helpful discussions and critical reading of the manuscript.
This study was supported by start up funds from Oregon National Primate Research Center, Oregon Stem Cell Center and grants from the National Institutes of Health. The authors have no financial affiliation that may be perceived as biasing this work.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 2.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 3.Zeviani M, et al. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 4.Solano A, Playan A, Lopez-Perez MJ, Montoya J. Genetic diseases of the mitochondrial DNA in humans. Salud publica de Mexico. 2001;43:151–161. [PubMed] [Google Scholar]
- 5.Keating DJ. Mitochondrial dysfunction, oxidative stress, regulation of exocytosis and their relevance to neurodegenerative diseases. J Neurochem. 2008;104:298–305. doi: 10.1111/j.1471-4159.2007.04997.x. [DOI] [PubMed] [Google Scholar]
- 6.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 7.Reeve AK, Krishnan KJ, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- 8.Trushina E, McMurray CT. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 9.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 10.Sutovsky P, et al. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 11.Majamaa K, et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Am J Hum Genet. 1998;63:447–454. doi: 10.1086/301959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer AM, et al. Prevalence of mitochondrial DNA disease in adults. Annals of neurology. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 14.Steffann J, et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J Med Genet. 2006;43:244–247. doi: 10.1136/jmg.2005.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorburn DR, Dahl HH. Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. American journal of medical genetics. 2001;106:102–114. doi: 10.1002/ajmg.1380. [DOI] [PubMed] [Google Scholar]
- 16.Byrne JA, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 17.Mitalipov SM, et al. Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. Hum Reprod. 2007;22:2232–2242. doi: 10.1093/humrep/dem136. [DOI] [PubMed] [Google Scholar]
- 18.Wolf DP, et al. Use of assisted reproductive technologies in the propagation of rhesus macaque offspring. Biol Reprod. 2004;71:486–493. doi: 10.1095/biolreprod.103.025932. [DOI] [PubMed] [Google Scholar]
- 19.Moraes CT, Atencio DP, Oca-Cossio J, Diaz F. Techniques and pitfalls in the detection of pathogenic mitochondrial DNA mutations. J Mol Diagn. 2003;5:197–208. doi: 10.1016/S1525-1578(10)60474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuboniwa N, et al. Safety evaluation of hemagglutinating virus of Japan--artificial viral envelope liposomes in nonhuman primates. Hum Gene Ther. 2001;12:469–487. doi: 10.1089/104303401300042366. [DOI] [PubMed] [Google Scholar]
- 21.Poulton J, Kennedy S, Oakeshott P, Wells D. Preventing transmission of maternally inherited mitochondrial DNA diseases. BMJ (Clinical research ed. 2009;338:b94. doi: 10.1136/bmj.b94. [DOI] [PubMed] [Google Scholar]
- 22.Roberts RM. Prevention of human mitochondrial (mtDNA) disease by nucleus transplantation into an enucleated donor oocyte. American journal of medical genetics. 1999;87:265–266. doi: 10.1002/(sici)1096-8628(19991126)87:3<265::aid-ajmg14>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 23.Cohen J, et al. Ooplasmic transfer in mature human oocytes. Mol Hum Reprod. 1998;4:269–280. doi: 10.1093/molehr/4.3.269. [DOI] [PubMed] [Google Scholar]
- 24.Lanzendorf SE, et al. Pregnancy following transfer of ooplasm from cryopreserved-thawed donor oocytes into recipient oocytes. Fertil Steril. 1999;71:575–577. doi: 10.1016/s0015-0282(98)00504-4. [DOI] [PubMed] [Google Scholar]
- 25.Cohen J, Scott R, Schimmel T, Levron J, Willadsen S. Birth of infant after transfer of anucleate donor oocyte cytoplasm into recipient eggs. Lancet. 1997;350:186–187. doi: 10.1016/S0140-6736(05)62353-7. [DOI] [PubMed] [Google Scholar]
- 26.Barritt JA, Brenner CA, Malter HE, Cohen J. Mitochondria in human offspring derived from ooplasmic transplantation. Hum Reprod. 2001;16:513–516. doi: 10.1093/humrep/16.3.513. [DOI] [PubMed] [Google Scholar]
- 27.Brenner CA, Barritt JA, Willadsen S, Cohen J. Mitochondrial DNA heteroplasmy after human ooplasmic transplantation. Fertil Steril. 2000;74:573–578. doi: 10.1016/s0015-0282(00)00681-6. [DOI] [PubMed] [Google Scholar]
- 28.Brown DT, et al. Transmission of mitochondrial DNA disorders: possibilities for the future. Lancet. 2006;368:87–89. doi: 10.1016/S0140-6736(06)68972-1. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi T, Neri QV, Katagiri Y, Rosenwaks Z, Palermo GD. Effect of treating induced mitochondrial damage on embryonic development and epigenesis. Biol Reprod. 2005;72:584–592. doi: 10.1095/biolreprod.104.032391. [DOI] [PubMed] [Google Scholar]
- 30.Wilding M, et al. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod. 2001;16:909–917. doi: 10.1093/humrep/16.5.909. [DOI] [PubMed] [Google Scholar]
- 31.Meirelles FV, Smith LC. Mitochondrial genotype segregation in a mouse heteroplasmic lineage produced by embryonic karyoplast transplantation. Genetics. 1997;145:445–451. doi: 10.1093/genetics/145.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meirelles FV, Smith LC. Mitochondrial genotype segregation during preimplantation development in mouse heteroplasmic embryos. Genetics. 1998;148:877–883. doi: 10.1093/genetics/148.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato A, et al. Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc Natl Acad Sci U S A. 2005;102:16765–16770. doi: 10.1073/pnas.0506197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cree LM, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 35.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.