Abstract
The Fat-Hippo signaling pathway plays an important role in the regulation of normal organ growth during development, and in pathological growth during cancer. Fat-Hippo signaling controls growth through a transcriptional co-activator protein, Yorkie. A Fat-Hippo pathway has been described in which Yorkie is repressed by phosphorylation, mediated directly by the kinase Warts and indirectly by upstream tumor suppressors that promote Warts kinase activity. We present here evidence for an alternate pathway in which Yorkie activity is repressed by direct physical association with three other pathway components: Expanded, Hippo, and Warts. Each of these Yorkie repressors contains one or more PPXY sequence motifs, and associates with Yorkie via binding of these PPXY motifs to WW domains of Yorkie. This direct binding inhibits Yorkie activity independently from effects on Yorkie phosphorylation, and does so both in vivo and in cultured cell assays. These results emphasize the importance of the relative levels of Yorkie and its upstream tumor suppressors to Yorkie regulation, and suggest a dual repression model, in which upstream tumor suppressors can regulate Yorkie activity both by promoting Yorkie phosphorylation and by direct binding.
Keywords: Fat, Hippo, Oncogene, Yorkie
INTRODUCTION
The Fat-Hippo signaling pathway plays an essential role in the regulation of organ growth from Drosophila to mammals (reviewed in Reddy and Irvine, 2008). Many of the components of this pathway were first identified as tumor suppressors genes, and the importance of this pathway in diverse cancers has been increasingly recognized. Fat-Hippo signaling controls growth through the regulation of transcription. The critical mediator of its effects on transcription is a transcriptional co-activator protein, called Yorkie (Yki) in Drosophila and YAP mammals (Huang et al., 2005). Yki acts as an oncogene, and in conjunction with the DNA-binding protein Scalloped (Sd), promotes the expression of genes that accelerate growth and cell cycle progression, whilst inhibiting apoptosis (reviewed in Reddy and Irvine, 2008).
Most of the known upstream components of Fat-Hippo signaling function as tumor suppressors, and normally inhibit Yki activity. This inhibition is effected most directly by Warts (Wts), a kinase that phosphorylates Yki (Huang et al., 2005). Phosphorylation of Yki or YAP inhibits Yki activity by promoting its retention in the cytoplasm (Dong et al., 2007; Hao et al., 2008; Oh and Irvine, 2008; Zhang et al., 2008; Zhao et al., 2007). Wts in turn is regulated by several upstream tumor suppressors that influence its levels and activity, including Mats, which acts as an essential co-factor for Wts, Hippo (Hpo), which is a kinase that phosphorylates Mats and Warts, Salvador, which binds to Wts and Hpo, Expanded (Ex) and Merlin, two FERM domain proteins that promote Hpo activity, and Fat, a protocadherin that influences the levels or localization of Wts and Ex via the unconventional myosin Dachs (reviewed in Reddy and Irvine, 2008).
Structure-function studies of Yki have led to the identification of several motifs important for its activity. A critical Wts phosphorylation site is Ser168, phosphorylation of which creates a binding site for 14-3-3 proteins (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008; Zhao et al., 2007). This contributes to the retention of phosphorylated Yki in the cytoplasm, as mutation of Ser168 to Ala hyperactivates Yki and increases its nuclear localization. Yki also has two additional Wts phosphorylation sites, at Ser111 and Ser250, each of which makes additional contributions to the repression of Yki activity (Oh and Irvine, 2009). Sd binds to a conserved region in the amino terminus of Yki (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008). In addition, Yki has two WW domains, which are 35–40 amino acid protein interaction domains that include two Trp residues (Macias et al., 2002). The WW domains of Yki were first identified for their role in binding to Wts, which suggested that they might promote Yki phosphorylation (Huang et al., 2005). However, subsequent studies have shown that the WW domains are not required for Yki phosphorylation (Oh and Irvine, 2008; Oh and Irvine, 2009). More recently, we and others have determined that the WW domains positively influence Yki/YAP activity, presumably because they interact with co-factors that are required for transcriptional activation by Yki (Oh and Irvine, 2009; Wu et al., 2008; Zhao et al., 2009). In this work, we identify an additional activity of the WW domains in repression of Yki, through a mechanism that does not depend on the phosphorylation of Yki by Wts.
The existence of this phosphorylation-independent mechanism was suggested by characterization of a mutant form of Yki (Yki:V5S111A,S168A,S250A, henceforth abbreviated as Yki:V53SA) in which all three Wts phosphorylation sites are mutant (Oh and Irvine, 2009). Expression of Yki:V53SA results in dramatic overgrowth phenotypes, whereas expression of a wild-type form of Yki:V5 at the same level results in no apparent phenotype, consistent with the conclusion that phosphorylation of Yki by Wts represses Yki activity. Yki:V53SA was also resistant to expression of upstream tumor suppressors implicated in promoting Yki phosphorylation, because some overgrowth occurs even when Yki:V53SA is co-expressed with Wts, Hpo, and Ex. However, Wts, Hpo, and Ex did have an effect on Yki:V53SA, in that they could reduce its nuclear localization, even though they could not affect its phosphorylation. The discovery of this phosphorylation-independent repression of Yki nuclear localization by upstream tumor suppressors prompted further explorations of its mechanism and significance.
Our results identify a phosphorylation-independent mechanism by which upstream tumor suppressors, including Ex, Wts, and Hpo, can repress Yki activity. This alternate mechanism involves interactions between the WW domains of Yki and PPXY sequence motifs on its binding partners. Recently, Badouel et al (2009) described the results of studies in which they identified an ability of Ex to repress Yki through a mechanism that depends on the PPXY motifs of Ex. As discussed below, our results are consistent with this study in that we also identify a phosphorylation-independent mechanism by which Ex can repress Yki, but differ in our additional identification of phosphorylation-independent activities of Wts and Hpo, and in our assessment of the respective contributions of different mechanisms of Yki repression.
MATERIALS AND METHODS
Fly stocks
UAS-yki: V5 lines employed were site-specific insertions in the attP2 site at 68A (UAS-yki:V5[attP-68A], or mutant variants) (Oh and Irvine, 2009). For hpo, wts, and ex expression we used UAS-ex[ex-3] (Boedigheimer et al., 1997), UAS-Myc:wts.2 (Jia et al., 2003), and UAS-hpo[dMst.3] (Jia et al., 2003). For ectopic expression clones, y w hs-FLP[122]; Act>y+>Gal4 (AyGal4) or y w hs-FLP[122]; Act>y+>Gal4 UAS-GFP (AyGal4-GFP) were crossed to UAS-yki:V5[attP-68A] (or mutant isoforms). For co-expression with hpo, wts, and ex in clones, UAS-ex was crossed to y w hs-FLP[122]; UAS-Myc:wts.2/CyO, GFP; UAS-yki:V5 UAS-hpo[dMst.3]/TM6b. Ectopic expression of UAS-yki:V5 isoforms, with or without UAS-hpo, UAS-wts, and UAS-ex was induced under GMR-Gal4 control in crosses of GMR-Gal4 or GMR-Gal4; UAS-ex/TM6b to y w hs-FLP[122]; UAS-Myc:wts.2/CyO, GFP; UAS-yki3SA:V5/TM6b or y w hs-FLP[122]; UAS-Myc:wts.2/CyO, GFP; UAS-yki3SA:V5 UAS-hpo[dMst.3]/TM6b. yki mutant clones were generated using FLP-FRT-mediated recombination with w; ykiB5 FRT42D/CyO-GFP and y w hs-FLP; FRT42D Ubi-GFP/CyO-GFP. wts mutant clones were generated using y w hs-FLP tub-Gal4 UAS-GFP; UAS-y+/CyO; FRT82B tub-Gal80/TM6b, y w hs-FLP; If/CyO; FRT82B wtsX1/TM6b and y w hs-FLP; If/CyO; UAS-Ex FRT82B wtsX1/TM6b. The site-specific insertions UAS-Myc:wts[attP-25C7] and UAS-Myc:wtsK743A[attP-25C7] (kinase dead) lines were created by inserting KpnI-XbaI fragments from UAS-Myc:Wts plasmids into pUAST-attB; the resulting plasmids were then inserted into the attP40 site at 25C7 by phiC31-medited integration (Genetic Services). Unless otherwise noted, all flies were cultured at 25°C.
Histology and imaging
Imaginal discs were fixed and stained as described previously (Cho and Irvine, 2004), using as primary antibodies rabbit anti-Yki (1:4000)(Oh and Irvine, 2008), mouse anti-V5 (1:400, Invitrogen), mouse anti-Diap1 (1: 600, gift from B. Hay) and guinea pig anti-Ex (1:2000, gift from R. Fehon) mouse anti-Myc (1:200, Babco) and goat anti-β-gal (1:1000, Biogenesis). Fluorescent stains were captured on a Leica TCS SP5 confocal microscope. For quantitative analysis of Yki localization in wing imaginal disc cells, ratios of the average flourescence intensity in the nucleus and cytoplasm of 30 cells were measured using NIH Image J.
Plasmid constructs
Mutations in the five PPXY motifs of Wts (Y286A, Y304A, Y410A, Y463A, Y550A), the PPXY motif of Hpo (Y591A) and Wts-KD (K743A) (kinase-dead) were introduced by primer-mediated site-directed mutagenesis using QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Flag-tagged Hpo, Myc-tagged Wts and HA-tagged Ex UAS expression plasmids have been described previously (Hamaratoglu et al., 2006; Jia et al., 2003). For some experiments these genes were cloned into pAC5.1 for actin promoter regulated expression, pAC5.1-V5:Wts has been described previously (Huang et al., 2005). pAC5.1-Gal4-DBD:Yki:V5 has been described previously, pAC5.1-Gal4-DBD:Yki:HA was created by exchanging a BamHI fragment from pAC5.1-yki:HA (Huang et al., 2005) into pAC5.1-Gal4-DBD:Yki:V5.
S2 cell assays and co-immunoprecipitation
S2 cells were cultured with Schneider’s Drosophila medium (Invitrogen) and 10% FBS (Sigma). For co-immunoprecipitations, transient transfections were performed with equal amounts of DNA (0.5 μg per construct) using Cellfectin (Invitrogen) in 6 well plates according to the manufacturer’s protocol, using plasmids pAW-Gal4 (S. Blair), pAct-GFP:V5-His (Cho, 2006), Yki:V5 isoforms, pUAS-Myc:Wts, pUAS-Flag:dMST (Jia et al., 2003), pAct-Ex:HA (Hamaratoglu et al., 2006), pAC5.1-3xFLAG:Hpo, and pAC5.1-3xFLAG:HpoPPXA. Co-immunoprecipitation assays were performed according to published protocols (Chen et al., 2003). In brief, cells were harvested 48 hours later in RIPA lysis buffer (50 mM Tris-HCL, pH 7.5; 150 mM NaCl; 1% NP40; 0.5% Sodium deoxycholate; 0.1% SDS; 1 mM EDTA) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Calbiochem). Cell lysates (500 μg for each sample) were incubated with 10 μl Anti-V5 or anti-FLAG beads (Sigma) at 4°C for 2 hours followed by five washes in RIPA buffer. Beads were then boiled in Laemmli sample buffer at 100°C for five minutes and loaded onto SDS-PAGE gels. Western blotted proteins were visualized using: rabbit Anti-Myc (1:2000, Santa Cruz), rabbit anti-Wts, rabbit anti-Yki (1:4000), rabbit anti-YkiS168P (gift of D. Pan), rabbit anti-Hpo (1:2000, gift of N. Tapon), mouse anti-V5 (1:5000, Invitrogen), IRdye 800 conjugated anti-Flag (1:100000, rabbit), IR dye 800 conjugated anti-HA (1:100000, rabbit) (Rockland), IR dye 800 conjugated anti-rabbit (goat, 1:10000), IR dye 680 conjugated anti-mouse (goat, 1:10000) (Odyssey). Blots were analyzed using the Odyssey Infrared Imaging system (Licor biosciences). Phos-tag gels (FMS Laboratory) were prepared as described previously (Oh and Irvine, 2008).
Luciferase reporter assays were performed using the Dual Luciferase Assay System (Promega) according to the manufacturer’s instructions. S2 cells were transfected in triplicate with pUAST-Luciferase (10 ng) and copia-renilla luciferase (0.2 ng) reporters (Lum et al., 2003) in 24-well plates together with Gal4DB-Yki constructs (100 ng) and upstream tumor suppressors (Hpo, Wts and/or Ex, 100 ng each, or 500 ng for a five fold increase) in a pAc5.1 vectors (Invitrogen) and incubated for 48 h after transfection.
RESULTS
Repression of unphosphorylatable Yki by upstream tumor suppressors
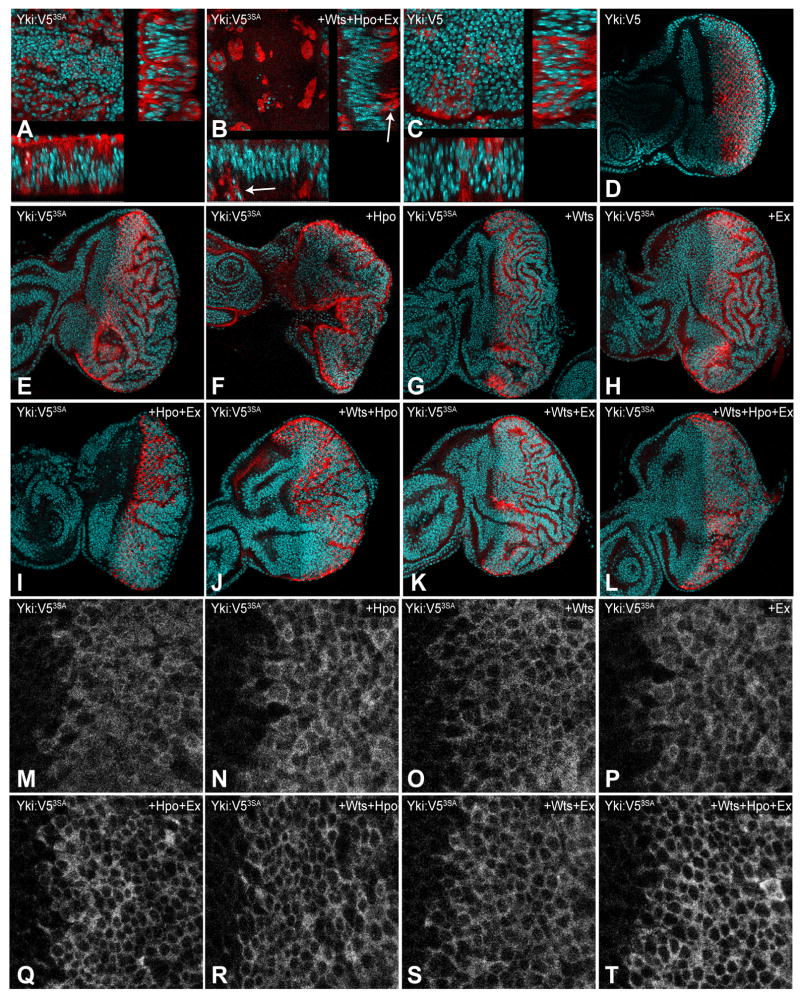
Clones of cells in which Wts, Hpo, and Ex are over-expressed together with Yki:V53SA generally overgrow compared to control clones, but can be less overgrown than Yki:V53SA-expressing clones. This is most evident within the wing pouch, in which clones of cells co-expressing Yki:V53SA together with Hpo, Wts and Ex can be small and appear apoptotic, as evidenced by the small, fragmented nuclei that appear basal to the disc epithelium (Fig. 1B). There also appeared to be a tendency of Yki:V53SA clones to overgrow to a lesser extent in other regions of the wing or eye disc in the presence of Hpo, Wts and Ex, but because this effect was hard to quantify relative to the normal variations in clone size, we turned to a system in which transgenes would be expressed in constant regions.
Fig 1. Phosphorylation-independent repression of Yki.
A–C) Horizontal (upper left) and vertical (bottom and right) sections of wing discs with flip-out clones expressing Yki:V5 (wild-type control) or Yki:V53SA (activated Yki), identified by elevated Yki (red), and with nuclei labeled by DAPI (cyan). A) act>y+>Gal4; UAS-yki:V53SA. B) act>y+>Gal4; UAS-Myc:wts.2;UAS-yki:V53SA UAS-ex[ex-3]/UAS-hpo[dMst.3]. In the presence of elevated Wts, Hpo, and Ex, Yki:V53SA-expressing cells become apoptotic and drop basally (arrows). C) act>y+>Gal4 UAS-yki:V5. D-L) Eye discs expressing Yki:V5 (UAS-yki:V5) or Yki:V53SA (UAS-yki:V53SA) under GMR-Gal4 control, stained for elevated Yki (red), and with nuclei labeled by DAPI (cyan). E) Expression of Yki:V53SA results in extensive overgrowth, which is mostly taken up in folds of tissue. F-L) Co-expression of UAS-ex[ex-3], UAS-Myc:wts.2 or UAS-hpo[dMst.3], or combinations thereof, with Yki:V53SA, as indicated. M-T) Close-ups of Yki:V53SA expression in eye discs under GMR-Gal4 control to show nuclear localization, using same genotypes as in E-L.
This was achieved by expressing Yki:V53SA in eye discs under GMR-Gal4 control, in the presence or absence of Wts, Hpo, and Ex. GMR-Gal4 drives expression in the posterior of the third instar eye disc, behind the morphogenetic furrow. Expression of UAS-Yki:V53SA transgenes using GMR-Gal4 results in overgrowth of the eye disc (Fig. 1E), and ultimately of the adult eye (Oh and Irvine, 2009), which is mostly taken up in extensive folds of tissue. This overgrowth and folding is largely suppressed by co-expression of Hpo, Wts and Ex (Fig. 1L). Moreover, co-expression with Hpo, Wts and Ex reduces nuclear accumulation of Yki3SA in the eye (Fig. 1T). We also examined all of the possible single and double combinations of Hpo, Wts and Ex co-expression with Yki3SA (Fig. 1). The repression of Yki3SA-mediated overgrowth was most effective in the presence of all three upstream tumor suppressors, although addition of Hpo and Wts, or Hpo and Ex also had obvious growth inhibitory effects (Fig. 1I–L), and an effect on nuclear localization could be detected for all of the pair-wise combinations (Fig. 1Q–S). These observations confirm that there is a phosphorylation-independent mechanism by which upstream tumor suppressors can inhibit Yki activity and reduce its nuclear localization in vivo.
Binding of Yki to Hpo, Wts and Ex
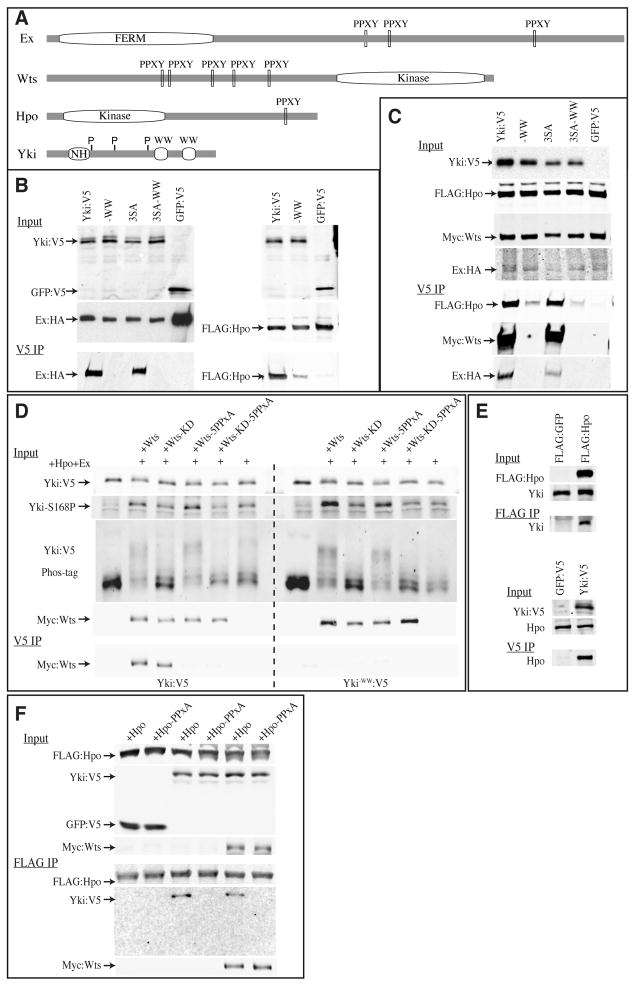
Yki was first identified through binding to Wts in a two-hybrid screen, mediated by WW domain – PPXY motif interactions (Huang et al., 2005). Yki has two WW domains, and Wts has five PPXY motifs (Fig. 2A). Interestingly, we found one PPXY motif in Hpo and three in Ex, raising the possibility that Yki might also bind to them (Fig. 2A). Indeed, when Hpo or Ex were co-expressed with Yki in cultured cells, they could be co-precipitated with Yki, and their association was abolished (Ex), or reduced (Hpo) by mutation of the WW domains (Fig. 2B). When all four proteins were co-transfected together into S2 cells, Yki was partitioned among them, as it could be co-precipitated with Wts, Ex, and Hpo (Fig. 2C). Examination of the Yki3SA mutant established that this association does not depend on the Wts phosphorylation sites of Yki (Fig. 2B, C). As the WW domains are not required for phosphorylation of Yki by Wts (Oh and Irvine, 2008; Oh and Irvine, 2009), the functional significance of Yki-Wts binding was unknown. The observations that Hpo, Wts and Ex can effect a phosphorylation-independent repression of Yki in vivo, and that these proteins can all bind directly to Yki, suggest that this binding mediates phosphorylation-independent repression.
Fig 2. Binding of Hpo, Wts, and Ex to Yki.
A) Schematic structures of Ex, Wts, Hpo, and Yki, depicting locations of FERM domain, PPXY motifs, kinase domains, WW domains, Sd binding region of Yki (NH) and Wts phosphorylation sites on Yki (P). B–F) Co-immunprecipitation experiments on proteins expressed in S2 cells. Upper panels (Input) show Western blots on lysates, lower panels (IP) show Western blots on material precipitated on anti-V5 or anti-FLAG beads, as indicated. B) Co-precipitation of tagged Yki and Ex (left) or Yki and Hpo (right) is eliminated or reduced by mutation of the WW domains (−WW) but is not affected by mutation of the Yki phosphorylation sites (3SA). 3SA-WW is a Yki isoform that includes both WW domain and phosphorylation site mutations. C) Co-precipitation of tagged Hpo, Wts, and Ex with Yki. When all four proteins are mixed together, Yki co-precipitates Hpo, Wts, or Ex, and this association is eliminated or reduced by mutation of the WW domains (−WW) but not by mutation of Yki phosphorylation sites (3SA). D) Binding between Yki and Wts requires the PPXY motifs, but not the kinase activity of Wts (Wts-KD). Wts kinase activity, but not the PPXY motifs, promotes phosphorylation of Yki, as evidenced by blotting with phospho-specific antisera (Yki-S168P)(Dong et al., 2007) and by Phos-tag gel analysis (Oh and Irvine, 2008). E) Upper panel shows that FLAG-tagged Hpo can co-precipitate endogenous Yki; lower panel shows that V5 tagged Yki can co-precipitate endogenous Hpo. GFP control proteins are not visible here because their mobilities are distinct from Hpo and Yki. F) Co-precipitation of HpoY591A,(PPXA), Wts, and Yki in S2 cells. Yki-Hpo binding requires the PPXY motif of Hpo, and is not affected by exogenous Wts; Wts-Hpo binding does not require the Hpo PPXY motif.
The ability of Yki to bind to Ex was also recently identified by Badouel et al (2009), but binding of Yki to Hpo has not been described previously. Available antibodies against endogenous proteins did not work well enough for us to detect a convincing signal above background in co-immunoprecipitation experiments. However, co-immunoprecipitation of endogenous Yki could be detected when exogenous FLAG:Hpo was precipitated from S2 cells, and co-immunoprecipitation of endogenous Hpo could be detected when exogenous Yki:V5 was precipitated from S2 cells (Fig. 2E). Moreover, mutation of the PPXY motif in Hpo (Y591A) abolished detectable binding to Yki, but did not affect binding to Wts (Fig. 2F). These observations suggest that Hpo and Yki can interact directly in vivo through their PPXY and WW domains.
Phosphorylation-independent repression of Yki in S2 cells
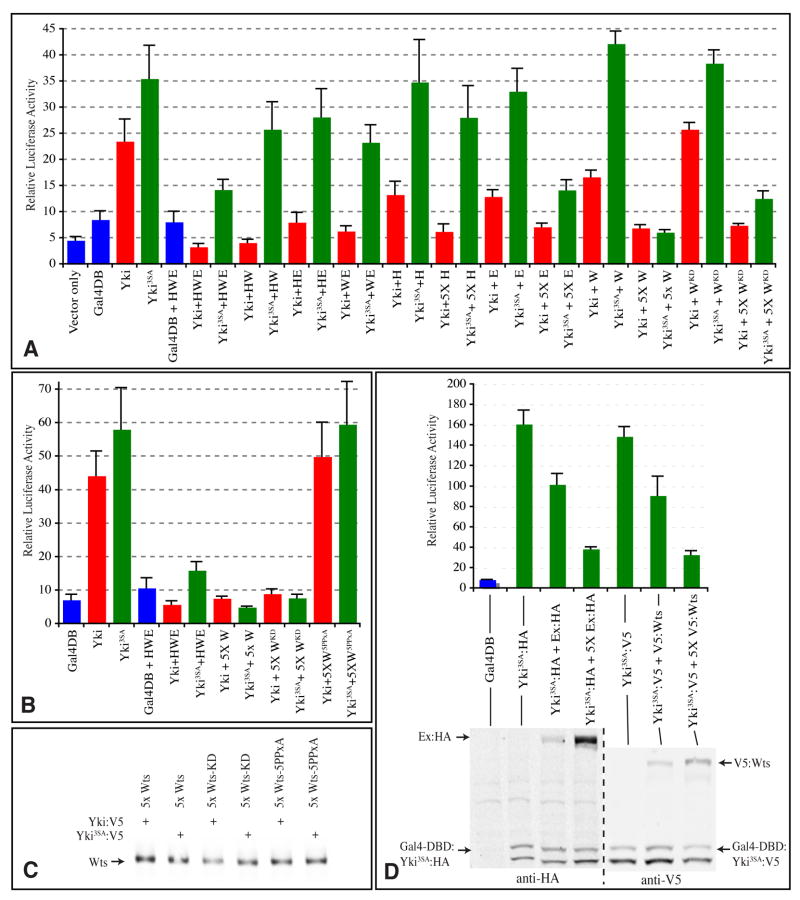
Yki’s activity as a transcriptional co-activator can be visualized by measuring the activity of a Yki-Gal4 DNA binding domain fusion (Gal4-DB:Yki) on a Gal4-responsive luciferase reporter (Huang et al., 2005). To investigate phosphorylation-independent repression using this transcriptional assay, Gal4-DB:Yki or Gal4-DB:Yki3SA were co-transfected with different combinations of upstream tumor suppressors into S2 cells. Gal4-DB:Yki3SA exhibited greater transcriptional activity than Gal4-DB:Yki under a range of conditions (Fig. 3A). Moreover, individual expression of Ex, Wts and Hpo on their own at moderate doses (i.e., using identical plasmid vectors and equal amounts of DNA for Gal4-DB:Yki and each of these repressors) partially repressed Gal4-DB:Yki-mediated transcriptional activation, but failed to repress Gal4-DB:Yki3SA-mediated transcriptional activation, consistent with the importance of Yki phosphorylation in Yki repression.
Fig 3. Phosphorylation-independent repression of Yki-mediated transcription.
Expression from a UAS-luciferase reporter is indicated by luciferase activity in lysates of S2 cells transfected to express wild-type (red) or 3SA mutant (green) Gal4-DBD:Yki:V5 fusions (Yki) (or a control Gal4-DBD protein, blue). In A, B, where indicated, Yki transgenes were co-expressed with Flag:Hpo (H), Ex:HA (E), Myc:Wts (W), or Myc:WtsKD or Myc:Wts5xPPxA mutants, using equal amounts of DNA, or, where indicated (5x), a five-fold excess of DNA. Histograms depict the average values from triplicate experiments; error bars indicate standard deviation. The results depicted in A and B are from separate experiments. C) Western blot on lysates from samples in B, showing similar expression of Wts. D) Upper panel shows luciferase activity, lower panel shows Western blot on the same cell lysates. The first three experimental (Yki) samples employed HA-tagged Ex and Gal4-DBD:Yki, and the last three employed V5-tagged Wts and Gal4-DBD:Yki, as indicated. The proteins are distinguished on the Western blots by their mobilities, Gal4-DBD:Yki runs as a doublet.
However, when multiple upstream tumor suppressors were combined (i.e. Hpo, Wts and Ex) then Gal4-DB:Yki3SA was also repressed (Fig. 3A), consistent with the repression of Yki3SA observed in vivo. In control experiments, transcription mediated by Gal4-DB alone, or by full length Gal4, was not affected (Fig. 3 and data not shown). To investigate whether phosphorylation-independent repression requires combinations of upstream tumor suppressors, or might instead be achieved by increasing the quantity of a single gene, we examined the ability of upstream tumor suppressors to repress Gal4-DB:Yki3SA when co-transfected in five-fold greater amounts. Under these conditions, Wts or Ex strongly repressed both Gal4-DB:Yki and Gal4-DB:Yki3SA, whereas Hpo strongly repressed Gal4-DB:Yki but not Gal4-DB:Yki3SA (Fig. 3A). In order to confirm that the levels of Ex or Wts need for phosphorylation-independent repression are only a few fold greater than the levels of Yki, we also constructed and characterized transgenes with the same epitope tags (Gal4-DBD:Yki3SA:HA and Ex:HA, Gal4-DBD:Yki3SA:V5 and V5:Wts), such that their levels could be directly compared by Western blotting. Phosphorylation-independent repression was also observed using these constructs, and, levels of Ex or Wts were only modestly greater than levels of Yki (Fig. 3D).
To investigate requirements for Wts kinase activity in Yki repression, we assayed a kinase dead mutant isoform (WtsKD)(Wei et al., 2007). At moderate levels, WtsKD had no effect on either Gal4-DB:Yki or Gal4-DB:Yki3SA, but at five-fold higher levels, WtsKD repressed their activity (Fig. 3A,B). Wild-type Wts and WtsKD were expressed at similar levels in S2 cells (Fig. 2D). However, when we compared the expression of wild-type Wts and WtsKD in imaginal discs, the levels of WtsKD were substantially lower (Supplementary Figure S1). Because the wts expression transgenes in this experiment were inserted at the same cytological location using phiC31 mediated integration (Groth et al., 2004), it appears that inactive Wts is less stable in disc cells, at least when over-expressed. Expression of WtsKD under GMR-Gal4 or en-Gal4 control did not result in evident phenotypes, but as the levels of expression are so low, the significance of this negative result is unclear.
The role of the WW domains in this repression cannot be directly assessed, because they are also positively required for Yki transcriptional activity (Oh and Irvine, 2009). However, when the five PPXY motifs of Wts were mutated by changing the Tyr residues to Ala (Wts5PPxA), the ability of Wts to effect phosphorylation independent repression was lost (Fig. 3B). As expected, Wts5PPxA also failed to bind to Yki in co-IP experiments, but still promoted Yki phosphorylation (Fig. 2D). Using a similar assay, Badouel et al (2009) observed that mutation of the PPXY motifs of Ex impaired its ability to repress Yki. Altogether, these results confirm the existence of phosphorylation-independent repression of Yki transcriptional activity, and show that it is effected by upstream tumor suppressors with PPXY motifs.
Influence of WW domain-PPXY interactions on Yki localization
Endogenous Yki is broadly distributed throughout the cytoplasm (Fig. 4C,D)(Oh and Irvine, 2008), whereas Ex is localized to the subapical membrane (Boedigheimer et al., 1997), so only a small fraction of endogenous Yki could normally be associated with Ex (Fig. 4C,D). Endogenous localization profiles for Hpo and Wts have not been described, but when epitope-tagged forms of Wts are over-expressed, they appear slightly enriched apically, but otherwise broadly distributed throughout the cytoplasm (Cho et al., 2006). Hence the Wts localization profile is potentially consistent with widespread Yki interaction.
Fig 4. Yki localization in imaginal cells.
Portions of wing discs, panels marked by prime symbols show individual channels of immunofluorescent staining. A,B) Expression of UAS-yki:V5 (A) or UAS-yki:V5−WW (B) under ptc-Gal4. C,D) ykiB5 mutant clones in the wing imaginal disc, marked by absence of Yki (green) and stained for Ex (magenta). yki clones are shown to emphasize that the staining in wild-type cells is not background, and that in wild-type Yki co-localization with Ex was not discernible. C) Shows a vertical section, D) Shows a horizontal section, because this is a thin section and the discs are not flat Ex staining is in focus in only part of the image; Ex is also down-regulated within yki mutant clones. E,F) Horizontal (E) and vertical (F) sections through a clone co-expressing Ex, Hpo and Yki:V53SA; (AyGal4; UAS-yki:V5 UAS-hpo[dMst.3]/UAS-ex[ex-3]) a fraction of Yki (green/white) co-localizes with Ex (magenta/white) at the sub-apical membrane, in the absence of Ex over-expression no specific accumulation of Yki at the membrane was detected. G,H) Wing imaginal discs with MARCM clones (marked by presence of GFP, green) of cells mutant for wtsX1 and stained for expression of Diap1 (red). Clones in H also over-express Ex from a UAS-ex transgene. Upregulation of Diap1 (arrows), clone size, and clone shape are similar in the presence and absence of exogenous Ex. I-L) Overlap of Ex (magenta/white) and Yki (green/white) at the sub-apical membrane in wtsX1 mutant clones. I,K show horizontal sections, J,L show vertical sections. Clones in I,J over-express endogenous Ex due to mutation of wts (Hamaratoglu et al., 2006), clones in K,L also over-express Ex from a UAS-ex transgene; at the confocal settings used to detect this elevated Ex, endogenous Ex in wild-type cells is not detected. By contrast to the situation in wild-type, where Ex levels are elevated, a distinct accumulation of Yki could be identified over-lapping Ex. Examples of this are highlighted by the yellow boxes.
To quantify the influence of the WW domains on Yki localization, Yki:V5 and Yki:V5−WW were expressed in identical locations, in a stripe of cells along the center of the wing imaginal disc under ptc-Gal4 control. A slight increase in Yki nuclear localization was detected (Fig. 4A,B; Mean nuclear/cytoplasmic ratio of 0.26 for Yki:V5−WW and 0.22 Yki:V5), and the difference was statistically significant (P value =0.013), consistent with the possibility that the WW domains influence Yki localization in vivo. The effect was slight, but this might be explained in part by a simultaneous decrease in binding to nuclear co-activators that interact with WW domains.
To further investigate the potential influence of Yki’s WW domains on its localization, we over-expressed Ex, because its localization is normally so different from Yki’s. Over-expression of Ex kills cells (Blaumueller and Mlodzik, 2000), but this lethality is suppressed by mutation of wts (Cho et al., 2006; Hamaratoglu et al., 2006). Indeed, as we have noted previously, clones of cells over-expressing Ex but mutant for wts appear very similar to wts mutant clones without Ex over-expression, both in terms of their size and shape, and the expression of downstream genes (Cho et al., 2006) (Fig. 4G,H). This differs from the recent report of Badouel et al (2009), but they appear to have used a different UAS-ex transgene, which, if it drives higher expression, might be able to bypass the normal requirement for wts, just as high level over-expression of Ex in cultured cells can effect a Wts-independent repression of Yki3SA-Gal4DBD mediated transcription (Fig. 3A).
When the localization of Yki in wts mutant clones over-expressing Ex was examined, we found that in addition to the weaker cytoplasmic and stronger nuclear staining characteristic of wts mutant clones (Dong et al., 2007; Oh and Irvine, 2008), a fraction of Yki now accumulated at the sub-apical membrane, where it overlapped Ex (Fig. 4K,L). A partial overlap between Ex and Yki at the subapical membrane could also be detected when the lethality of clones expressing Ex and Hpo was rescued by expression of Yki:V53SA (Fig. 4E,F).
In the course of doing these experiments we realized that a visible accumulation and overlap of Yki with Ex could also be observed in wts mutant clones in which Ex was not over-expressed from UAS transgenes (Fig. 4I,J). Because Ex is a transcriptional target of Fat-Hippo signaling, Ex is over-expressed from the endogenous ex locus in wts mutant clones (Hamaratoglu et al., 2006)(Fig. 4I,J). Importantly, this accumulation of Yki with Ex observed in wts clones indicates that tethering of Yki outside the nucleus at sites of Ex accumulation is not solely a consequence of over-expression of exogenous Ex, but rather can be effected by endogenous Ex.
DISCUSSION
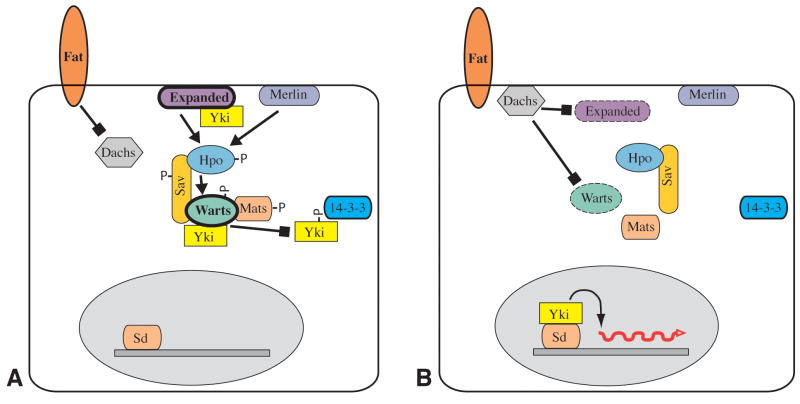
The discovery of Yki as an oncogene that is genetically downstream of wts and a substrate of Wts kinase activity led to a model in which Fat-Hippo signaling regulates Yki by phosphorylation (Huang et al., 2005). However, the results described here, together with the recent study of Badouel et al (2009), identify an alternate mechanism that does not involve phosophorylation (Fig. 5). Since they can visibly affect the sub-cellular localization of Yki3SA in disc cells (Figs 1,4)(Oh and Irvine, 2009), and directly bind to Yki in cultured cell assays (Fig. 2)(Badouel et al., 2009), the simplest explanation would be that Wts, Ex and Hpo exclude Yki from the nucleus by acting directly as cytoplasmic anchoring proteins. Phosphorylation-independent repression is expected to require the WW domains, but analysis of their importance to Yki repression is complicated by their positive role in transcriptional activation by Yki (Oh and Irvine, 2009; Zhao et al., 2009). Indeed, the dual role of the WW domains in both repressing and activating Yki suggests a model in which positive and negative factors compete for association with Yki at its WW domains. Mutation of the WW domains did increase Yki nuclear localization, although the effect was subtle. However, even when the most critical Wts phosphorylation site on Yki (Ser168) is mutated, Yki localization is only modestly affected (Oh and Irvine, 2008). Thus Yki localization is affected not only by its phosphorylation, but also by interaction with other factors.
Fig 5. Model of Yki regulation.
A) In the Yki “off” state, Wts is active, and levels of Wts and Ex are high (thick outline). Active Warts phosphorylates Yki, which inhibits Yki by promoting its association with 14-3-3 proteins in the cytoplasm, thereby excluding it from the nucleus. In addition, Wts and Ex can directly bind Yki to exclude it from the nucleus B) In the Yki “on” state, Wts is inactive, and levels of Wts and/or Ex are lower (dashed outline). Components of the Hippo kinase cassette are unphosphorylated, and interactions between them are reduced. Yki is not phosphorylated, and enters the nucleus where it complexes with Sd to promote the transcription of downstream target genes.
The discovery of this alternate Yki repression mechanism raises the question of its relevance to normal Yki regulation in vivo. The critical regulatory step in phosphorylation-dependent repression is the amount of active Wts, which is reflected in the phosphorylation status of Yki. Conversely, the critical regulatory step in phosphorylation-independent repression is expected to be the total amount of Wts, Ex and Hpo relative to Yki. Most published analysis of Yki regulation, including both loss-of-function and gain-of-function experiments, in vivo and in cultured cells, have involved manipulations of Fat-Hippo signaling that could potentially affect both of these mechanisms, and thus haven’t distinguished between them. Indeed, since they involve the same upstream tumor suppressors, both phosphorylation-dependent and phosphorylation-independent mechanisms could normally function in parallel to repress Yki, and there are observations that support the relevance of both types of regulation.
The most direct evidence in support of the importance of phosphorylation-dependent repression is provided by mutation of Wts phosphorylation sites on Yki. In the context of over-expressed Yki, the Wts phosphorylation sites are crucial, as UAS-yki:V5 transgenes inserted at identical chromosomal locations have no phenotype in their wild-type form, but dramatic overgrowth phenotypes when the Wts phosphorylation sites are mutant (Oh and Irvine, 2009), and similar results have been reported for YAP and Yki phosphorylation site mutants using a variety of approaches (Dong et al., 2007; Hao et al., 2008; Oh and Irvine, 2008; Zhang et al., 2008; Zhao et al., 2007). Since over-expression of kinase dead Hpo has a mild dominant negative phenotype, one can also infer that, at least for Hpo, its role in phosphorylation-dependent repression outweighs its contribution to phosphorylation-independent repression. In the context of endogenous yki the evidence is more circumstantial. Gain-of-function alleles of yki with mutations surrounding the phosphorylation site at Ser168 have been identified (Zhao et al., 2007). Their phenotypes are quite mild, but it is possible that they do not completely block phosphorylation and 14-3-3 binding at this site. In further support of the importance of phosphorylation-dependent repression, we note that both Ex and Fat have been reported to influence Wts activity in cultured cells (Hamaratoglu et al., 2006; Silva et al., 2006; Willecke et al., 2006), and that Ex and Fat also both influence Yki phosphorylation in vivo (Oh and Irvine, 2008).
The clearest evidence in support of phosphorylation-independent repression is the observation that Wts, Ex and Hpo can repress the activity of Yki3SA, both in vivo and in cultured cell assays (Figs 1,3). This repression was readily detected when Wts, Ex and Hpo were expressed in combination at the same levels as Yki (i.e., under the same promoter), or when a single one of these proteins was expressed at higher levels. Although the absolute levels of Yki versus these upstream tumor suppressors in vivo are not known, they can vary substantially. Ex is subject to negative feedback regulation in Fat-Hippo signaling, as elevated transcription of ex is a consequence of Yki activation (Hamaratoglu et al., 2006). Indeed, under the elevated Ex levels that exist in wts mutants, a fraction of Yki became visibly co-localized with Ex, suggesting that direct binding between Ex and Yki at least contributes to negative feedback regulation in Fat-Hippo signaling. Conversely, mutation or inactivation of Fat can decrease levels of Ex at the membrane, which might decrease phosphorylation-independent repression of Yki by Ex. Fat also has a strong effect on the total amount of Wts protein in cells (Cho et al., 2006). Moreover, the total amount of Wts is critical for the normal regulation of Yki, as over-expression of Wts can suppress fat, ex or dco3 mutations (Feng and Irvine, 2007; Feng and Irvine, 2009). Finally, we note that a marked elevation in total YAP levels, as opposed to a simple change in the YAP phosphorylation state, has been observed in many solid tumors (Steinhardt et al., 2008). Taken together, these observations suggest that phosphorylation-independent repression could be important in some contexts in vivo. Moreover, our observations clearly establish that phosphorylation-independent repression can occur when this pathway is experimentally manipulated, and hence must be taken into consideration, both in evaluating past results and designing future experiments.
Supplementary Material
Wing imaginal discs expressing Myc:Wts transgenes in posterior cells (right side), stained for expression of Myc:Wts. A) en-Gal4 UAS-Myc:Wts[attP-25C7]. B) en-Gal4 UAS-Myc:WtsKD[attP-25C7]. This kinase dead transgene is inserted at the identical location as the wild-type transgene shown in A, but protein expression is much lower, presumably because kinase dead Wts is less stable.
Acknowledgments
We thank B. Hay, R. Fehon, J. Jiang, D. Pan, N. Tapon, Z. Lai, and the Bloomington Stock Center for antibodies, plasmids, and Drosophila stocks, and C. Rauskolb for comments on the manuscript. This research was supported by HHMI and NIH post-doctoral fellowship 1F32GM079817 to H.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–20. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Mlodzik M. The Drosophila tumor suppressor expanded regulates growth, apoptosis, and patterning during development. Mech Dev. 2000;92:251–62. doi: 10.1016/s0925-4773(00)00246-x. [DOI] [PubMed] [Google Scholar]
- Boedigheimer MJ, Nguyen KP, Bryant PJ. Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev Genet. 1997;20:103–10. doi: 10.1002/(SICI)1520-6408(1997)20:2<103::AID-DVG3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–68. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–50. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci U S A. 2007;104:20362–7. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Processing and phosphorylation of the Fat receptor. Proc Natl Acad Sci U S A. 2009;106:11989–94. doi: 10.1073/pnas.0811540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–41. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–82. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–9. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–45. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–7. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–8. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–27. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–38. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The Tumor-Suppressor Gene fat Controls Tissue Growth Upstream of Expanded in the Hippo Signaling Pathway. Curr Biol. 2006;16:2081–9. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. Embo J. 2007;26:1772–81. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The Fat Cadherin Acts through the Hippo Tumor-Suppressor Pathway to Regulate Tissue Size. Curr Biol. 2006;16:2090–100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–98. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–87. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–98. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wing imaginal discs expressing Myc:Wts transgenes in posterior cells (right side), stained for expression of Myc:Wts. A) en-Gal4 UAS-Myc:Wts[attP-25C7]. B) en-Gal4 UAS-Myc:WtsKD[attP-25C7]. This kinase dead transgene is inserted at the identical location as the wild-type transgene shown in A, but protein expression is much lower, presumably because kinase dead Wts is less stable.